ABSTRACT
La and La-related proteins (LARPs) are characterized by a common RNA interaction platform termed the La module. This structural hallmark allows LARPs to pervade various aspects of RNA biology. The metazoan LARP7 protein binds to the 7SK RNA as part of a 7SK small nuclear ribonucleoprotein (7SK snRNP), which inhibits the transcriptional activity of RNA polymerase II (Pol II). Additionally, recent findings revealed unanticipated roles of LARP7 in the assembly of other RNPs, as well as in the modification, processing and cellular transport of RNA molecules. Reduced levels of functional LARP7 have been linked to cancer and Alazami syndrome, two seemingly unrelated human diseases characterized either by hyperproliferation or growth retardation. Here, we review the intricate regulatory networks centered on LARP7 and assess how malfunction of these networks may relate to the etiology of LARP7-linked diseases.
Introduction
The eukaryotic La-related protein (LARP) family comprises members with various functions in RNA metabolism. LARPs typically exploit a combination of a winged helix domain, termed La-motif (LaM), and an adjacent RNA recognition motif (RRM1) to contact their RNA substrates [Citation1–6], whereby the LaM and the RRM1 are collectively referred to as ‘La module’ (reviewed in [Citation7]).
Target interactions are nevertheless not restricted to this characteristic structure. Instead, LARPs possess additional RNA binding motifs, which are also critical for substrate binding [Citation8–11]. The presence of such specialized building blocks together with a versatile La module allow LARPs to target specific subsets of RNAs applying a fascinating mechanistic diversity (reviewed in [Citation12]). While LARP1, the two paralogs LARP4 and LARP4B, as well as LARP6 mainly localize to the cytoplasm where they bind to mRNAs [Citation11,Citation13–16], LARP7 and the genuine La protein/SSB are mostly nuclear and both interact with non-coding RNAs transcribed by RNA polymerase III (Pol III). These transcripts terminate on an oligo-uridine stretch, which is the primary binding site for the La module of the two closely related proteins [Citation4,Citation5]. LARP7 and La also share a similar overall structural organization as they both contain a second RRM, referred to as RRM2 [Citation17,Citation18].
Despite these analogies, LARP7 and La fulfill unrelated and non-overlapping cellular functions [Citation19,Citation20]. Even though both gene products are essential in mouse, the homozygous Larp7 knockouts survive until embryonic day 16.5 [Citation21], whereas disruption of the La gene impairs early embryonic development and is lethal already at the pre-implantation stage [Citation22]. In addition, distinct symptomatic spectra of human pathologies are linked to these two homologous proteins. For example, La, but not LARP7, is a common autoantigen found in autoimmune diseases such as Lupus erythematosus [Citation23].
La is known to bind virtually all primary Pol III transcripts immediately upon transcription termination [Citation24]. However, the interaction with La is usually transient and restricted to the maturation of the respective RNA targets [Citation25] and its release is mandatory for the assembly of the targeted RNAs into functional RNPs [Citation26]. In contrast, LARP7 interacts with and promotes the stability of its main target, the non-coding 7SK RNA [Citation20,Citation27,Citation28]. This highly structured transcript folds into four hairpin domains and serves as a scaffold for the binding of several proteins assembling into 7SK snRNP complexes (reviewed in [Citation29]). More than a decade ago, LARP7 was first observed to co-purify with such particles together with the positive transcription elongation factor b (P-TEFb) [Citation30]. P-TEFb functions as important regulator of RNA polymerase II (Pol II) activity and it has been well established meanwhile that it can be tethered in an inactive state to the 7SK RNA (reviewed in [Citation31]).
This review aims to provide an overview of our current understanding of LARP7-associated pathways and processes. In the first part, we focus on recent structural and biochemical studies providing novel insights into the assembly and function of LARP7-7SK RNA complexes. We will pinpoint common and distinguishing structural properties between LARP7 and the homologous La protein as well as LARP7 orthologs from unicellular eukaryotes lacking the 7SK RNA [Citation32–36]. The central part of this review will focus on recent studies that uncovered thus far unknown 7SK RNA-independent functions of LARP7 in different aspects of RNA metabolism, including RNA modification. In the last sections, we will discuss the implications of 7SK-dependent and -independent LARP7 functions for human malignancies.
A clamp and a clip to grab the 7SK RNA
The LARP7 protein consists of the N-terminal La module and the C-terminal RRM2, which are connected by a central region predicted to be largely unstructured [Citation5] (). Studies on the RNA binding properties of LARP7 focused on the interaction with its main target, the 7SK RNA and in particular on its 3ʹ terminal hairpin (often referred to as HP4 or SL4). As any other Pol III transcript, the 7SK RNA possesses a 3ʹ terminal single-stranded oligo-uridine tail, which is, together with the preceding HP4, essential for LARP7 binding in vivo [Citation26]. Recent structural studies have resolved the details of the protein-RNA interactions, demonstrating that the HP4 is contacted by both, the La module as well as the RRM2 [Citation5,Citation9,Citation10].
Figure 1. Two RNA-binding platforms within the LARP7 protein mediate contacts to RNA. (A) Schematic overview of the domain organization of human LARP7. The N-terminal La motif (LaM, blue) and the adjacent RNA recognition motif (RRM1, green) form the so-called La module, the common hallmark of LARPs. A flexible linker (dashed line, grey) connects it to the C-terminal RRM2 (salmon). (B) Graphical representation of the binding of LARP7 to terminal hairpin of the 7SK RNA (HP4, yellow) according to the structural studies by Uchikawa et al. (2015) and Eichhorn et al. (2018). The La module is represented as a clamp pinching the single-stranded 3ʹ end of HP4, the RRM2 as a clip inserted on top of the apical loop. The corresponding structures are shown at the sides and were depicted using PyMOL (https://pymol.org/2/) and the data deposited in the Protein Data Bank (PDB): 4WKR for the La module and 6D12 for the RRM2, starting from residue 456. Colours as in (A) with the α3-helix highlighted by darker colouring
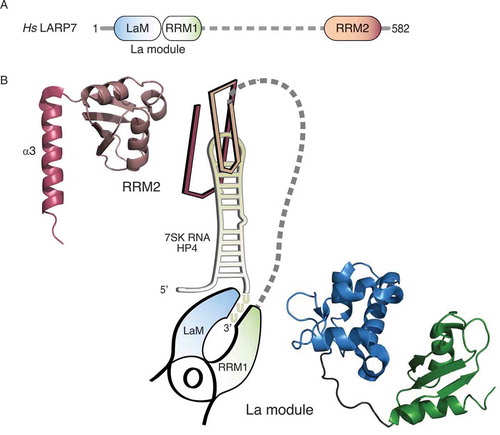
The structure of the isolated La module of LARP7 in complex with terminal fragments of the HP4 revealed a synergistic interplay of the LaM and the RRM1 to clamp the single stranded oligo-uridine substrate [Citation5] (). This arrangement had been previously observed also for the La module of human La in complex with oligo-uridine RNA [Citation4,Citation6]. Remarkable differences between the La modules of both proteins are nevertheless found within the RRM1 [Citation5], suggesting an adaptation to different functional requirements. Indeed, replacement of the RRM1 of LARP7 with the RRM1 of La abolishes the interaction with the 7SK RNA in vivo [Citation20]. Classical RRMs consist of a four-stranded β-sheet packed against two α-helices, whereby canonical RNA contacts are formed at the surface of the β-sheet (reviewed in [Citation37]). In addition, the RRM1 of La contains a C-terminal α-helix, which, together with one β-strand, is apparently lacking in LARP7 [Citation5]. Importantly, the cleft formed by the LaM and the RRM1, tailored to bind the single-stranded oligo-uridine end of LARP7 and La targets, does not involve the canonical RNA binding surface of the RRM1. This suggests that the RRM1 of LARP7 and La is available for additional RNA contacts [Citation4–6,Citation38] and may explain the different target selectivity between the two proteins (reviewed in [Citation12]).
Despite the ability of the La module of LARP7 to bind RNA strongly in vitro [Citation5,Citation10], it is not sufficient for 7SK RNA binding and stabilization in vivo [Citation20], suggesting that other segments are necessary for the efficient LARP7-7SK RNA interaction. Experimental evidences indicate that the long central linker of LARP7 might contribute to such additional contacts, in particular via several stretches of basic residues [Citation5,Citation10].
While further investigations are required to fully understand the rather enigmatic role of the central linker, it is meanwhile clear that, in addition to the La module, the C-terminal RRM2 constitutes a second fundamental RNA binding region on LARP7 [Citation5,Citation9,Citation10,Citation20]. Isothermal titration calorimetry (ITC) measurements, for example, demonstrated that the RRM2 has an additive effect on the binding of the 7SK HP4 in the context of the full-length LARP7 protein [Citation10]. The same study also solved the crystal structure of the isolated RRM2 in complex with an artificial RNA encompassing parts of the HP4 and revealed that the RRM2 is placed like a cap on top of HP4 [Citation10] (). These data also uncover the sequence-specific binding of the RRM2 to the apical loop of HP4, confirming previous biochemical observations [Citation5,Citation9,Citation26]. Of note, a two-nucleotide bulge within the stem of HP4 has also been shown to impact on the interaction with LARP7 [Citation26,Citation39]. Since the RNA substrate co-crystallized in complex with RRM2 did not encompass the region containing the bulge [Citation10], LARP7 binding to this region cannot be fully excluded. Such interaction could occur, for example, in solution during the assembly or remodelling of LARP7-7SK RNA complexes.
Despite this ongoing debate, the structures of the RNA-free [Citation9] and RNA-bound RRM2 [Citation10] of the human LARP7 protein clearly uncovered several peculiarities shared with the RRM2 of genuine La [Citation35,Citation40], as well as p65, an ortholog of LARP7 in the ciliate Tetrahymena [Citation35]. Canonical RRMs possess two conserved consensus motifs, referred to as RNP1 and RNP2, and both participate in RNA interactions (reviewed in [Citation37]). The RRM2 of LARP7 family members, however, lacks these motifs. Instead, the RRM2 has acquired a novel consensus motif (RNP3), which relocated to a different region of the β-sheet surface but still contributes to RNA binding. An even more distinguishing feature of this atypical RRM is the presence of an α-helical extension (α3) at its C-terminus, which is positioned in a perpendicular orientation, across the β-sheet surface [Citation9,Citation10,Citation35,Citation40,Citation41]. In the case of human LARP7, the RRM2 resembles a clip and inserts on top of the HP4 of the 7SK RNA providing additional contacts to the double-stranded stem of HP4 [Citation10] (). Due to these characteristic structural features, the RRM2 of La and LARP7 family members have been grouped into a distinct subclass of the RRM domain which was named ‘xRRM’ [Citation9,Citation10,Citation35,Citation41].
The comparison of the RNA-free NMR structure [Citation9] and the RNA-bound X-ray crystal structure [Citation10] of the LARP7 RRM2 further revealed that the C-terminus of the domain becomes more structured in the presence of RNA. This results in the extension of half a turn of the α3-helix (referred to as α3x), a peculiarity which is found also in the xRRM of p65 [Citation35]. Finally, the very C-terminus of LARP7 appears to be rather unstructured, although an additional α-helix (α4) might be present [Citation9]. A recent study conducted by our labs indicates that this region of LARP7 might play a critical role for the interaction with RNA [Citation42], but also a function unrelated to RNA binding has been reported and will be described in one of the following sections [Citation39].
Taken together, LARP7 uses two distinct RNA binding modules to grab the 3ʹ terminal hairpin of the 7SK RNA. While the La module pinches the 3ʹ end, the RRM2 clips onto the apical loop of HP4 (). However, these studies were restricted either to the isolated La module or RRM2 and they applied short or artificial RNA constructs. It is possible that the interaction of LARP7 to the 7SK RNA in the context of full-length protein and RNA still bears additional surprises.
LARP7 and MePCE form the core of the 7SK snRNP
LARP7 binding stabilizes the 7SK RNA in vivo and hence ensures 7SK RNP formation and function. However, 7SK RNA stabilization is not accomplished solely by LARP7, which indeed acts in concert with the methylphosphate capping enzyme (MePCE), also known as BCDIN3 [Citation20,Citation28,Citation30,Citation43–45] and both are core components of the 7SK snRNP [Citation44] (). An interdependency of these two proteins is also supported by the fact that decreased MePCE levels are accompanied by downregulation of LARP7 [Citation44,Citation46]. Inversely, however, LARP7 knockdown does not affect MePCE expression [Citation43].
Figure 2. Maturation of the 7SK snRNP. The upper panels depict the maturation events according to the current model. The 7SK RNA is immediately bound by La (blue) upon transcription by Pol III. While the La module of La binds the 3ʹ end of the 7SK RNA, other regions of La might contact the 5ʹ end of bound substrates as well [Citation147,Citation148]. Binding of MePCE (green) to the 7SK RNA and the following methylation of the cap were proposed to allow replacement of La with LARP7 [Citation49]. The resulting core 7SK snRNP is composed of the 7SK RNA, MePCE and LARP7 and is boxed in yellow. The C-terminus of LARP7 was shown to inhibit the catalytic activity of MePCE (represented by the red dashed line) and was thus named MePCE inactivating domain (MID). As an alternative to this La-dependent 7SK snRNP maturation pathway, chromatin bound MePCE might directly sequester and methylate the 5ʹ end of nascent 7SK RNA transcripts precluding the access by La and recruiting LARP7 (La-independent pathway, lower left panel)
![Figure 2. Maturation of the 7SK snRNP. The upper panels depict the maturation events according to the current model. The 7SK RNA is immediately bound by La (blue) upon transcription by Pol III. While the La module of La binds the 3ʹ end of the 7SK RNA, other regions of La might contact the 5ʹ end of bound substrates as well [Citation147,Citation148]. Binding of MePCE (green) to the 7SK RNA and the following methylation of the cap were proposed to allow replacement of La with LARP7 [Citation49]. The resulting core 7SK snRNP is composed of the 7SK RNA, MePCE and LARP7 and is boxed in yellow. The C-terminus of LARP7 was shown to inhibit the catalytic activity of MePCE (represented by the red dashed line) and was thus named MePCE inactivating domain (MID). As an alternative to this La-dependent 7SK snRNP maturation pathway, chromatin bound MePCE might directly sequester and methylate the 5ʹ end of nascent 7SK RNA transcripts precluding the access by La and recruiting LARP7 (La-independent pathway, lower left panel)](/cms/asset/114d1926-3002-46bc-adbd-033fd94e75c2/krnb_a_1767952_f0002_c.jpg)
Importantly, MePCE contains a methyltransferase domain accounting for the transfer of a methyl group from S-adenosyl methionine (SAM) to the γ phosphate at the 5ʹ end of the 7SK RNA. This catalytic activity is involved in the stabilization of the RNA [Citation30,Citation45] and the acquisition of the cap modification might also govern the maturation of 7SK RNA particles.
A small fraction of cellular 7SK RNA has been found associated with the La protein [Citation26,Citation47,Citation48], which might reflect a pool of newly transcribed 7SK RNA immediately bound by La upon transcription termination. Since it is very likely that the interaction of the 7SK RNA with La or LARP7 is mutually exclusive, La needs to be replaced by LARP7 for maturation of 7SK snRNPs. It has been thus hypothesized that the methylation of the 5ʹ end of the 7SK RNA by MePCE might induce the dissociation of La from the transcript and could prime newly synthetized RNAs for the assembly into 7SK snRNPs [Citation49,Citation50] (, La-dependent pathway, upper panel).
In contrast, this proposed order of maturation events during the biogenesis of 7SK snRNPs is questioned by the fact that capping of the 7SK RNA by MePCE seems to occur co-transcriptionally [Citation43]. Taking this into consideration, nascent 7SK transcripts might be pre-bound by MePCE resulting in the rapid acquisition of the 5ʹ γ-monomethyl phosphate cap. In this La-independent maturation pathway, La binding to the 3ʹ end of the fully transcribed RNA would be impeded by the cap modification [Citation50] and most 7SK RNA transcripts could directly assemble with LARP7 (, lower panel). Concomitantly, the spurious interaction of La with the 7SK RNA might be indicative for an alternative minor La-dependent maturation pathway ().
Of note, MePCE also modifies the 5ʹ end of the spliceosomal U6 snRNA [Citation30,Citation51]. In this case, however, the enzyme dissociates from the RNA upon catalysis while it remains as stable component in the 7SK RNP. Indeed, protein factors assembling on 7SK snRNP complexes might further strengthen the association to MePCE accounting for its selective retention on the 7SK RNA [Citation39]. Structural models suggest that specific regions in LARP7, together with elements on the 3ʹ segment of the 7SK RNA, cooperatively form a binding surface allowing docking of MePCE [Citation10]. This model is further supported by the finding that the presence of LARP7 is sufficient to tether MePCE on 7SK RNA variants lacking RNA segments directly bound by MePCE [Citation26]. Inversely, MePCE alone is not able to recruit LARP7 to 3ʹ terminally truncated 7SK RNA variants highlighting the importance of this region for the formation of a stable trimeric complex [Citation26,Citation43].
In a current model, protein-protein contacts within the core 7SK snRNP were proposed to be formed between the catalytic center of MePCE and the very C-terminal part of LARP7 [Citation39,Citation43]. This region of LARP7, which follows the RRM2 and comprises the aforementioned α4 helix, was shown in in vitro assays to be critically required for the recruitment of MePCE to the 7SK RNA [Citation39]. Intriguingly, it also confers LARP7 the ability to repress the methylation activity of MePCE [Citation39,Citation43] and has been thus referred to as MePCE inactivating domain (MID), albeit the physiological relevance of this property has not yet been entirely understood [Citation39].
The interaction between MePCE and LARP7, while assembled on the 7SK RNA, has important structural implications. Taken into consideration that MePCE primarily associates at the 5ʹ end of the RNA, whereas LARP7 grasps its 3ʹ end, it is appealing to speculate that these protein-protein contacts might promote a non-covalent circularization of the 7SK RNA. Indeed, such a ‘closed’ conformation of the 7SK RNA [Citation52] has been proposed as an alternative to the ‘open’ structure model suggested by Wassarman and Steitz [Citation48]. Native 7SK snRNP complexes are believed to be dynamic and to undergo substantial remodelling in terms of protein composition [Citation28,Citation53] and RNA structure [Citation39,Citation54,Citation55]. The latter involves the formation of alternative intramolecular base-pairing interactions within the 7SK RNA (). The concept that such reshaping occurs and might even mediate the 7SK snRNP function, reconciled the two opposing models suggesting that the ‘open’ and the ‘closed’ conformations of the 7SK RNA cycle between each other and co-exist in a native environment [Citation39].
Figure 3. Models for the conformation of the 7SK RNA within the core 7SK snRNP. (A) In the ‘open’ structure model, the initial segment of the 7SK RNA is involved in basepairing interactions, which extend the stem of the 5ʹ terminal hairpin. In this conformation, the MID of LARP7 is not able to repress the catalytic activity of MePCE (represented by the red dashed lines). (B) In the ‘closed’ structure model, the 5ʹ end of the 7SK RNA basepairs with the basal region of HP4, joining the ends of the RNA. Consequently, LARP7 and MePCE are also in close spatial proximity and the MID of LARP7 inhibits MePCE by direct contacts (red lines)
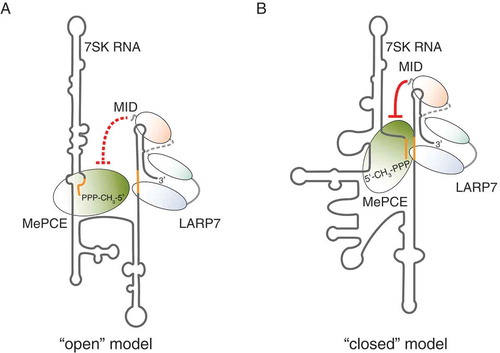
Direct interactions between MePCE and LARP7 in the ‘closed’ state of the 7SK RNA can be easily inferred. In contrast, it is more tempting to assume that MePCE and LARP7 are physically separated in the ‘open’ configuration, where the ends of the 7SK RNA are not joined by RNA-RNA contacts [Citation39]. The possible switching between the two conformations of the 7SK RNA consequently implies breaking or, at least, rearranging of MePCE-LARP7 interactions. Further studies are required to provide more details concerning this interesting structural aspect.
The 7SK snRNP controls availability of active P-TEFb
As a core component of the 7SK snRNP, LARP7 participates in all processes regulated by this complex. Although it is not the scope of this review to provide a detailed overview of the latest discoveries related to 7SK snRNP functions (reviewed in [Citation56–58]), some emerging concepts and in particular those implying a direct role of LARP7 will be discussed in the following section.
In general, within the 7SK snRNP, different protein components are tethered to the RNA scaffold. The most extensively studied example is the binding and inactivation of P-TEFb, which consists of the cyclin-dependent kinase 9 (CDK9) and a cyclin T (either T1 or T2) [Citation59,Citation60]. This is achieved by the repressor protein HEXIM1 or its paralog HEXIM2, which are also found within 7SK snRNP complexes [Citation61–65]. Interestingly, the 7SK RNA itself works as an allosteric co-factor inducing conformational changes in the HEXIM proteins [Citation64,Citation65], which in turn allows the formation of an inhibitory interaction between HEXIMs and P-TEFb [Citation66,Citation67].
P-TEFb plays an essential role during gene expression as it phosphorylates Ser2 in the C-terminal domain (CTD) of Pol II as well as subunits of two repressive complexes (DSIF and NELF) that maintain the polymerase in a promoter-proximal paused state (reviewed in [Citation68]). The establishment of the post-translation modification on these targets ultimately allows Pol II to transit into the productive elongation phase. Thus, the current model implies that LARP7 and the 7SK snRNP are important regulators of P-TEFb activity (reviewed in [Citation31]). During the last years of research, the captivating concept arose that the 7SK snRNP does not simply buffer the P-TEFb reservoir broadly attenuating Pol II activity. Instead, it is now thought that inactive P-TEFb is required to be delivered via 7SK snRNPs to the sites of request (reviewed in [Citation36,Citation56,Citation58,Citation69]). In support of this notion, cumulative reports argue that 7SK snRNPs associate with chromatin and in particular with promoter-proximal regions [Citation70–77], albeit a broader occupancy across the entire length of transcribed genes has been also reported for the 7SK RNA as determined by sequencing of chromatin isolation by RNA purification (ChIRP-seq) experiments [Citation55]. The triggered release of P-TEFb from the chromatin-associated inhibitory complexes would subsequently relieve Pol II pausing directly ‘on site’ (reviewed in [Citation36,Citation56,Citation58,Citation69]).
The question then arises of how the 7SK snRNP is tethered on genes with paused Pol II. Several scenarios have been envisioned to address this point, whereby the 7SK RNA was proposed to directly bind to a specific set of histones [Citation72]. 7SK snRNP-associated proteins such as HMGA1 [Citation78] or WDR43 [Citation73] were also shown to link the complex to chromatin and act thereby as adaptor molecules. Intriguingly, LARP7 itself might also act as an anchor point through direct binding of KAP1 (also known as TRIM28), a protein associated with chromatin-bound transcription factors [Citation75].
How does the controlled release of P-TEFb occur? Diverse stress factors, e.g., inhibition of transcription or UV-light [Citation59,Citation60] as well as genotoxic stress induced by DNA-damaging agents [Citation79] were found to liberate P-TEFb and some of the underlying molecular mechanism have been revealed (reviewed in [Citation57,Citation58]). These involve, for example, the post-translational modification of selected 7SK snRNP-associated proteins [Citation57,Citation80]. Importantly, a specific 7SK snRNP complex has been identified where HEXIM and P-TEFb are replaced by a set of heterogeneous nuclear ribonucleoproteins (hnRNPs) [Citation28,Citation81] and binding of these proteins appears to be a prerequisite for efficient P-TEFb release [Citation53,Citation82]. Thus, the 7SK snRNP was proposed to exert its regulatory function by cycling between the alternative P-TEFb-bound or hnRNP-bound states [Citation39,Citation82].
In contrast to this scenario, which would allow re-cycling of the 7SK snRNP components, the release of P-TEFb can be also obtained by more drastic strategies triggering the irreversible disassembly of the whole 7SK snRNP. This might for example occur upon removal of the monomethyl cap at the 5ʹ end of the 7SK RNA by the JMJD6 demethylase, which is expected to cause the destabilization of the RNA [Citation72]. Furthermore, during the differentiation of megakaryocytes, the blood cells accounting for platelets production, the protease calpain 2 was found to cleave MePCE leading to a pronounced reduction of its steady-state levels. LARP7 expression was also downregulated in megakaryocytes compared to the progenitor cells, but this effect occurred in a calpain 2-independent manner as it was not reversed by its inhibition. Together, the severe and global depletion of the core 7SK snRNP components causes the activation of P-TEFb and guarantees the correct megakaryocytic morphogenesis and perturbation of this pathways are associated with a subset of human megakaryocytic leukemias [Citation83].
In the mechanisms unravelled so far, the 7SK snRNP is manipulated in a way that weakens its interaction with P-TEFb, which apparently simply falls off the 7SK snRNP. According to other studies, however, specific factors extract P-TEFb more actively from the repressive complex [Citation54,Citation70,Citation72–74,Citation79].
One of these factors was shown to be the WDR43 protein, which is not only implicated in the recruitment of the 7SK snRNP to nascent RNA transcripts but also stimulates the release of P-TEFb from the complex [Citation73]. Although the underlying molecular mechanism is yet unknown, the authors of this work hypothesize that it might occur in phase-separated ‘transcription bodies’. They further provide first evidences suggesting that the formation of such structures is promoted by low-complexity sequences within the WDR43 protein, as well as DDX21, another known 7SK snRNP interactor [Citation73]. It will be interesting to follow, whether such a dependency on a phase-transited environment for the function of the 7SK snRNP will be further corroborated. Of note, the RNA helicase DDX21 itself also promotes the release of P-TEFb and this turned out to strictly require its ATP-dependent catalytic activity [Citation74].
The involvement of an RNA helicase [Citation53,Citation74,Citation84], as well as the structural changes observed upon hnRNP binding to the 7SK RNA [Citation39,Citation54], suggest that the remodelling of the RNA scaffold might be a prerequisite for the dissociation of P-TEFb from the 7SK snRNP. The contribution of RNA remodellers to this process might thus be more significant than so far appreciated. Intriguingly, LARP7 possesses RNA chaperone activity [Citation19] similarly to its paralog La [Citation85–89] and it is thus tempting to speculate, that this intrinsic property might mediate at least some of the dynamic rearrangements within the life-cycle of the 7SK snRNP. Furthermore, the ability to induce conformational changes of the bound RNA substrate has been attributed as well to the LARP7-related p65 protein in Tetrahymena [Citation90–92].
Unfortunately, direct experimental evidences supporting the idea that metazoan LARP7 proteins might actively reshape the structure of the 7SK RNA are still missing. Nevertheless, it is remarkable that RBM7, a factor recently described to promote the release of P-TEFb upon genotoxic stress, directly interacts with LARP7 and MePCE [Citation79]. Still, it is unclear, how this accounts for the observed effects on the composition of the 7SK snRNP. Might any of the identified P-TEFb release factors exert its function by inducing the RNA chaperone activity of LARP7?
LARP7 in P-TEFb-unrelated 7SK RNPs
7SK snRNP complexes with diverse protein composition have been identified over the last years and it became more and more clear that P-TEFb is not a constitutive component of all these particles. Some of them reflect intermediates within the cycle of P-TEFb regulation, whereby P-TEFb alternates with hnRNPs for binding to the 7SK snRNP [Citation53,Citation82].
However, dedicated 7SK snRNPs exist and they act in pathways unrelated to the regulation of P-TEFb activity (reviewed in [Citation55]). Genome-wide analyses using ChIRP-seq and ChIP-seq techniques provided indications for this idea by revealing distinct occupancy profiles for the 7SK RNA and other canonical 7SK snRNP components, as well as for selected transcription factors and chromatin marks [Citation55]. The intersection of these datasets has driven the hypothesis that the 7SK RNA forms different complexes which might fulfill dedicated functions at distinct genomic locations. Biochemical experiments confirmed that the 7SK RNA not only co-localizes but also interacts with the BAF chromatin remodelling complex suggesting an additional function of specific 7SK snRNPs in the re-positioning of nucleosomes [Citation55].
Biochemical investigations demonstrated that the 7SK RNA, together with MePCE and LARP7, are integral components of the little elongation complex (LEC) and that they are required for its structural integrity [Citation76]. This multi-subunit transcription factor is specifically involved in the Pol II-dependent synthesis of snRNAs and of small nucleolar (sno)RNAs transcribed from independent promoters. Indeed, knockdown of LARP7 impaired the recruitment of the LEC and consequently of Pol II to the promoters of such genes. This ultimately lead to the reduced production and accumulation of the snRNAs transcribed by Pol II and of those snoRNAs, which are not processed from the introns of host genes [Citation76].
Finally, a distinct 7SK snRNP has been also recently identified in the cytoplasm of motoneurons and this localization depends on the interaction with hnRNP R, which directly binds the 7SK RNA [Citation93]. Both were detected in close proximity of each other along the axons and in the growth cones of motoneurons. Depletion of hnRNP R or the 7SK RNA caused a reduction in axon growth and both components were required for the axonal translocation or stabilization of a subset of transcripts [Citation93]. Given its vital role in the stabilization of the 7SK RNA, it is likely that also LARP7 might be a component of this cytoplasmic neuronal 7SK RNA-hnRNP R complex but this aspect has not been addressed experimentally.
Moonlighting function of LARP7 proteins
During the process of DNA replication, linear chromosomal ends are progressively shortened, which, however, is prevented by the action of the telomerase complex. This RNP consists of a non-coding telomerase RNA (named TR, TER or TERC), a reverse transcriptase subunit (TERT) and several accessory factors (reviewed in [Citation94]). Among them, members of the LARP7 protein family were found to be integral components of the mature telomerase holoenzyme in ciliates [Citation95–97]. TER is transcribed by Pol III in these organisms, and in Tetrahymena, for example, the LARP7 ortholog p65 [Citation17,Citation18] binds the 3ʹ terminal oligo-uridine stretch and additional elements of the RNA [Citation98–100]. The interaction with p65 stabilizes TER, mediates the hierarchical and efficient assembly of the telomerase complex and promotes its catalytic activity [Citation90,Citation91,Citation97,Citation100,Citation101].
Despite the ancestral function of the telomerase complex, an astonishing diversity in the origin and biogenesis of this RNP is found across eukaryotes (reviewed in [Citation94]). In the yeast Schizosaccharomyces pombe, TER is produced by Pol II and its maturation requires an elaborated processing strategy, including an unconventional single cleavage reaction performed by the spliceosome. This event is stimulated by the binding of the Sm proteins, which are replaced in the mature telomerase complex by the Sm-like (LSm) complex [Citation102]. Sm and LSm proteins are structurally related proteins, which assemble in heteroheptameric ring structures enclosing the bound target RNAs (reviewed in [Citation103,Citation104,Citation105]). Surprisingly, three independent groups reported that the S. pombe LARP7-like protein Pof8 (or Lar7) promotes the loading of the LSm complex on TER. Similarly to p65 in Tetrahymena, Pof8 was identified as a constitutive component of the telomerase holoenzyme, which also depends on this protein to exert its full catalytic activity [Citation32–34]. The involvement of a LARP7 family member in the maturation of telomerase complexes containing Pol II-transcribed TER was rather unexpected and might be interpreted as a hint for a more general function of LARPs in the assembly of this RNP. While no evidence has been found to date for a possible incorporation of LARP7 within the human holoenzyme [Citation106–108], telomerase activity was recently reported to be reduced upon depletion of LARP7 and cell lines carrying LARP7 loss-of-function mutations had shorter telomeres [Citation109] indeed suggesting a role of LARP7 family members in this process. This model, however, needs to be further corroborated.
In addition to the binding to non-coding RNAs, Larp7 overexpressed in mouse embryonic stem cells (mESCs) has been found to co-immunoprecipitate the mRNA of the developmental regulator Lin28. Apparently, this association accounts for the recruitment of the terminal nucleotidyl transferase Tut1/Star-PAP to the Lin28 mRNA and is critical for the stability of this transcript. Albeit the molecular mechanism how Tut1/Star-PAP preserves the expression of Lin28 has not been elucidated, perturbation of this axis was found to prime mESC for precocious differentiation [Citation110].
Intriguingly, TUT1/Star-PAP also extends the primary transcript of the U6 snRNA with non-templated uridines [Citation111,Citation112]. In addition, a number of other LARP7 interactors have been implicated in U6 snRNP biogenesis as well [Citation28,Citation110]. Most prominently, MePCE catalyzes the same 5ʹ γ-monomethyl phosphate cap modification on the U6 and the 7SK snRNA [Citation51]. In contrast to the 7SK snRNP, however, the interaction with the U6 snRNA is restricted to its maturation process and MePCE is not found in mature U6 particles. More recently, LARP7 was co-immunoprecipitated from cell lysates together with METTL16 [Citation113], which was identified as the methyltransferase responsible for the m6A modification of an internal base within the U6 snRNA [Citation113,Citation114]. In order to join the splicing reaction, U6 snRNA-containing particles are assembled step-wise into the U4/U6 di-snRNP and finally into the U4/U6.U5 tri-snRNPs, whereby the former process is assisted by SART3 [Citation115–117]. SART3 was among the first factors identified within 7SK snRNP complexes [Citation30] and it was repeatedly detected in LARP7 affinity purifications [Citation28,Citation75], again linking LARP7 with U6 biogenesis. The U6 snRNA itself was observed to interact with LARP7, but was initially considered as an unspecific binding event [Citation26,Citation28]. Two studies have recently challenged this interpretation and demonstrated that LARP7 fulfils a yet unappreciated role for the biogenesis of the U6 snRNP. Indeed, the depletion of LARP7 resulted in the specific loss of 2ʹ-O-methylated nucleosides within the U6 snRNA [Citation42,Citation118]. These post-transcriptional modifications are introduced with the help of C/D box snoRNAs, which undergo base-pairing interactions with target RNAs and, by that, direct snoRNPs containing the methyltransferase fibrillarin to selected sites (reviewed in [Citation119]). In this context, LARP7 was shown to use the La module to bind to the U6 snRNA, while contacts to the U6-specific snoRNAs occur via the RRM2 [Citation42,Citation118]. This model describes LARP7 as the first auxiliary factor supporting efficient snoRNA-mediated snRNA modification. Furthermore, the two RNA binding platforms are proposed to act as distinct modules bridging the interaction between two different RNAs, which is, according to the current knowledge, a unique characteristic among LARPs.
Evidences for the requirement of LARP7 during bacterial infection
LARP7 has been linked to several human pathologies, most recently to infectious diseases caused by intracellular bacterial pathogens [Citation120]. Members of the Legionella genus, which are responsible for the Legionnaires’ disease, secrete so-called effector proteins into the infected cells to target and to manipulate molecular pathways of the host (reviewed in [Citation121]). For example, in L. pneumophila the ankyrin repeats-containing protein Ankyrin H (AnkH, alternatively also known as LegA3 or Lpg2300), supports efficient intracellular replication of the pathogen [Citation122]. Intriguingly, AnkH is among the very restricted set of effector proteins with homologs across all Legionella species, as well as other human and animal pathogenic bacteria, like e.g., the Q fever-causing Coxiella burnetii [Citation123–125]. In order to define the molecular role of AnkH, interaction studies were performed and LARP7 was identified as the top AnkH binding partner. Confirming the relevance of this interaction, efficient infection of human cells with L. pneumophila depend on the presence of LARP7. AnkH was thus proposed to hijack LARP7 and to partially prevent its function in the context of the 7SK snRNP. It was further hypothesized that the assumed perturbations of P-TEFb activity would be responsible for transcriptional reprogramming of infected cells, which ultimately favours bacterial replication [Citation120]. While this model needs to be further solidified, the striking occurrence of AnkH homologs across several intracellular pathogens, hints towards a pivotal and widespread mechanism of LARP7 targeting by bacterial proteins with important implications for infection.
LARP7: a putative tumor suppressor for different cancer types
The involvement of LARP7 within the 7SK-mediated sequestration of P-TEFb and the resulting inhibition of Pol II activity supported the obvious concept that LARP7 might act as a tumor suppressor [Citation20]. Indeed, a link between LARP7 mutations and human diseases was established in gastric cancer by a study analysing the frequency of mutations occurring within microsatellite repeats [Citation126]. Microsatellites are short repetitive sequences across the genome that are particularly prone to acquire length variations, a phenomenon that is termed microsatellite instability [Citation127]. Of note, two distinct microsatellites within the LARP7 genes were reported to be frequently mutated in different cancers [Citation126,Citation128,Citation129]. Such events can have detrimental consequences if the short repeat is located within a protein coding sequence, as insertions or deletions can lead to frameshift mutations. This is the case for one of the microsatellites embedded within the LARP7 gene where a stretch of eight consecutive adenosines was found to be altered in gastric tumors [Citation126]. The location of this microsatellite repeat would likely induce non-sense mediated decay (NMD) of LARP7 mRNAs with a shifted coding frame. Nevertheless, even if these transcript variants would be translated, the resulting LARP7 proteins would lack the RRM2 and, consequently, would be unable to stabilize the 7SK RNA in vivo [Citation20].
Reduced LARP7 expression was later confirmed in gastric cancer [Citation130], in invasive breast cancer [Citation131] as well as in papillary thyroid cancer [Citation132]. While the relevance of LARP7 microsatellite polymorphisms has not been addresses in these studies, it is conceivable that deregulated LARP7 expression could not only be dependent on such genomic mutations but also be caused by other pathways.
Importantly, several studies report that reduced LARP7 levels promote tumorigenesis, cancer progression and metastasis formation [Citation20,Citation130–132] corroborating the assumption that LARP7 is a tumor suppressor gene. While this might hold true for invasive breast cancer [Citation131], data obtained from other systems point towards the existence of a compensatory mechanism counteracting the possible hyperproliferative outcome expected upon LARP7 reduction. Indeed, prolonged knockdown of Larp7 in mESCs [Citation110] or similar experiments performed in human cell lines [Citation28] were accompanied by a reduction in the protein levels of the P-TEFb subunit Cdk9 [Citation110]. This or similar, but yet unidentified reactions, are possible reasons why a general enhancement of Pol II activity was absent despite the expected increase in P-TEFb availability following breakdown of 7SK snRNPs [Citation42,Citation118].
Loss-of-function mutations in the LARP7 gene cause the Alazami syndrome
Mutations in the LARP7 gene have been associated over the last years with a rare developmental disease termed Alazami syndrome [Citation133]. To date, around 20 different patients have been reported carrying homozygous or compound heterozygous LARP7 loss-of-function mutations [Citation42,Citation109,Citation133–140]. This severe disease manifests in developmental defects, most prominently, primordial dwarfism, which is characterized by pre- and postnatal growth retardation. A broad spectrum of additional skeletal and in particular facial dysmorphisms, including microcephaly, contribute to the complex phenotype of the disease. Severe intellectual disability with delayed speech development, as well as delayed motor development are additional characteristic symptoms [Citation134,Citation136]. Most of the Alazami-associated mutations identified so far introduce frameshifts in the coding sequence of the LARP7 mRNA and likely cause NMD. However, this has been experimentally demonstrated only in one study showing a complete loss of LARP7 in patients-derived cells [Citation133]. A possible exception might occur in a recently reported case of Alazami syndrome, whereby a patient has a genotype of biallelic LARP7 variants, one being a frameshift mutation and the other one an intronic point mutation [Citation139]. The latter variant possibly leads to compromised splicing, although residual expression of functional LARP7 protein might occur and could account for the milder clinical phenotype manifested by this specific patient [Citation139]. More recently, a short deletion in the LARP7 gene has been described. This mutation destroys the last intron-exon boundary and, in a homozygous state, causes the Alazami syndrome in two siblings of a consanguineous family. Importantly, this specific mutation does not lead to NMD of the transcript, but results in a LARP7 protein variant with a slightly different C-terminus and impaired functionality [Citation42]. The pleiotropic nature of the Alazami syndrome and the broad expression of LARP7 across tissues [Citation21,Citation44,Citation133] are indicative of its widespread physiological relevance. Nevertheless, the exact function of LARP7, which becomes critical for the pathogenesis of the Alazami syndrome, remains elusive. Concomitantly with the first reports of Alazami patients, it has been critically questioned, whether the etiology of the disease could be explained in a satisfying way by the canonical role of LARP7 in promoting P-TEFb-dependent transcriptional control through the stabilization of the 7SK snRNP [Citation133]. A major concern is the fact that the affected patients do not display any phenotype that might be readily explained by the hyper-activation of Pol II transcription, a consequence that can be envisioned due to LARP7 deficiency. Nevertheless, a single Alazami patient has been reported to date, who has been additionally diagnosed with papillary thyroid carcinoma. Analysis of a tumor biopsy identified the somatic V600E mutation in the BRAF gene, which is the most common mutation found in this type of cancers. Driven by the surprisingly young age of the patient affected by this malignancy, the authors of the study speculated that loss of LARP7 in Alazami patients might cause an increased susceptibility for the acquisition of cancer-related mutations [Citation140].
Differing from this report, the vast majority of Alazami patients do not provide indications for uncontrolled cellular proliferation or tumorigenesis. Instead, the syndrome is characterized by the opposite effect, such as reduced growth [Citation21,Citation44,Citation133]. Similarly, Larp7 depletion caused severe developmental defects and reduced growth of zebrafish [Citation44] and mouse embryos [Citation21]. In contrast to LARP7 deficiency in human, homozygous Larp7 knockout mouse embryos died before birth. The reason for this discrepancy is unclear, but a possible explanation might be found in the presence of the microRNA (miRNA) cluster miR-302/367 within an intronic sequence of the Larp7 gene, which could be affected in these animals. Importantly, these miRNAs were demonstrated to control the differentiation of neural progenitor cells and depletion of the miR-302/367 cluster had detrimental consequences during the murine embryonic development leading to prenatal lethality [Citation141,Citation142]. Of note, miR-302/367 knockout embryos displayed defects in the closure of the neural tube, a phenotype which was also observed in some of the Larp7 knockouts [Citation21]. Since it has not been assayed whether the generation of the knockout mice had an influence on the miRNA cluster embedded within the targeted Larp7 gene, it cannot be excluded that the reported embryonic lethality is due to impaired miR-302/367 expression.
Still, the mutations described in patients affected by the Alazami syndrome are not expected to compromise the correct biogenesis of the miRNAs encoded within the miR-302/367 cluster. Furthermore, the developmental defects, which are consistent between the patients and the phenotypes observed in different model systems, strongly indicate the primary cause for the disease is the loss-of-function of the LARP7 protein. Due to the role of LARP7 orthologs in the assembly of the telomerase complex in ciliates and yeast (see above), it has been hypothesized that a similar function might be conserved in human. In support of this idea, reduced telomere lengths were detected in the DNA isolated from leukocytes of Alazami patients, but the molecular mechanism underlying this finding are yet unclear. Intriguingly, knockdown of LARP7 affected the alternative splice pattern of the mRNA coding for the catalytic subunit of the telomerase complex and less functional transcript was produced [Citation109]. Perturbations of alternative splicing upon knockdown of LARP7 were also reported previously. In this case, the effects were attributed to the P-TEFb-dependent regulation of transcription, which is tightly linked to pre-mRNA processing [Citation44]. Two recent studies further indicate that LARP7 is an essential factor required for the post-transcriptional modification of the spliceosomal U6 snRNA and, by that, LARP7 might affect splicing robustness [Citation42,Citation118]. Taken together, several evidences point towards a possible role of aberrant splicing in the etiology of the Alazami syndrome.
Concluding remarks and perspectives
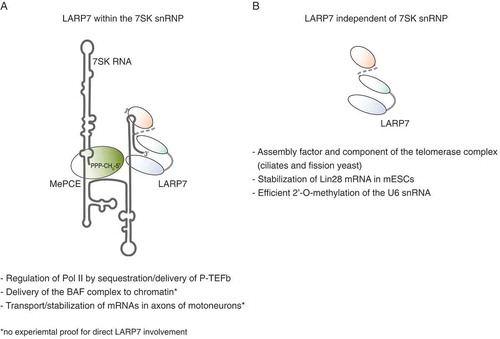
LARP7 plays key roles in various RNA pathways, including transcription, modification, processing and transport, which are summarized in . In order to fulfill this broad spectrum of functions, LARP7 assembles in distinct RNPs. A future challenge will be to isolate specific particles in order to determine their individual composition and, to acquire structural insights into such LARP7-containing complexes. The advanced studies of the 7SK snRNP in the context of P-TEFb sequestration have already provided clear mechanistic evidences for the dynamic nature of the LARP7 interactome. Even though different complexes might be related to the same pathway, they might undergo substantial remodelling in terms of protein composition and secondary structure of the bound RNA. This notion adds an additional layer of complexity to the attempt of defining specific functions for selected RNP subpopulations.
Figure 4. Versatile functions of LARP7. Summary of the 7SK snRNP-dependent (A) and – independent (B) roles of LARP7
A growing number of studies have revealed that the action of LARP7 has specific implications for certain cell and tissue types [Citation21,Citation110,Citation118]. In particular, Larp7 was recently shown to be essential for male fertility, which was attributed to its role in the 2ʹ-O-methylation of the U6 snRNA [Citation118]. Furthermore, varying subnuclear localization has been observed for Larp7 across embryonic tissues in flies, albeit the implications of this finding are not clear [Citation143]. In human cells, LARP7 was mainly detected in the nucleoplasm [Citation27,Citation75] but it was also identified in nucleoli [Citation144] as well as in Cajal bodies [Citation76]. Indirect evidences also exist for the presence of LARP7 in nuclear speckles [Citation145] and a small portion might even find its way to the cytoplasm [Citation93]. Careful investigations are thus required to determine the mechanisms governing the subcellular trafficking of LARP7 and further address the consequences of this multifarious localization.
The variety of processes affected by LARP7 also complicates the interpretation of knockdown or knockout experiments, which impair all LARP7-related pathways at the same time. Complementation with LARP7 mutants that retain only selected activities will be a very helpful tool to resolve the intricate network of LARP7 functions. Successful examples for such an approach have been reported recently [Citation42,Citation118].
Albeit the role of the 7SK snRNP in the inhibition of P-TEFb has been well established, the exact consequences for the regulation of Pol II transcription are still rather puzzling. According to the current model, destruction of the 7SK snRNP is expected to result in increased P-TEFb activity, enhanced Ser2-phosphorylation of Pol II and, ultimately, increased general transcriptional output. However, this was neither the case in cell lines derived from an Alazami patient or HEK293 LARP7 knockout cells [Citation42], nor in conditional Larp7 knockout mice [Citation118] and also not upon knockdown of MePCE in breast cancer cells [Citation45]. Only subsets of selected transcripts reacted to the loss of physiological amounts of 7SK snRNPs [Citation42,Citation118,Citation146] and, even more surprisingly, reduced amounts of Ser2-phosphorylated Pol II were found at selected genes [Citation45]. In light of the growing evidences indicating that the 7SK snRNP acts as a delivery system for P-TEFb (reviewed in [Citation56,Citation58]) and for other factors [Citation53,Citation55,Citation70,Citation74,Citation76,Citation82], more studies need to clarify how this targeted delivery is achieved and why only specific subsets of transcription units are susceptible to perturbation.
Acknowledgments
We thank Giriram Kumar Mohana and Jean-Yves Roignant for discussions.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Al-Ashtal HA, Rubottom CM, Leeper TC, et al. The LARP1 La-Module recognizes both ends of TOP mRNAs. RNA Biol. 2019;1–11. DOI:10.1080/15476286.2019.1669404
- Martino L, Pennell S, Kelly G, et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2015;43(1):645–660.
- Yang R, Gaidamakov SA, Xie J, et al. La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w Motif, and can promote mRNA stability. Mol Cell Biol. 2011;31(3):542–556.
- Alfano C, Sanfelice D, Babon J, et al. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol. 2004;11(4):323–329.
- Uchikawa E, Natchiar KS, Han X, et al. Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7. Nucleic Acids Res. 2015;43(6):3373–3388.
- Teplova M, Yuan Y-R, Phan AT, et al. Structural basis for recognition and sequestration of UUU(OH) 3ʹ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85.
- Maraia RJ, Mattijssen S, Cruz-Gallardo I, et al. The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. Wiley Interdiscip Rev RNA. 2017;8:e1430.
- Cruz-Gallardo I, Martino L, Kelly G, et al. LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA. Nucleic Acids Res. 2019;47:4272–4291.
- Eichhorn CD, Chug R, Feigon J. hLARP7 C-terminal domain contains an xRRM that binds the 3ʹ hairpin of 7SK RNA. Nucleic Acids Res. 2016;44:9977–9989.
- Eichhorn CD, Yang Y, Repeta L, et al. Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc Natl Acad Sci. 2018;115(28):E6457–66.
- Lahr RM, Fonseca BD, Ciotti GE, et al. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife. 2017;6:e24146.
- Dock-Bregeon A-C, Lewis KA, Conte MR. The La-related proteins: structures and interactions of a versatile superfamily of RNA-binding proteins. RNA Biol. 2019;1–16. DOI:10.1080/15476286.2019.1695712
- Merret R, Martino L, Bousquet-Antonelli C, et al. The association of a La module with the PABP-interacting motif PAM2 is a recurrent evolutionary process that led to the neofunctionalization of La-related proteins. RNA. 2013;19(1):36–50.
- Küspert M, Murakawa Y, Schäffler K, et al. LARP4B is an AU-rich sequence associated factor that promotes mRNA accumulation and translation. RNA. 2015;21(7):1294–1305.
- Cai L, Fritz D, Stefanovic L, et al. Binding of LARP6 to the conserved 5′ stem–loop regulates translation of mRNAs encoding type i collagen. J Mol Biol. 2010;395(2):309–326.
- Lahr RM, Mack SM, Héroux A, et al. The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5′TOP sequence. Nucleic Acids Res. 2015;43(16):8077–8088.
- Bousquet-Antonelli C, Deragon J-M. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15(5):750–764.
- Deragon J-M. Distribution, organization an evolutionary history of La and LARPs in eukaryotes. RNA Biol. 2020;1–9. DOI:10.1080/15476286.2020.1739930
- Hussain RH, Zawawi M, Bayfield MA. Conservation of RNA chaperone activity of the human La-related proteins 4, 6 and 7. Nucleic Acids Res. 2013;41(18):8715–8725.
- He N, Jahchan NS, Hong E, et al. A la-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29(5):588–599.
- Okamura D, Maeda I, Taniguchi H, et al. Cell cycle gene-specific control of transcription has a critical role in proliferation of primordial germ cells. Genes Dev. 2012;26(22):2477–2482.
- Park J-M, Kohn MJ, Bruinsma MW, et al. The multifunctional RNA-binding protein la is required for mouse development and for the establishment of embryonic stem cells. Mol Cell Biol. 2006;26(4):1445–1451.
- Didier K, Bolko L, Giusti D, et al. Autoantibodies associated with connective tissue diseases: what meaning for clinicians? Front Immunol. 2018;9:541.
- Fairley JA, Kantidakis T, Kenneth NS, et al. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc Natl Acad Sci U S A. 2005;102(51):18350–18355.
- Rinke J, Steitz JA. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La Lupus antibodies. Cell. 1982;29(1):149–159.
- Muniz L, Egloff S, Kiss T. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res. 2013;41(8):4686–4698.
- Markert A, Grimm M, Martinez J, et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575.
- Krueger BJ, Jeronimo C, Roy BB, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36(7):2219–2229.
- Borkar AN, D’Orso I. Structural basis for assembly and function of the 7SK snRNP complex. Non-coding RNA Investig. 2018;2:70.
- Jeronimo C, Forget D, Bouchard A, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27(2):262–274.
- Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA. 2012;3(1):92–103.
- Páez-Moscoso DJ, Pan L, Sigauke RF, et al. Pof8 is a La-related protein and a constitutive component of telomerase in fission yeast. Nat Commun. 2018;9(1):587.
- Mennie AK, Moser BA, Nakamura TM. LARP7-like protein Pof8 regulates telomerase assembly and poly(A)+TERRA expression in fission yeast. Nat Commun. 2018;9(1):586.
- Collopy LC, Ware TL, Goncalves T, et al. LARP7 family proteins have conserved function in telomerase assembly. Nat Commun. 2018;9(1):557.
- Singh M, Wang Z, Koo B-K, et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme la family protein p65. Mol Cell. 2012;47(1):16–26.
- D’Orso I. 7SKiing on chromatin: move globally, act locally. RNA Biol. 2016;13(6):545–553.
- Afroz T, Cienikova Z, Cléry A, et al. One, two, three, four! How multiple RRMs read the genome sequence. Methods Enzymol. 2015;558:235–278.
- Huang Y, Bayfield MA, Intine RV, et al. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol. 2006;13(7):611–618.
- Brogie JE, Price DH. Reconstitution of a functional 7SK snRNP. Nucleic Acids Res. 2017;45(11):6864–6880.
- Jacks A, Babon J, Kelly G, et al. Structure of the C-terminal domain of human la protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11(7):833–843.
- Singh M, Choi CP, Feigon J. xRRM: a new class of RRM found in the telomerase La family protein p65. RNA Biol. 2013;10(3):353–359.
- Hasler D, Meduri R, Bąk M, et al. The Alazami syndrome-associated protein LARP7 guides U6 small nuclear RNA modification and contributes to splicing robustness. Mol Cell. 2020;77(5):1014–1031.e13.
- Xue Y, Yang Z, Chen R, et al. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38(2):360–369.
- Barboric M, Lenasi T, Chen H, et al. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci. 2009;106(19):7798–7803.
- Shelton SB, Shah NM, Abell NS, et al. Crosstalk between the RNA methylation and histone-binding activities of MePCE regulates P-TEFb activation on chromatin. Cell Rep. 2018;22(6):1374–1383.
- Schneeberger PE, Bierhals T, Neu A, et al. de novo MEPCE nonsense variant associated with a neurodevelopmental disorder causes disintegration of 7SK snRNP and enhanced RNA polymerase II activation. Sci Rep. 2019;9(1):12516.
- Chambers JC, Kurilla MG, Keene JD. Association between the 7 S RNA and the lupus La protein varies among cell types. J Biol Chem. 1983;258(19):11438–11441.
- Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol. 1991;11(7):3432–3445.
- Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta. 2010;1799(5–6):365–378.
- Bhattacharya R, Perumal K, Sinha K, et al. Methylphosphate cap structure in small RNAs reduces the affinity of RNAs to La protein. Gene Expr. 2002;10:243–253.
- Yang Y, Eichhorn CD, Wang Y, et al. Structural basis of 7SK RNA 5′-γ-phosphate methylation and retention by MePCE. Nat Chem Biol. 2019;15(2):132–140.
- Marz M, Donath A, Verstraete N, et al. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol. 2009;26(12):2821–2830.
- Van Herreweghe E, Egloff S, Goiffon I, et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. Embo J. 2007;26(15):3570–3580.
- Krueger BJ, Varzavand K, Cooper JJ, et al. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One. 2010;5(8):e12335.
- Flynn RA, Do BT, Rubin AJ, et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat Struct Mol Biol. 2016;23(3):231–238.
- Egloff S, Studniarek C, Kiss T. 7SK small nuclear RNA, a multifunctional transcriptional regulatory RNA with gene-specific features. Transcription. 2018;9(2):95–101.
- Quaresma AJC, Bugai A, Barboric M. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res. 2016;44(16):7527–7539.
- Li Y, Liu M, Chen L-F, et al. P-TEFb: finding its ways to release promoter-proximally paused RNA polymerase II. Transcription. 2018;9(2):88–94.
- Yang Z, Zhu Q, Luo K, et al. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414(6861):317–322.
- Nguyen VT, Kiss T, Michels AA, et al. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414(6861):322–325.
- Blazek D, Barboric M, Kohoutek J, et al. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res. 2005;33:7000–7010.
- Li Q, Price JP, Byers SA, et al. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280(31):28819–28826.
- Byers SA, Price JP, Cooper JJ, et al. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005;280(16):16360–16367.
- Michels AA, Fraldi A, Li Q, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. Embo J. 2004;23:2608–2619.
- Barboric M, Kohoutek J, Price JP, et al. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. Embo J. 2005;24:4291–4303.
- Dames SA, Schonichen A, Schulte A, et al. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc Natl Acad Sci. 2007;104:14312–14317.
- Kobbi L, Demey-Thomas E, Braye F, et al. An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb. Proc Natl Acad Sci. 2016;113:12721–12726.
- Bacon CW, D’Orso I. CDK9: a signaling hub for transcriptional control. Transcription. 2019;10:57–75.
- McNamara RP, Bacon CW, D’Orso I. Transcription elongation control by the 7SK snRNP complex: releasing the pause. Cell Cycle. 2016;15:2115–2123.
- Ji X, Zhou Y, Pandit S, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868.
- McNamara RP, McCann JL, Gudipaty SA, et al. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep. 2013;5:1256–1268.
- Liu W, Ma Q, Wong K, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595.
- Bi X, Xu Y, Li T, et al. RNA Targets Ribogenesis Factor WDR43 to chromatin for transcription and pluripotency control. Mol Cell. 2019;75:102–116.e9.
- Calo E, Flynn RA, Martin L, et al. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature. 2015;518:249–253.
- McNamara RP, Reeder JE, McMillan EA, et al. KAP1 recruitment of the 7SK snRNP complex to promoters enables transcription elongation by RNA polymerase II. Mol Cell. 2016;61:39–53.
- Egloff S, Vitali P, Tellier M, et al. The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. Embo J. 2017;36:934–948.
- D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17:815–821.
- Eilebrecht S, Le Douce V, Riclet R, et al. HMGA1 recruits CTIP2-repressed P-TEFb to the HIV-1 and cellular target promoters. Nucleic Acids Res. 2014;42:4962–4971.
- Bugai A, Quaresma AJC, Friedel CC, et al. P-TEFb activation by RBM7 shapes a pro-survival transcriptional response to genotoxic stress. Mol Cell. 2019;74:254–267.e10.
- Sun Y, Liu Z, Cao X, et al. Activation of P-TEFb by cAMP-PKA signaling in autosomal dominant polycystic kidney disease. Sci Adv. 2019;5:eaaw3593.
- Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880.
- Barrandon C, Bonnet F, Nguyen VT, et al. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol. 2007;27:6996–7006.
- Elagib KE, Rubinstein JD, Delehanty LL, et al. Calpain 2 activation of P-TEFb drives megakaryocyte morphogenesis and is disrupted by leukemogenic GATA1 mutation. Dev Cell. 2013;27:607–620.
- Mück F, Bracharz S, Marschalek R. DDX6 transfers P-TEFb kinase to the AF4/AF4N (AFF1) super elongation complex. Am J Blood Res. 2016;6:28–45.
- Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. Embo J. 1998;17:7442–7453.
- Chakshusmathi G, Do KS, Rubinson DA, et al. A La protein requirement for efficient pre-tRNA folding. Embo J. 2003;22:6562–6572.
- Belisova A, Semrad K, Mayer O, et al. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11:1084–1094.
- Kucera NJ, Hodsdon ME, Wolin SL. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc Natl Acad Sci U S A. 2011;108:1308–1313.
- Naeeni AR, Conte MR, Bayfield MA. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. J Biol Chem. 2012;287:5472–5482.
- Berman AJ, Gooding AR, Cech TR. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol. 2010;30:4965–4976.
- Stone MD, Mihalusova M, O’Connor CM, et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461.
- Akiyama BM, Loper J, Najarro K, et al. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA. 2012;18:653–660.
- Briese M, Saal-Bauernschubert L, Ji C, et al. hnRNP R and its main interactor, the noncoding RNA 7SK, coregulate the axonal transcriptome of motoneurons. Proc Natl Acad Sci. 2018;115:E2859–68.
- Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759.
- Aigner S, Lingner J, Goodrich KJ, et al. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. Embo J. 2000;19:6230–6239.
- Lingner J, Cech TR. Purification of telomerase from euplotes aediculatus: requirement of a primer 3ʹ overhang. Proc Natl Acad Sci. 1996;93:10712–10717.
- Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev. 2004;18:1107–1118.
- Jiang J, Miracco EJ, Hong K, et al. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192.
- Jiang J, Chan H, Cash DD, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070.
- O’Connor CM, Collins K. A novel RNA binding domain in tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol. 2006;26(6):2029–2036.
- Prathapam R, Witkin KL, O’Connor CM, et al. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol. 2005;12(3):252–257.
- Tang W, Kannan R, Blanchette M, et al. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484(7393):260–264.
- Weichenrieder O. RNA binding by Hfq and ring-forming (L)Sm proteins: a trade-off between optimal sequence readout and RNA backbone conformation. RNA Biol. 2014;11(5):537–549.
- Gruss OJ, Meduri R, Schilling M, et al. UsnRNP biogenesis: mechanisms and regulation. Chromosoma. 2017;126(5):577–593.
- Páez-Moscoso DJ, Pan L, Sigauke RF, et al. Pof8 is a La-related protein and a constitutive component of telomerase in fission yeast. Nat Commun. 2018;9(1):587.
- Nguyen THD, Tam J, Wu RA, et al. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature. 2018;557:190–195.
- Venteicher AS, Abreu EB, Meng Z, et al. A human telomerase holoenzyme protein required for cajal body localization and telomere synthesis. Science. 2009;323(5914):644–648.
- Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28(5):773–785.
- Holohan B, Kim W, Lai T-P, et al. Impaired telomere maintenance in Alazami syndrome patients with LARP7 deficiency. BMC Genomics. 2016;17(S9):749.
- Dai Q, Luan G, Deng L, et al. Primordial dwarfism gene maintains Lin28 expression to safeguard embryonic stem cells from premature differentiation. Cell Rep. 2014;7:735–746.
- Trippe R. A highly specific terminal uridylyl transferase modifies the 3ʹ-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126.
- Trippe R. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12(8):1494–1504.
- Warda AS, Kretschmer J, Hackert P, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014.
- Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m 6 A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–835.e14.
- Bell M, Schreiner S, Damianov A, et al. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. Embo J. 2002;21:2724–2735.
- Medenbach J, Schreiner S, Liu S, et al. Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol Cell Biol. 2004;24:7392–7401.
- Novotný I, Malinová A, Stejskalová E, et al. SART3-dependent accumulation of incomplete spliceosomal snrnps in cajal bodies. Cell Rep. 2015;10(3):429–440.
- Wang X, Li Z-T, Yan Y, et al. LARP7-mediated U6 snRNA modification ensures splicing fidelity and spermatogenesis in mice. Mol Cell. 2020;77(999–1013.e6). DOI:10.1016/j.molcel.2020.01.002.
- Lui L, Lowe T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem. 2013;54:53–77.
- Von Dwingelo J, Chung IYW, Price CT, et al. Interaction of the Ankyrin H core effector of legionella with the host LARP7 component of the 7SK snRNP complex. MBio. 2019;10. DOI:10.1128/mBio.01942-19
- Qiu J, Luo Z-Q. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol. 2017;15(10):591–605.
- Habyarimana F, Al-khodor S, Kalia A, et al. Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol. 2008;10(6):1460–1474.
- Burstein D, Amaro F, Zusman T, et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet. 2016;48(2):167–175.
- Gomez-Valero L, Rusniok C, Carson D, et al. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci. 2019;116(6):2265–2273.
- Pan X, Luhrmann A, Satoh A, et al. Ankyrin repeat proteins comprise a diverse family of bacterial type iv effectors. Science. 2008;320(5883):1651–1654.
- Mori Y, Sato F, Selaru FM, et al. Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res. 2002;62:3641–3645.
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–346.
- Fujimoto A, Fujita M, Hasegawa T, et al. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res. 2020;30(3):334–346.
- Kondelin J, Gylfe AE, Lundgren S, et al. Comprehensive evaluation of protein coding mononucleotide microsatellites in microsatellite-unstable colorectal cancer. Cancer Res. 2017;77(15):4078–4088.
- Cheng Y, Jin Z, Agarwal R, et al. LARP7 is a potential tumor suppressor gene in gastric cancer. Lab Investig. 2012;92(7):1013–1019.
- Ji X, Lu H, Zhou Q, et al. LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis. Elife. 2014;3:e02907.
- Sui X, Sui Y, Wang Y. LARP7 in papillary thyroid carcinoma induces NIS expression through suppression of the SHH signaling pathway. Mol Med Rep. 2018;17:7521–7528.
- Alazami AM, Al-Owain M, Alzahrani F, et al. Loss of function mutation in LARP7, chaperone of 7SK ncRNA, causes a syndrome of facial dysmorphism, intellectual disability, and primordial dwarfism. Hum Mutat. 2012;33(10):1429–1434.
- Dateki S, Kitajima T, Kihara T, et al. Novel compound heterozygous variants in the LARP7 gene in a patient with Alazami syndrome. Hum Genome Var. 2018;5:18014.
- Ling TT, Sorrentino S. Compound heterozygous variants in the LARP7 gene as a cause of Alazami syndrome in a Caucasian female with significant failure to thrive, short stature, and developmental disability. Am J Med Genet Part A. 2016;170(1):217–219.
- Imbert-Bouteille M, Mau Them FT, Thevenon J, et al. LARP7 variants and further delineation of the Alazami syndrome phenotypic spectrum among primordial dwarfisms: 2 sisters. Eur J Med Genet. 2019;62:161–166.
- Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478(7367):57–63.
- Hollink IH, Alfadhel M, Al-Wakeel AS, et al. van de Laar IM. Broadening the phenotypic spectrum of pathogenic LARP7 variants: two cases with intellectual disability, variable growth retardation and distinct facial features. J Hum Genet. 2016;61(3):229–233.
- Wojcik MH, Linnea K, Stoler JM, et al. Updating the neurodevelopmental profile of Alazami syndrome: illustrating the role of developmental assessment in rare genetic disorders. Am J Med Genet A. 2019;179:1565–1569.
- Ivanovski I, Caraffi SG, Magnani E, et al. Alazami syndrome: the first case of papillary thyroid carcinoma. J Hum Genet. 2020;65(2):133–141.
- Yang S-L, Yang M, Herrlinger S, et al. MiR-302/367 regulate neural progenitor proliferation, differentiation timing, and survival in neurulation. Dev Biol. 2015;408(1):140–150.
- Parchem RJ, Moore N, Fish JL, et al. miR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 2015;12:760–773.
- Nguyen D, Krueger BJ, Sedore SC, et al. The drosophila 7SK snRNP and the essential role of dHEXIM in development. Nucleic Acids Res. 2012;40(12):5283–5297.
- Slomnicki LP, Malinowska A, Kistowski M, et al. Nucleolar enrichment of brain proteins with critical roles in human neurodevelopment. Mol Cell Proteomics. 2016;15(6):2055–2075.
- Prasanth KV, Camiolo M, Chan G, et al. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell. 2010;21(23):4184–4196.
- Castelo-Branco G, Amaral PP, Engström PG, et al. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013;14(9):R98.
- Fan H, Goodier JL, Chamberlain JR, et al. 5′ processing of tRNA precursors can be modulated by the human la antigen phosphoprotein. Mol Cell Biol. 1998;18(6):3201–3211.
- Marrella SA, Brown KA, Mansouri-Noori F, et al. An interdomain bridge influences RNA binding of the human La protein. J Biol Chem. 2019;294(5):1529–1540.
