ABSTRACT
Previously, we have demonstrated that transgenic Arabidopsis and barley plants, expressing a 791 nucleotide (nt) dsRNA (CYP3RNA) that targets all three CYP51 genes (FgCYP51A, FgCYP51B, FgCYP51C) in Fusarium graminearum (Fg), inhibited fungal infection via a process designated as host-induced gene silencing (HIGS). More recently, we have shown that spray applications of CYP3RNA also protect barley from fungal infection via a process termed spray-induced gene silencing (SIGS). Thus, RNAi technology may have the potential to revolutionize plant protection in agriculture. Therefore, successful field application will require optimization of RNAi design necessary to maximize the efficacy of the RNA silencing construct for making RNAi-based strategies a realistic and sustainable approach in agriculture. Previous studies indicate that silencing is correlated with the number of siRNAs generated from a dsRNA precursor. To prove the hypothesis that silencing efficiency is correlated with the number of siRNAs processed out of the dsRNA precursor, we tested in a HIGS and SIGS approach dsRNA precursors of increasing length ranging from 400 nt to 1500 nt to assess gene silencing efficiency of individual FgCYP51 genes. Concerning HIGS-mediated disease control, we found that there is no significant correlation between the length of the dsRNA precursor and the reduction of Fg infection on CYP51-dsRNA-expressing Arabidopsis plants. Importantly and in clear contrast to HIGS, we measured a decrease in SIGS-mediated Fg disease resistance that significantly correlates with the length of the dsRNA construct that was sprayed, indicating that the size of the dsRNA interferes with a sufficient uptake of dsRNAs by the fungus.
Introduction
Crop plants are challenged by a multitude of different pathogens, insects, animals and weeds that cause many different plant diseases and constitute a constant threat to the food supply. It is estimated that those organisms cause yield losses up to 40% of the global agricultural production, which constitutes extremely high costs for our growing world population [Citation1,Citation2]. Besides the yield losses, mycotoxin contamination of foods and feedstuffs caused by phytopathogenic fungi, such as Fusarium graminearum (Fg), poses an almost intractable problem in agricultural production [Citation3,Citation4]. Current plant protection strategies rely on fungicide application for both Fusarium disease control and limitation of mycotoxin accumulation. The most commonly used fungicides are azoles, which target the cytochrome P450 sterol 14α-demethylase encoded by CYP51 genes. Inhibition of CYP51 causes depletion of ergosterol which results in loss of membrane integrity followed by growth inhibition and death of fungal cells [Citation5]. However, as a consequence of continuous fungicide applications, an increasing rate of azole insensitivity was observed in several plant pathogenic fungi, including Fusarium species [Citation6–Citation8]. Thus, the development of alternative control strategies has become one of the biggest challenges for plant pathologists these days.
RNA interference (RNAi) is a mechanism of gene regulation that utilizes small RNAs (sRNAs) to direct the silencing of gene expression at the transcriptional or posttranscriptional level. Over the last decade, RNAi-based gene silencing has emerged as a powerful genetic tool in various fields of applied research, such as agriculture [for a review see: Citation9; Citation10, Citation11, Citation12, Citation13, Citation14]. The mechanism of RNAi-mediated plant protection strategies relies on the expression of the transgene that provokes the formation of a double-stranded (ds)RNA precursor molecule in planta. This dsRNA subsequently triggers the plant’s RNAi machinery to process small interfering (si)RNAs that in the case of an infection exhibit target gene silencing of a certain pathogen [Citation9].
In a previous study, we demonstrated that transgenic Arabidopsis and barley (Hordeum vulgare) plants, expressing a 791 nucleotide (nt) dsRNA (CYP3RNA) that targets all three CYP51 genes (FgCYP51A, FgCYP51B, FgCYP51C) in Fg, inhibited fungal infection via a process known as host-induced gene silencing (HIGS) [Citation15,Citation16]. In addition to the generation of RNA silencing signals in planta, plants can be protected from pathogens and pests by exogenously applied RNA biopesticides, known as spray-induced gene silencing (SIGS) [Citation17–Citation20,Citation21–Citation22].
Regardless of how target-specific inhibitory RNAs are applied (i.e. endogenously or exogenously), the use of HIGS and SIGS technologies to control Fusarium species has been shown to be a potential alternative to conventional pesticides [Citation15,Citation19,Citation23,Citation24,Citation25–Citation27,Citation28,Citation30–Citation32], supporting the notion that RNAi strategies may improve food safety by controlling the growth of phytopathogenic, mycotoxin-producing fungi [reviewed by Citation33; Citation34].
Given the ease of dsRNA design, its high specificity, and applicability to diverse pathogens, the use of target-specific dsRNA as an anti-fungal agent offers an unprecedented potential as a new plant protection strategy. However, successful field application will require optimization of RNAi design necessary to maximize the efficacy of the RNA silencing construct. Recently, we compared the efficiencies of HIGS and SIGS dsRNA delivery strategies to assess the activity of novel dsRNA species that were designed to target one or two FgCYP51 genes [Citation19]. Using barley as a cereal model, we found that dsRNA constructs targeting two FgCYP51 genes inhibited fungal growth more efficiently than single constructs, although both types of dsRNAs decreased fungal infections [Citation19]. Based on these findings, we anticipate that constructs that target two genes in parallel were more efficient because the number of effective siRNAs derived from those double constructs is higher.
Therefore, we tested in a HIGS and SIGS approach dsRNA precursors of increasing length ranging from 400 nt to 1500 nt to assess gene silencing efficiency of individual FgCYP51 genes.
We found that SIGS efficiencies depend on the length of the dsRNA that was sprayed, indicating that the size of the sprayed dsRNA interferes with sufficient uptake mechanisms of the fungus. Interestingly, we found that HIGS-mediated disease resistance was independent of the length of the dsRNA constructs. Our findings suggest that HIGS and SIGS approaches differ concerning their mechanistic basis, thus leading to different silencing efficiencies and disease resistance phenotypes depending on RNAi construct design.
Results and discussion
Host-induced gene silencing by CYP51-dsRNAs of different length confers resistance to Fg in transgenic arabidopsis
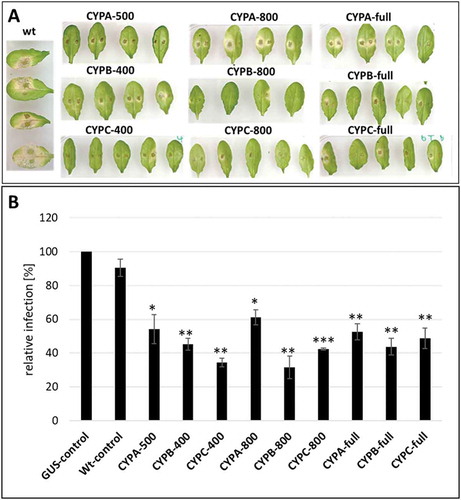
To assess whether silencing efficiency correlates with dsRNA length, dsRNA constructs of 400 nt to 500 nt and 800 nt were generated targeting single FgCYP51 genes (CYPA-500/800, CYPB-400/800, CYPC-400/800). In addition, the full-length cDNA of each FgCYP51 gene (CYPA-full, CYPB-full, CYPC-full) was cloned without the start and the stop codon to avoid protein expression. The constructs were inserted into the vector p7U10-RNAi (Fig. S1) and transgenic Arabidopsis plants were generated. Resistance to Fg was analysed on detached leaves inoculated with 5 × 10–4 Fg conidia per ml and incubated at room temperature (RT). At 5 days post-inoculation (dpi) untransformed wild-type (wt) plants showed water-soaked spots with chlorotic and necrotic lesions representing typical symptoms of a successful Fg infection (). In clear contrast, plants expressing CYP51-dsRNA of different length showed significantly reduced necrotic lesions compared with wt (). Notably, there were no clear phenotypic differences between 400 nt and 800 nt or full-length constructs () and the reduction of the infection area was for nearly all constructs to a similar extent of about 50–60% in comparison to the control (). The only exceptions were CYPC-400 and CYPB-800, which showed a higher resistance and reduced infection area by 66% and 69%, respectively. Concerning HIGS-mediated disease control, we found that there is no significant correlation between the length of the dsRNA precursor and the reduction of Fg infection on CYP51-dsRNA-expressing Arabidopsis plants.
Figure 1. Host-induced gene silencing in Fg on leaves of transgenic Arabidopsis expressing CYP51-dsRNAs of different lengths. (A), 15 detached rosette leaves of CYP51-dsRNA-expressing Arabidopsis plants (T2 generation) were drop-inoculated with 5 × 104 conidia ml–1. Infection symptoms were evaluated at 5 dpi. (B), quantification of the visibly infected area at 5 dpi shown as per cent of the total leaf area. Error bars represent SE of four independent experiments each using 15 leaves of 10 different plants for each transgenic line. Asterisks indicate statistical significance (*p < 0.05; **p < 0.01; ***p < 0.001; Student’s t test).
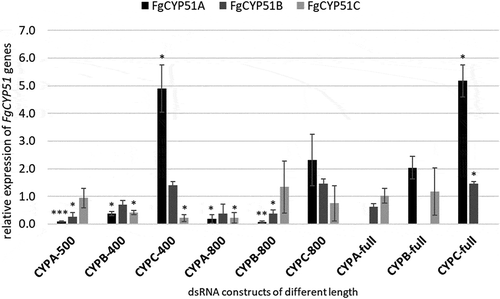
Previously, we found that CYP51-dsRNA activity involved co-suppression in the respective non-targeted paralogous FgCYP51 genes [Citation19]. To analyse whether the observed phenotypes were provoked by co-silencing effects in the non-targeted CYP51 genes, we measured the transcript levels of FgCYP51 genes in the infected leaf tissue using quantitative real-time polymerase chain reaction (qRT-PCR). As anticipated, the relative transcript levels of targeted genes FgCYP51A, FgCYP51B, and FgCYP51C were reduced after inoculation of leaves expressing the respective CYP-dsRNA constructs ().
Figure 2. Silencing of FgCYP51 genes of infected transgenic Arabidopsis leaves. Gene-specific expression of FgCYP51A, FgCYP51B, and FgCYP51C was measured by qRT-PCR and normalized to fungal EF1-α (FGSG_08811) as a reference gene. cDNA was generated after total RNA extraction from infected leaves at 5 dpi. The reduction in CYP51 gene expression in the Fg-inoculated dsRNA-expressing leaves compared with the wt control was statistically significant. Error bars represent SE of four independent experiments each using 15 leaves of 10 different plants for each transgenic line. Asterisks indicate statistical significance (*p < 0.05; **p < 0.01; ***p < 0.001; Student’s t test).
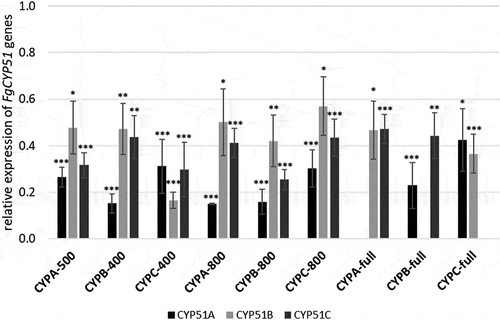
Most interestingly, we found that silencing of FgCYP51A was more efficient than for FgCYP51B and FgCYP51C (). This is consistent with our latest finding, that siRNAs complementary to CYP51A sequence showed the highest abundance among CYP3RNA-transgene-derived siRNAs revealed by RNA-seq analysis (unpublished). Based on this result, we speculate that the amount of siRNAs targeting FgCYP51A that reach the fungus is higher, thus, explaining the better gene silencing efficiency. Interestingly, more efficient silencing of FgCYP51A, in consequence, did not lead to higher Fg disease resistance because CYPA-500 and CYPA-800 showed the lowest resistance by reducing infection areas by only 46% and 39%, respectively (). Furthermore, we found that constructs targeting FgCYP51B exhibited higher gene silencing concerning all three FgCYP51 genes than constructs targeting FgCYP51A. This observation is consistent with our off-target prediction that showed the most off-targets for FgCYP51B (). Consistent with these findings, we have recently shown that even strong silencing of FcCYP51A by almost 90% did not lead to growth inhibition and morphological changes of Fusarium culmorum (Fc) in vitro cultures compared with only 40% silencing of FcCYP51B, which resulted in 30% retardation of fungal growth as well as abnormal hyphal phenotypes [Citation30]. These results further support previous reports that have shown that FgCYP51B is the most important gene for ergosterol biosynthesis and thus survival of the fungus [Citation19,Citation30,Citation33,Citation35,Citation36].
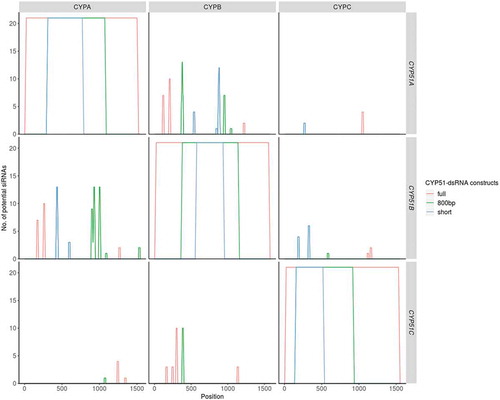
Figure 3. Off-target prediction for single CYP-dsRNA constructs with different length. Sequences of CYP51-dsRNAs were split into k-mers of 21 nt. These were targeted against the corresponding complementary DNA sequences (cDNA) of FgCYP51A, FgCYP51B, and FgCYP51C. For each position within the cDNA (x-axis) the k-mers that target the respective gene is plotted. The plot is facetted along the x-axis by the dsRNA constructs and along the y-axis by the target gene. The colours indicate the dsRNA length. While siRNAs deriving from the short dsRNAs (400 bp/500 bp) are shown in blue. logically also the longer dsRNAs produce these siRNAs. Accordingly, the siRNAs deriving from the 800 bp constructs are shown in blue and siRNAs derived exclusively from the full-length dsRNAs are shown in red.
However, this finding is specific for transgene-derived dsRNAs and their processing by plant Dicers (DCLs) because we did not observe similar results for SIGS. Quite the contrary, as predicted by our off-target analysis, spraying of dsRNA constructs targeting FgCYP51C (CYPC-400, CYPC-800, and CYPC-full) did not provoke silencing of non-targeted FgCYP51A and FgCYP51B genes (; see next section).
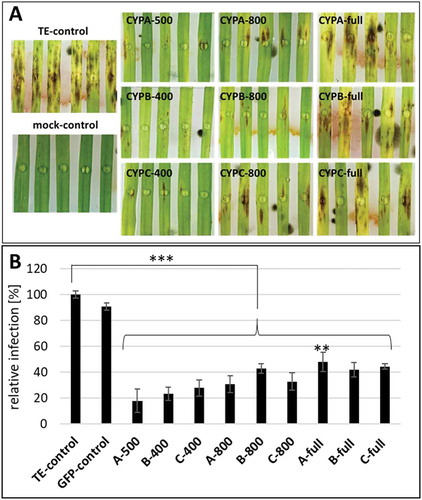
Figure 4. Fg infection of barley leaves that were spray treated with CYP51-dsRNAs of different lengths. (A), Detached leaves of 3-week old barley plants were sprayed with CYP51-dsRNAs or TE buffer. After 48 h, leaves were drop-inoculated with 5 × 104 conidia ml–1 and evaluated for infection symptoms at 5 dpi. (B), Infection area, shown as per cent of the total leaf area for 10 leaves for each dsRNA and the TE control. Error bars indicate SE of four independent experiments. Asterisks indicate statistical significance (**p < 0.01; ***p < 0.001; Student’s t test).
Spray-induced gene silencing efficiency correlates with the length of the sprayed dsRNA
Spraying barley with dsRNA constructs of 200–300 nt in length targeting single CYP51 genes of Fg was superior to HIGS-mediated Fg disease control [Citation19]. Encouraged by these findings, we assessed whether 400 nt, 800 nt, as well as full-length CYP51-dsRNAs are also active in spray experiments. Moreover, whether gene silencing efficiency and Fg disease resistance can be increased by spraying longer dsRNAs. Towards this, detached barley leaves were sprayed with 20 ng µl–1 dsRNA and drop-inoculated 48 h later with a suspension of Fg conidia. After 5 dpi, necrotic lesions were visible at the inoculation sites of leaves sprayed with TE buffer (control) or GFP-dsRNA (control to exclude unspecific dsRNA effects). All CYP51-dsRNAs reduced the infection symptoms as revealed by significantly smaller lesions (). We found the strongest resistance after spraying with 400–500 nt constructs because the infection symptoms were reduced by 82% for CYPA-500, 77% for CYPB-400, and 72% for CYPC-400 (). Interestingly, infected areas of 800 nt constructs were reduced on average by 65% compared with the control (), suggesting that the efficiency slightly decreased by spraying 800 nt dsRNA. Recently, we have demonstrated that spraying a 791 nt long non-coding dsRNA (CYP3RNA), which targets the three CYP51 genes FgCYP51A, FgCYP51B, and FgCYP51C of Fg, strongly inhibited fungal growth on barley leaves [Citation18]. Moreover, we have shown that SIGS was mediated by the uptake of the 791 nt dsRNA in the fungus and required the fungal RNAi machinery for the processing of siRNAs [Citation18]. Consistent with these findings, we found that 800 nt CYP51-dsRNAs inhibited Fg infection by almost 65% (). However, 800 nt dsRNAs targeting individual FgCYP51 genes were less efficient than the 791 nt CYP3RNA that targets all three FgCYP51 genes in parallel which led to 93% disease resistance [Citation19]. This difference can be partly explained by co-silencing of FgCYP51 genes.
Table 1. Growth inhibition of Fg during different RNAi-based silencing setups. Growth inhibition is shown as reduction in % of the infected leaf area in comparison to the control*.
Previously we found that HIGS- as well as SIGS-mediated targeting of individual FgCYP51 genes resulted in co-silencing of the non-targeted FgCYP51 genes [Citation19]. To explore co-silencing effects further, we calculated possible off-targets in FgCYP51 genes for all tested CYP51-dsRNA constructs (; ). Sequences of the different dsRNA constructs were split into k-mers of 21 bases and targeted against the complementary DNAs of the three FgCYP51 genes, with the plant miRNA target prediction tool TAPIR [Citation37]. Based on the default parameters, we calculated off-targets for all constructs in the respective non-target FgCYP51 genes () which is consistent with the observed gene silencing of all three FgCYP51 genes (; ).
Table 2. siRNA prediction of CYP51-dsRNA constructs using TAPIR.
Figure 5. Silencing of FgCYP51 genes of infected spray treated barley leaves. Gene-specific expression of FgCYP51A, FgCYP51B, and FgCYP51C was measured using qRT-PCR and normalized to fungal EF1-α as a reference gene. Detached leaves of 3-week-old barley plants were sprayed with CYP51-dsRNA or TE buffer. After 48 h, leaves were drop-inoculated with 5 × 104 macroconidia ml–1. cDNA was generated at 5 dpi after total RNA extraction from infected leaves. Error bars represent SE of two independent experiments. Asterisks indicate statistical significance. (*p < 0.05; **p < 0.01; ***p < 0.001; Student’s t test).
Consistent with our previous results, we observed that the number of off-targets increased with the increasing length of the different CYP51-dsRNAs (; ). Thereby, we found that dsRNA constructs targeting FgCYP51A and FgCYP51B had the highest number of off-target hits in the non-targeted FgCYP51 genes (). These observations can be explained based on the closer phylogenetic relationship between FgCYP51A and FgCYP51B, which leads to a higher amount of potential siRNAs targeting both genes (; Fig. S3). In contrast, FgCYP51C as phylogenetically more distant predicts fewer siRNAs complementary to FgCYP51A and FgCYP51B and vice versa (; Fig. S3). Thus, phylogenetic relationship correlates with the number of predicted siRNAs causing potential off-target hits. Notably, less stringent prediction criteria increase the predicted off-targeting siRNAs (Table S1; Fig. S4). Thereby, the number of off-targets per construct increased with the length of the precursor RNA showing a maximum in the full-length constructs, as expected (). This was regardless of whether FgCYP51A, FgCYP51B, or FgCYP51C was the actual target. Given this correlation, we observed the strongest silencing efficiencies for CYPA- and CYPB-dsRNA constructs (). Notably, FgCYP51C-derived constructs seem to have fewer off-targets in respective non-target genes than FgCYP51A- and FgCYP51B-derived constructs, reflected by qRT-PCR results (). Even more important, we observed that CYPC-400 and CYPC-800 showed similar Fg disease resistance phenotypes compared with CYPA- and CYPB-dsRNA constructs (; ). Based on this result, we speculate that silencing of FgCYP51C is sufficient to provoke fungal inhibition at least in SIGS. Of note and in clear contrast to the SIGS data, HIGS plants expressing CYPC-dsRNA showed silencing of the non-targeted genes FgCYP51A and FgCYP51B although this was not expected by the stringent off-target prediction (; ). Together, these findings further prove that the molecular mechanisms underlying the transgene-based (HIGS) or spray-mediated (SIGS) delivery strategy are different, either requiring the plant’s or the fungal RNA silencing machinery (), thus leading to different silencing phenotypes.
Figure 6. Molecular mechanisms underlying the transgene-based (HIGS) or spray-mediated (SIGS) delivery strategy. (A), the molecular mechanism of HIGS is based on the plant’s silencing machinery. The integration of our findings supports the view that HIGS involves the processing of transgene-derived dsRNA in the plant (plant DCLs) and extracellular vesicle (EV)-mediated delivery into the interacting fungus. (B), the molecular mechanism of SIGS is controlled by the fungal silencing machinery. In summary, our findings support the model that SIGS involves the uptake of sprayed dsRNA by the plant (via stomata) and subsequent uptake of apoplastic dsRNA by the fungus resulting in fungal DCL-dependent production of siRNAs.
However, these data were obtained from in silico predictions; therefore, their accuracies remain unknown. As such, RNA sequencing (RNA-seq) must be performed to quantify, analyse, and map the HIGS-derived siRNAs to their target genes as well as their dsRNA precursors to prove whether the generation of secondary siRNAs can explain the obtained HIGS results. In addition, RNA-seq of SIGS plants would help to clarify whether the production of siRNAs in SIGS and/or barley plants occurred at all. To prove further whether the longer dsRNAs result in higher numbers of siRNAs, we predicted the number of siRNA hits for each CYP51-dsRNA construct using SiFi (https://sourceforge.net/projects/sifi21) as a prediction tool. Similar to what we observed for the off-target prediction, we found a strong correlation between the length of the dsRNA precursor and the precursor-derived siRNAs (). However, these off-target-based co-silencing effects were more obvious for SIGS than for HIGS (compare with ) further supporting our previous finding that SIGS involves the uptake of dsRNA by the fungus and requires the fungal RNAi machinery [Citation18,Citation38]. Consistent with this idea, uptake of unprocessed dsRNAs from leaf tissue and processing by fungal DCLs might be more efficient than application of siRNAs because this would increase the amount of efficient siRNAs in the target organism. Moreover, in contrast to using only one specific siRNA, processing of long dsRNA into many different inhibitory siRNAs by the fungus may reduce the chance of pathogen resistance under field test conditions. Thus, we assume that SIGS can be more efficient than HIGS depending on the lifestyle of the targeted pathogen. Importantly, until now SIGS was demonstrated against necrotrophic fungal pathogens, such as Fg and Botrytis cinerea [Citation18,Citation22]. However, further research must show whether uptake of SIGS-derived siRNAs by biotrophic pathogens would also increase SIGS-mediated disease resistance.
Table 3. siRNA prediction of CYP51-dsRNA constructs using siFi21. Total and efficient siRNAs of 21 nt were predicted with the siFi21 software for the cDNA of the respective FgCYP51 target genes FgCYP51A, FgCYP51B and FgCYP51C.
Inhibition of fungal infection negatively correlates with the length of the sprayed dsRNA
Spraying with full-length CYP51-dsRNA resulted in the lowest decrease in Fg infection by at least 50% (). Thus, we observed a decrease in Fg infection that correlates with the length of the sprayed dsRNA (). However, expression analysis of FgCYP51 genes in infected leaves showed target gene silencing and an overall strong co-silencing (). All constructs led to downregulation of respective non-targeted CYP51 genes except for constructs targeting FgCYP51C (CYPC-400, CYPC-800, and CYPC-full), where upregulation of the non-targeted FgCYP51A and FgCYP51B genes was measured (). This was consistent with the off-target prediction, where CYPC constructs showed the lowest number of siRNA that match to the FgCYP51A and FgCYP51B genes (; ). Overall, these data show a strong correlation between resistance phenotypes induced by CYP51-dsRNA constructs and reduced expression of FgCYP51 genes. Notably, we observed that co-suppression was completely lost when leaves were sprayed with full-length CYP51-dsRNA (). Unfortunately, analysis of target gene silencing was not possible because there were no primers available that would not amplify the sprayed RNA as well.
However, the decrease in efficiency from 65% for 800 nt CYP51-dsRNAs to only 50% with the full-length dsRNAs indicates that the size of the dsRNA interferes with a sufficient uptake of dsRNAs by the fungus. To prove this idea further, we treated Fg with full-length CYP51-dsRNAs in vitro. We found that there was no silencing of the non-targeted FgCYP51 genes, indicating that Fg was not able to take up >1500 nt dsRNA from liquid cultures (Fig. S2). Unfortunately, we were not able to analyse target gene silencing directly because there were no primers available that would distinguish between dsRNA that was applied to the medium and silencing of FgCYP51 target genes. However, because we observed 50% fewer infection symptoms after spraying barley leaves with full-length CYP51-dsRNA (), we anticipate that there are differences in the mechanism of fungal dsRNA uptake in SIGS compared with in vitro culture treatments. In Aspergillus nidulans as well as in Aspergillus fumigatus uptake of siRNAs resulted in gene silencing [Citation39–Citation41]. In Candida albicans a structure-dependent uptake efficiency could be observed because linear nucleic acids were taken up more efficiently than hairpin structures [Citation42].
Congruent with the idea of a different uptake/translocation mechanism, we demonstrated that HIGS, even though 200–500 nt CYP51-dsRNA constructs were less efficient than SIGS, is not limited by the size of the dsRNA precursor the plants were transformed with. Because dsRNAs are expressed in planta and subsequently processed by DCLs enzymes, we would expect that increasing the length of the dsRNA results in more siRNAs that were taken up by the fungus (). Unfortunately, and in contrast to our expectation, the data did not show such a correlation (). However, even for HIGS, it must be considered that the host produced dsRNA precursor could be taken up and processed by the fungus itself. Consistent with our findings, a size-dependent efficiency of dsRNA-mediated silencing of target genes has been observed in insects [Citation43,Citation44]. Thereby, the most efficient dsRNA length varies hardly among different species whereas most studies show successful gene silencing with dsRNA lengths from 140 to 520 nt [Citation29,Citation45]. In the western corn rootworm, silencing efficiency increased with increasing dsRNA lengths (60–200 nt) whereas a minimum of 60 nt dsRNA is required for successful gene silencing [Citation43]. By targeting the same gene in aphid feeding studies, only dsRNA and not siRNAs were able to achieve gene silencing [Citation46]. In Leptinotarsa decemlineata even a 1842 nt dsRNA resulted in mortality in feeding experiments [Citation47,Citation48]. Interestingly, in the same study also shorter dsRNAs of 134 nt and 300 nt conferred gene silencing, suggesting that target gene selection is superior to dsRNA length. Summarizing, our results together with the studies mentioned before suggest that the efficiency of RNA-based disease control depends on the uptake and processing of efficient siRNAs that reach the pathogen/pest. Even more important, the mechanisms underlying uptake and processing of dsRNAs are different for HIGS and SIGS (), thus influencing design criteria of the RNAi trigger compound.
Conclusion
Plants can be protected from pathogens/pests mediated by lethal RNA silencing signals generated in planta [for a review on HIGS, see Citation9, Citation10, Citation11, Citation12, Citation13, Citation14] as well as by exogenously supplied RNA biopesticides [for a review on SIGS, see Citation10, Citation49, Citation50, Citation51, Citation21, Citation52]. Despite the promising potential of RNA silencing-based disease control the mechanisms underlying HIGS and SIGS technologies are inadequately understood.
Given these knowledge gaps, questions on the design of new and potentially promising dsRNAs arise. Whereas bioinformatics provides a variety of tools for the design, analysis, and evaluation of sRNA agents, little is known about the influence of dsRNA design on its processing and target gene silencing efficiency. Therefore, we attempt to identify the design principles of an optimal dsRNA trigger to improve/guarantee efficacy and specificity of HIGS- and SIGS-based plant protection approaches. By comparing efficiencies of infection reduction of HIGS with SIGS we observed a strong bias between those two approaches (). For example, CYP51-dsRNA construct CYPC-238 [Citation19, ] exhibited 78% reduction of infection in SIGS compared with only 31% for HIGS (). Another example is CYPA-500 which showed 82% efficiency in SIGS but only 46% decrease in Fg infection in HIGS. In other words, for dsRNA constructs of 200–500 nt, HIGS is around 30% less efficient than SIGS (under lab conditions) (). We assume that differences in silencing and disease resistance are due to processing and delivery of dsRNAs rather than depending on the concentration/amount of dsRNA and/or siRNA because we cannot compare the amount of transgene-derived siRNAs generated by plant DCLs with spray-applied dsRNA that were processed by fungal DCLs (). However, we observed that with increasing length of the CYP51-dsRNA construct, differences in efficiencies between HIGS and SIGS approaches became less obvious. If we compared dsRNA constructs of 400 nt length, SIGS was 20% more efficient than HIGS (). Notably, if we then compared efficiencies of CYP51-dsRNA constructs that were generated out of the full-length sequence (>1500 nt) of the individual FgCYP51 genes, HIGS and SIGS showed the same level of around 50% reduction of Fg infection (). Even more important and in clear contrast to HIGS, we measured a decrease in SIGS-mediated Fg disease resistance that is probably correlated with the length of the dsRNA construct that was sprayed. More explicitly, SIGS-based efficiencies decreased from 80% for 200–500 nt constructs to 65% for 800 nt constructs to 50% for >1500 nt dsRNA constructs.
Taken together, our results strongly support the notion that uptake of dsRNA of 200–500 nt in length and procession of siRNA by the fungus is more efficient in terms of disease control than uptake of siRNA via HIGS. However, little is known about how and which silencing signals (i.e. siRNAs and/or dsRNA precursors) are transferred from plant into fungal cells. Moreover, research into the mechanistic basis underlying HIGS and SIGS technologies is needed to integrate e.g. DCL-processing patterns that might help to define principles for RNAi vector design, producing effective siRNAs [Citation18,Citation23,Citation53].
Therefore, further research must address the question: What is the optimal dsRNA design for uptake, processing, translocation, and silencing efficiency of the RNAi trigger compound?
Methods
Construction of CYP51 containing p7U10 RNAi vectors
CYP51-dsRNA constructs CYPA-500/800/full, CYPB-400/800/full, and CYPC-400/800/full were amplified from Fusarium graminearum IFA65 cDNA using a gene-specific primer (Tab. S1) and inserted into the HindIII and XmaI restriction sites of p7U10 RNAi (Fig. S1).
Generation of transgenic arabidopsis thaliana plants
p7U10 plasmids for transformation of Arabidopsis were introduced into the A. tumefaciens strain AGL1 by electroporation. Transformation of Arabidopsis was performed with the floral dip method as described [Citation54] and transgenic plants were selected on ½ MS agar plates containing BASTA (7 μg/ml).
Plant infection assays and spray application of dsRNA
Fg IFA65 was grown on SNA agar plates at 22°C in an incubator (BINDER). For all leaf inoculation assays, Fg-IFA65 conidia concentration was adjusted to 5 × 104 macroconidia ml–1 in ddH2O containing 0.002% Tween-20. After inoculation, plates were stored at RT and infection symptoms were assessed at 5 dpi. To evaluate infection severity, fungal growth was determined by measuring the size of chlorotic and necrotic lesions using the ImageJ software (https://imagej.nih.gov/ij/index.html).
For the Arabidopsis–Fusarium infection, 15 rosette leaves of 10 different 5-wk-old plants of each transgenic line and control plants [Col-0 wild-type (wt)] were detached and transferred in square Petri dishes containing 1% agar. Inoculation of Arabidopsis was done by wound inoculation of detached leaves with 5 μl Fusarium conidia suspension on each leaf side. Wounding was performed by scratching the leaf surface with a pipette tip. At 5 dpi, leaves were frozen in liquid nitrogen and subjected to RNA extraction and cDNA synthesis.
For spray application, dsRNA was generated using MEGAscript RNAi Kit (Invitrogen) following the manufacturer’s instructions. p7U10 plasmids containing CYP51-dsRNA constructs were used as a template. Primer pairs with T7 promoter sequences at the 5′-end of both forward and reverse primers were designed for amplification of dsRNA (Tab. S1). The dsRNA, eluted in TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA), was diluted in 500 μl water to a final concentration of 20 ng μl–1. For the TE control, TE buffer was diluted in 500 μl water corresponding to the amount that has been used for dilution of the dsRNA. Typical RNA concentration after elution was 500 ng μl–1, representing a buffer concentration of 400 μM Tris-HCl and 40 μM EDTA in the final dilution. Detached barley leaves were covered with a plastic tray before spraying, leaving only the upper part (approximately 1 cm) uncovered. After spraying, dishes were kept open until the surface of each leaf was dried. After 48 h, leaves were drop-inoculated as described above.
Quantification of fungal transcripts by quantitative real-time PCR (qRT-PCR)
Before cDNA synthesis, remaining DNA was digested by DNAse I (Thermo Scientific) using RiboLock RNAse Inhibitor (Thermo Scientific) for 30 min at 37°C. For cDNA synthesis, 1 μg digested RNA was used. cDNA synthesis was performed using qScriptTM cDNA synthesis kit (Quanta). qRT-PCR was performed with freshly synthesized cDNA in the QuantStudio 5 Real-Time PCR system (Applied Biosystems) in 384-well plates using SYBR® green JumpStart Taq ReadyMix (Sigma-Aldrich). For each sample, three replicates were performed, and target transcript levels were determined using a gene-specific primer (Tab. S1) via the 2–ΔΔCt method [Citation55] by normalizing the amount of target transcript to the amount of reference transcript.
Bioinformatic off-target analysis
The precursor sequences of CYP51-dsRNAs were split into k-mers of 21 bases. These sequences were targeted against the complementary DNA sequences of FgCYP51 genes (cDNA) of Fusarium graminearum strain PH-1 (GCA_000240135.3) with TAPIR [Citation37] using the following settings: score cut-off 4 and minimum free energy ratio cut-off 0.7. For each target, the position in the respective cDNA was plotted with RStudio [Citation56] and the ggplot2 package [Citation57].
Phylogenetic analysis of FgCYP51 genes
FgCYP51 genes were aligned with the muscle algorithm with default settings and a maximum likelihood tree was generated using the MEGA7 software [Citation58].
Author Contributions
A.K. and L.H. wrote the manuscript; L.H. and A.K. designed the study; L.H., D.B. and A.S. conducted the experiments; A.K. and L.H. analyzed all data. A.K., L.H and B.T.W. drafted the figures. B.T.W. and L.J. performed bioinformatics analysis. All authors reviewed the final manuscript.
Supplemental Material
Download Zip (681.6 KB)Acknowledgments
We thank C. Birkenstock, U. Schnepp and V. Weisel for excellent plant cultivation. This work was supported by Deutsche Forschungsgemeinschaft to AK (RTG:2355).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Alexander P, Brown C, Arneth A, et al. Losses, inefficiencies and waste in the global food system. Agric Syst. 2017;153:190–200.
- Oerke E-C, Dehne H-W. Safeguarding production—losses in major crops and the role of crop protection. Crop Prot. 2004;23:275–285.
- Doll S, Danicke S. The Fusarium toxins deoxynivalenol (DON) and zearalenone (ZON) in animal feeding. Prev Vet Med. 2011;102:132–145.
- Ismaiel A, Papenbrock J. Mycotoxins: producing fungi and mechanisms of phytotoxicity. Agriculture. 2015;5(3):492–537.
- Yoshida Y. Cytochrome P450 of fungi: primary target for azole antifungal agents. Curr Topics Med Mycol. 1988;2:388–418.
- Becher R, Hettwer U, Karlovsky P, et al. Adaptation of Fusarium graminearum to tebuconazole yielded descendants diverging for levels of fitness, fungicide resistance, virulence, and mycotoxin production. Phytopathology. 2010;100:444–453.
- Spolti P, Del Ponte EM, Dong Y, et al. Triazole sensitivity in a contemporary population of fusarium graminearum from New York wheat and competitiveness of a tebuconazole-resistant isolate. Plant Dis. 2014;98:607–613.
- Yin Y, Liu X, Li B, et al. Characterization of sterol demethylation inhibitor-resistant isolates of Fusarium asiaticum and F. graminearum collected from wheat in China. Phytopathology. 2009;99(5):487–497.
- Koch A, Kogel KH. New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotech J. 2014;12:821-831.
- Gaffar FY, Koch A. Catch me if you can! RNA silencing-based improvement of antiviral plant immunity. Viruses. 2019;11:673.
- Qi T, Guo J, Peng H, et al. Host-induced gene silencing: a powerful strategy to control diseases of wheat and barley. Int J Mol Sci. 2019;20:206.
- Yin C, Hulbert S. Host Induced Gene Silencing (HIGS), a promising strategy for developing disease resistant crops. Gene Technol. 2015;4:130.
- Zhang J, Khan SA, Heckel DG, et al. Next-generation insect-resistant plants: rNAi-mediated crop protection. Trends Biotechnol. 2017;35:871–882.
- Liu S, Jaouannet M, Dempsey D’M A, Imani J, Coustau C, Kogel KH. RNA-based technologies for pest control in plant production. Biotechnol Adv. 2019. doi:10.1016/j.biotechadv.2019.107463
- Koch A, Kumar N, Weber L, et al. Host-induced gene silencing of cytochrome P450 lanosterol C14alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci U.S.A. 2013;110: 19324–19329.
- Nowara D, Gay A, Lacomme C, et al. HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 2010;22:3130–3141.
- Kaldis A, Berbati M, Melita O, et al. Exogenously applied dsRNA molecules deriving from the Zucchini yellow mosaic virus (ZYMV) genome move systemically and protect cucurbits against ZYMV. Mol Plant Pathol. 2018;19:883–895.
- Koch A, Biedenkopf D, Furch A, et al. An RNAi-based control of fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016;12:e1005901.
- Koch A, Höfle L, Werner BT, et al. SIGS vs HIGS: A study on the efficacy of two dsRNA delivery strategies to silence Fusarium FgCYP51 genes in infected host and non-host plants. Mol Plant Pathol. 2019;20:1636–1644.
- Konakalla NC, Kaldis A, Berbati M, et al. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta. 2016;244:961–969.
- Mitter N, Worrall EA, Robinson KE, et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plants. 2017;3:16207.
- Wang M, Weiberg A, Lin F-M, et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants. 2016;2:16151.
- Baldwin T, Islamovic E, Klos K, et al. Silencing efficiency of dsRNA fragments targeting Fusarium graminearum TRI6 and patterns of small interfering RNA associated with reduced virulence and mycotoxin production. PloS One. 2018;13(8):e0202798.
- Bharti P, Jyoti P, Kapoor P, et al. Host-induced silencing of pathogenicity genes enhances resistance to Fusarium oxysporum wilt in tomato. Mol Biotechnol. 2017;59(8):343–352.
- Chen W, Kastner C, Nowara D, et al. Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J Exp Bot. 2016;67(17):4979–4991.
- Cheng W, Song XS, Li HP, et al. Host‐induced gene silencing of an essential chitin synthase gene confers durable resistance to F usarium head blight and seedling blight in wheat. Plant Biotechnol J. 2015;13(9):1335–1345.
- Ghag SB, Shekhawat UK, Ganapathi TR. Host‐induced post‐transcriptional hairpin RNA‐mediated gene silencing of vital fungal genes confers efficient resistance against sufficient uptake mechanisms wilt in banana. Plant Biotechnol J. 2014;12(5):541–553.
- Hu Z, Parekh U, Maruta N, et al. Down-regulation of Fusarium oxysporum endogenous genes by host-delivered RNA interference enhances disease resistance. Front Chem. 2015;3:1.
- Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9:90–95.
- Koch A, Stein E, Kogel KH. RNA-based disease control as a complementary measure to fight fusarium fungi through silencing of the azole target cytochrome P450 lanosterol C-14 α-demethylase. Eur J Plant Pathol. 2018;152:1003–1010.
- Pareek M, Rajam MV. RNAi-mediated silencing of MAP kinase signalling genes (Fmk1, Hog1, and Pbs2) in Fusarium oxysporum reduces pathogenesis on tomato plants. Fungal Biol. 2017;121(9):775–784.
- Werner BT, Gaffar FY, Schuemann J, et al. RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. BioRxiv. 2019. DOI:10.1101/821868
- Machado AK, Brown NA, Urban M, et al. RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Manag Sci. 2018;74(4):790–799.
- Majumdar R, Rajasekaran K, Cary JW. RNA interference (RNAi) as a potential tool for control of mycotoxin contamination in crop plants: concepts and considerations. Front Plant Sci. 2017;8:200.
- Fan J, Urban M, Parker JE, et al. Characterization of the sterol 14α-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol. 2013;198:821–835.
- Liu X, Yu F, Schnabel G, et al. Paralogous cyp51 genes in Fusarium graminearum mediate differential sensitivity to sterol demethylation inhibitors. Fungal Genet Biol. 2011;48:113–123.
- Bonnet E, He Y, Billiau K, et al. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26(12):1566–1568.
- Gaffar FY, Imani J, Karlovsky JP, et al. Various components of the RNAi pathway are required for conidiation, ascosporogenesis, virulence, DON production and SIGS-mediated fungal inhibition by exogenous dsRNA in the Head Blight pathogen Fusarium graminearum. Front Microbiol. 2019. DOI:10.3389/fmicb.2019.01662
- Jöchl C, Loh E, Ploner A, et al. Development-dependent scavenging of nucleic acids in the filamentous fungus Aspergillus fumigatus. RNA Biol. 2014;6:179–186.
- Kalleda N, Naorem A, Manchikatla RV. Targeting fungal genes by diced siRNAs: a rapid tool to decipher gene function in Aspergillus nidulans. PloS One. 2013;8:e75443.
- Khatri M, Rajam MV. Targeting polyamines of Aspergillus nidulans by siRNA specific to fungal ornithine decarboxylase gene. Med Mycol. 2007;45:211–220.
- Disney MD, Haidaris CG, Turner DH. Uptake and antifungal activity of oligonucleotides in Candida albicans. Proc Natl Acad Sci USA. 2003;100:1530–1534.
- Bolognesi R, Ramaseshadri P, Anderson J, et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PloS One. 2012;7:e47534.
- Saleh M-C, van Rij RP, Hekele A, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802.
- de Andrade EC, Hunter WB. RNA interference – natural gene-based technology for highly specific pest control (HiSPeC). In: (Abdurakhmonov IY, editor. RNA interference. InTech; 2016. doi:10.5772/61612
- Mulot M, Boissinot S, Monsion B, et al. Comparative analysis of RNAi-based methods to down-regulate expression of two genes expressed at different levels in myzus persicae. Viruses. 2016;8(11):316.
- Baum JA, Bogaert T, Clinton W, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326.
- Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J Insect Physiol. 2010;56:227–235.
- Cai Q, He B, Kogel KH, Jin H. Cross-kingdom RNA trafficking and environmental RNAi — nature’s blueprint for modern crop protection strategies. Curr Opin Microbiol. 2018;46:58–64.
- Cagliari D, Dias NP, Galdeano DM, EÁ Dos Santos, Smagghe G, Zotti MJ. Management of pest insects and plant diseases by non-transformative RNAi. Front Plant Sci. 2019;10:1319.
- Dubrovina A, SKiselev KV. ExogenousRNAs for gene regulation and plant resistance. Int J Mol Sci. 2019;20(9):2282.
- Dalakouras, A, Wassenegger M, Dadami E, Ganopoulos I, Pappas ML, Papadopoulou K. Genetically Modiied Organism-Free RNA Interference: Exogenous. Plant Physiol. 2020;182.
- Yang Y, Jittayasothorn Y, Chronis D, et al. Molecular characteristics and efficacy of 16D10 siRNAs in inhibiting root-knot nematode infection in transgenic grape hairy roots. PloS One. 2013;8(7):e69463.
- Bechtold N, Pelletier G. In planta agrobacteriummediated transformation of adult arabidopsis thaliana plants by vacuum infiltration. In: (Martinez-Zapater JM, Salinas J, editors. Arabidopsis protocols. Totowa, NJ: Humana Press; 1998. p. 259–266.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408.
- Team R. RStudio: integrated development for R. RStudio, Inc. Vol. 42. Boston, MA; 2015. p. 14. Available from: http://www.rstudio.com.
- Wickham H. ggplot2: elegant graphics for data analysis. Switzerland: Springer; 2016. doi:10.1007/978-0-387-98141-3
- Kumar S, Stecher G, Tamura K. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874.
