ABSTRACT
The emergence of New Delhi metallo-beta-lactamase (NDM-1) has become a major health threat to clinical managements of gram-negative bacteria infections. A novel incompatibility group X3 plasmid (IncX3) pNDM-HN380 carrying blaNDM-1 has recently been found to epidemiologically link with multiple geographical areas in China. In this paper, we studied the metabolic responses of host bacteria E. coli J53 upon introduction of pNDM-HN380. A reduction of bacterial motility was observed in J53/pNDM-HN380. We profiled the RNA repertoires of the transconjugants and found a downregulation of genes involved in flagella and chemotaxis metabolic pathways at logarithmic (log) phase. We also identified a novel intragenic region (IGR) small RNA plas2. The plasmid-transcribed sRNA IGR plas2 was further characterized as a regulator of fucRwhich controls the fucose metabolism. By knockdown of IGR plas2 using an antisense decoy, we managed to inhibit the formation of bacterial biofilm of the host. Our study demonstrated a potential way of utilizing plasmid-transcribed sRNA against infectious bacteria.
Introduction
The New Delhi metallo-β-lactamase (NDM-1) is a novel carbapenemases initially identified in Klebsiella pneumoniaeisolated from a Swedish traveller who acquired urinary tract infections during his visit in New Delhi [Citation1]. The original clinical isolate containing NDM-1 was a ‘super-bug’ and was found resistant to almost all antibiotics including but not limited to β-lactams, fluoroquinolones and aminoglycoside [Citation2]. NDM-1 can destroy carbapenems, the known last resort of drugs for the treatment of extended-spectrum β-lactamases (ESBLs) producing bacterial infections and has become the biggest clinical challenge in controlling gram-negative infections [Citation1–4]. Today, researchers noticed that the NDM-1 coding gene blaNDM-1has been distributed to various plasmids in different incompatibility groups (IncF, IncA/C, IncH, IncL/Mand IncN) with narrow or broad host ranges among Enterobacteriaceae, but seldom found on bacterial chromosome [Citation2,Citation5–11], suggesting mobile genetic elements (MGEs) is the major contributor of disseminating NDM-1. Indeed, NDM-1 has disseminated globally to four continents, in particular the most popular countries such as China, India, United States and Kenya.
One concerning NDM-1 carrying plasmid, pNDM-HN380, was recently identified from K. pneumoniae isolates in a Hong Kong hospital. The complete sequencing of pNDM-HN380 revealed a 54kb IncX3 plasmid with 52 open reading frames whereas blaNDM-1 gene was found in a transposon-like structure flanked by ISAba125 and IS26 which are identical to plasmids from Acinetobacter isolates carrying blaNDM-1–positive Tn125 [Citation12,Citation13]. In addition to blaNDM-1, two other drug-resistance genes, blaMBL and blaSHV-12, were also found on pNDM-HN380 as well as a type IV conjugation secretion system, suggesting the plasmid could be an important vehicle for the dissemination NDM-1 to different geographical areas in China [Citation11].
In the last decade, studies of multidrug resistance (MDR) bacteria were mainly focused on their resistance and dissemination mechanisms [Citation3,Citation14,Citation15]. Nevertheless, more attention has been drawn to the beneficial phenotypic traits attributed by plasmid-mediated gene regulations of host bacteria in different ecological niches [Citation16–18]. Biofilm, for example, is one of the most important characteristics of bacteria. The formation of biofilm tends to trigger relapse after medical treatments and antibiotic tolerance in chronic infections, from which several lines of evidence have indicated that plasmid plays an important role in biofilm formation [Citation19–21]. Plasmid-transcribed small RNAs (sRNAs) which act as mass regulators are one of the most effective ways to control gene regulatory network of multidrug resistance bacteria [Citation22]. In addition, increasing evidences of utilizing sRNAs as potential weapons against bacterial resistance were found in different bacteria strains recently. In E. coli, overexpression of sRNA MtvRled to enhancement of bacterial sensitivities to multiple antibiotics, including ampicillin, chloramphenicol, ciprofloxacin and tetracycline [Citation23]. In P. aeruginosa, overexpression of the same sRNA could significantly inhibit the bacterial growth in medium with the mentioned antibiotics, while deletion of the sRNA reversed all induced antibiotic susceptibilities [Citation24]. Another sRNA, RyhB, was also found closely related to antibiotic resistance through the tricarboxylic acid cycle (TCA cycle) [Citation25]. Antibiotics like ampicillin employ the bacterial TCA cycle to produce deleterious hydroxyl radicals. The sRNA targets AcnA which encodes a TCA cycle-related enzyme. Thus, overexpression of the sRNA may lead to TCA cycle dysfunction and subsequently cause increases of resistance to β-lactam [Citation26]. In conclusion, sRNA has a great potential to be used as a new type of drug against drug-resistance bacteria.
In this work, we investigated the metabolic impacts of the bacterial host E. colicarrying pNDM-HN380. In RNA-seq result, we found that the expression of flagella and chemotaxis genes was down-regulated at the logarithmic phase and consequently caused a reduction of host bacterial motility. Moreover, a novel plasmid-transcribed sRNA IGR plas2 was identified and characterized as a regulator of fucR gene which involves in the fucose metabolism. In addition, biofilm formation of the bacteria carrying pNDM-HN380 was found to be mediated by IGR plas2 sRNA.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in . Escherichia coli (E. coli) J53 was used as a host for conjugation of pNDM-HN380 [Citation27] and E. coli DH5α was used for molecular cloning [Citation28]. The pNDM-HN380 plasmid isolated from clinical K. pneumoniae strain was kindly given by Dr. P. L. Ho. E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium with shaking at 250 rpm overnight as starter and then 1:1000 diluted in fresh LB medium. For RNA extraction and next-generation sequencing, bacteria were incubated in LB medium and harvested at mid-log (OD600 nm = 0.6) and early stationary phase (OD600 nm = 1.2), respectively. For growth curve experiments of sRNA IGR plas2, 0.5 mL overnight cultured bacteria (OD600 nm = 4.0 ± 0.2) were inoculated into 50 mL M9 medium supplemented with 0.4% glucose or fucose.
Table 1. Strains and plasmids used in this study
Antimicrobial susceptibility test
Antimicrobial susceptibility tests were performed using the Kirby-Bauer disk diffusion method according to the Clinical Laboratory Standards Institute (CLSI) 2012 guideline. Implanted antibiotics are listed in Supplementary Table S1. The interpretation of susceptibility result was conducted according to the CLSI guidelines. E. coli ATCC25922 was used as a reference strain as recommended.
RNA-sequencing
E. coli J53 carrying pNDM-HN380 (J53/pNDM-HN380) were grown in LB medium and harvested at mid-log and early stationary phases. Total RNAs were extracted from harvested cell pellets using the standard TRIZOL method (Thermo Fisher Scientific, Waltham, MA, USA). The TURBO DNA-free™ Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to remove host genomic DNA (gDNA). Small RNA (sRNA) and message RNA (mRNA) were isolated from the total RNA using the MICROExpress™ Bacterial mRNA Enrichment Kit (Thermo Fisher Scientific, Waltham, MA, USA) followed by the Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA). Both kits were used to remove ribosomal RNA (rRNA). Sequencing libraries were constructed using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina (Illumina, New England Biolabs, Ipswich, MA, USA) from the enriched sRNA and mRNA. Finally, RNA-sequencing was performed by the Illumina NextSeq system (NextSeq 500/550 Mid OutputKit v2, Illumina, San Diego, CA, USA).
Bioinformatics analysis
SRA files generated from the Illumina NextSeq500 were converted to fastq files using the SRA Toolkit. The Bowtie2 alignment tool (version 2.3.1) was employed to perform sequence-alignments of the converted fastq files to the Bowtie-indexes of E. coli MG1655 genome (GenBank accession:NC 000913.3). SAMtools (version 0.1.19) was used to sort and compress aligned sequence files into BAM files. The sorted and compressed BAM files were then used for downstream processes in the Integrative Genomics Viewer (IGV) and the R software (version 3.4.0) [Citation29]. Two R packages, GenomicAlignments and GenomicFeatures were used to analyze and generate transcript expression results. Gitools was used to perform Sample Level Enrichment Analysis (SLEA) with KEGG pathways. Circos was used to present transcriptome data [Citation30].
Swimming motility test
E. coli J53 and J53/pNDM-HN380 transconjugants were grown at 37°C with 250 rpm shaking overnight. Then, cultures were spread onto freshly prepared LB soft-agar plates (0.3% Bacto Agar) and incubated at 37°C until cells swam out. Cultures were picked from the leading edge of the swimming cells and inoculated in LB for overnight growth. Adjusted volume of the overnight culture was centrally spotted on soft-agar plates. After 8 h, the diameter of the spreading colony was measured from at least 10 plates for each experiment. The swimming motility tests were performed in three independent experiments.
Northern blot
Northern blot was employed to validate the presence of sRNA. Probes of 35–45 nucleotides that are specific for target sRNA were designed. Total RNA was separated by denaturing polyacrylamide-urea gel electrophoresis, transferred to a Hybond-XL membrane (Amersham) and cross-linked under ultra-violet (UV) radiation. Oligo probes specific for each sRNA candidate were labelled with [γ-P32]-ATP or alternatively labelled with digoxygenin using digoxygenin-dUTP (DIG DNA labelling and detection kit, Roche), and hybridized with the RNA on the membrane. Hybridization signals were exposed to a phosphor screen overnight ([γ-P32]-ATP labelling) and visualized with a phosphorimager (Typhoon TRIO, Amersham Biosciences) or were detected with an anti-DIG-alkaline phosphatase conjugate and the substrates NBT (Nitroblue Tetrazolium Salt) and BCIP (5-bromo-4-chloro-3-indolyl phosphate, toluidinium salt), which give a light-blue precipitate.
Construction of sRNA expression vector
Polymerase chain reactions (PCR) were used to amplify target DNA fragments from pNDM-HN380. The original promoter of IGR plas2 (TTAACG) was included in the amplified DNA fragment for IGR plas2 overexpression. For expression of the decoy of IGR plas2 (dplas2), promoter pTac was used. The amplified fragments were then cloned to two expression vectors, pET-28a+ (for IGR plas2) and pTL134 (for dplas2). For pET-28a+, XbaI and HindIII restriction cutting sites were used. For pTL134, PacII and AatIIrestriction cutting sites were used. The constructed vectors were then transformed into DH5α competent cells and then selected in LB Agar plates containing Kanamycin (50 μg/mL) or chloramphenicol (20 μg/mL). Colony PCR and sanger sequencing were used to confirm the correct construction of the fragment of interest. The constructed vectors pET28a_plas2 and pTL134_dplas2 were then extracted from DH5α transconjugants and transformed into J53 competent cells and then selected in LB Agar plates containing Kanamycin (50 μg/mL) or chloramphenicol (20 μg/mL).
sRNA target prediction and quantitative real-time PCR (qRT-PCR)
IntaRNA software (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp) was used to predict sRNA target genes. Predicted target genes were validated by qRT-PCR using the Quant Studio 3 Real-Time PCR System in 96-well plates. One microlitre of 10 ng/μLcDNA was added to 9 μL PCR mixture (5 μL SYBR green mix and 2 μL of2μMtarget gene primers). Primers were designed using the Primer3. (Supplementary Table S2). The following thermal cycling program was used: an initial denaturation step at 98°C for 30 s followed by 40 cycles of 98°C for 5 s and 63°C for 10 s (annealing/synthesis step), and then followed by a melting curve from 65°C to 95°C in 0.5°C and 5 s increments. gapA was used as internal control gene. All experiments were performed in three biological replicates, with three technical replicates each. Results were analysed using the comparative critical threshold (ddCT) method, in which the relative quantities of the genes of interest are normalized to the relative quantities of the reference genes across samples.
Biofilm assay
Bacterial cells were grown in LB medium at 37°C with shaking at 250 rpm overnight. The final optical density (OD) at 600 nm of the overnight culture was 4.0 ± 0.2, and 1.5 μL of the overnight culture was then added to the 96-wells plate containing 150 μL (100-fold) per well M9 minimal medium in the presence and absence of fucose or glucose (0.4%). To induce expression of dplas2, 0.4 mM final concentration of IPTG was added to each well after 4 h of static culture. The 96-wells plate was further incubated at 37°C for 24 h without shaking. Media were then removed, and wells were stained by 0.1% crystal violet for 10 min at room temperature. The crystal violet stains were rinsed off thoroughly with distilled water. Biofilms were dislodged by adding 100 μL 30% acetic acid to all wells. Pipet up and down until homogenous solution was formed in each well. The solutions were transferred into a new 96-wells plate. The absorbance at 595 nm of each well were measured by the plate reader.
RNA-RNA Gel-shift
The RNA-RNA gel-shift experiment was performed according to a previously reported protocol [Citation31] with minor modification. Briefly, DNA temples of IGR plas2 and fucR were amplified from pNDM-HN380 and J53 using specific primers attached with T7 promoter, respectively. RNAs were synthesized using the T7 RiboMAX™ Large-Scale RNA Production System, according to the manufacturer’s specific cations. Briefly, 40 ng of template DNA for RNA transcripts of ~200 nt in 20 μL of reaction volume was used. The reaction was incubated at 37°C for 2–4 h. DNase (1 U per μg of template DNA) was added and incubated at 37°C for 15 min for eliminating template DNA. Equivalent volumes of 2xRNA loading buffer were mixed with samples and ran on a 5% polyacrylamide gel with 7 M urea in 1xTBE for purifying RNA transcripts of the correct size from the mixture. Electrophoresis was performed at a constant voltage of 200 V for 2 h. The correct-sized RNA band was cut from the gel with a sterilized scalpel. The gel piece was transferred to a micro-centrifuge tube and mixed with 3–5 times volume of RNA elution buffer and mixed by inverting continuously overnight at 4°C. TRIZOL was added to the tube to extract the RNAs (procedures are the same as described in the RNA-sequencing section above). The following steps of RNA-RNA gel shift was conducted exactly as described in the protocol [Citation31].
Statistics
Paired Student’s t-test was performed to calculate the p-values. p-Values < 0.05 were considered as significant.
Results
Physiologicalstudy of E. coli J53 carrying pNDM-HN380
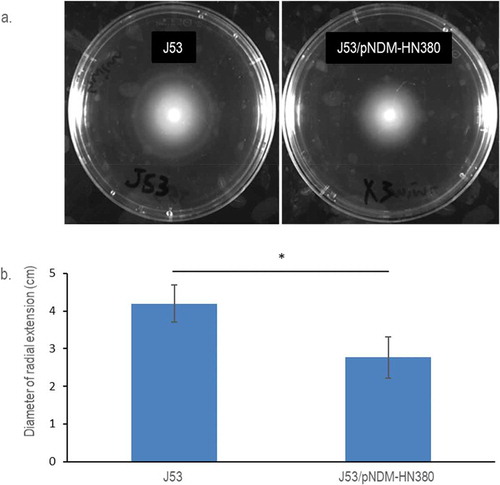
The plasmid pNDM-HN380 was conjugated to E. coli J53 in this study. The growth curves of E. coli J53 carrying pNDM-HN380 (J53/pNDM-HN380) and isogenic J53 were measured and compared as shown in Supplementary Figure S1, and no significant difference was observed. To investigate antibiotic resistance conferred by pNDM-HN380, the Kirby–Bauer disk diffusion test was used. A shorter zone diameter of inhibition indicates a higher antibiotic resistance in the test. As shown in Supplementary Table S1, J53/pNDM-HN380 was resistant to β-lactams including cephems (Cefotaxime, CTX) and carbapenems (Meropenem, Ertapenem and Imipenem) but not to Tetracycline, Kanamycin or Chloramphenicol. To investigate the effect of pNDM-HN380 on the mobility of bacteria, we performed swimming motility tests on soft-agar plates. As shown in , the swimming distance of J53/pNDM-HN380 was shorter than that of J53, suggesting a lower motility of transconjugants. These results demonstrated the physiological impacts of bacteria carrying plasmidpNDM-HN380, which confer multidrug resistance and attenuate the mobility of the cell.
Figure 1. Motility of E. coli J53 and J53/pNDM-HN380. (a). Representative pictures of radial extension in 0.3% soft LB agar. Bacterial strains were labelled accordingly. (b). Diameters of the radial extension halo. The experiments were conducted in biological triplicates. Bars indicate the standard deviation. * p-Value <0.05
Transcriptomes profiling of E.colicarrying pNDM-HN380
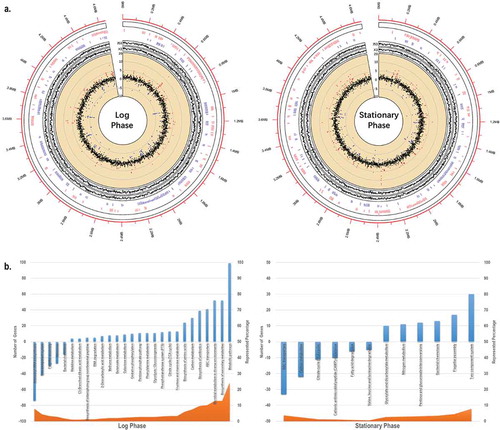
We sequenced and analysed the transcriptomes of E.coli J53 and J53/pNDM-HN380 at both log and stationary phases. Among 4469 genes on the E.coli J53 chromosome, 330 (7.6% at log phase) and 296 (6.6% at stationary phase) genes differentially expressed with log2-fold change bigger than two when the bacteria carry pNDM-HN380 (). To understand the physiological influence attributed by the pNDM-HN380, we performed the Sample Level Enrichment Analysis (SLEA) with KEGG pathways. Genes with log2-fold changes larger than one were counted as the pathway changes. As shown in , about 100 genes in metabolic pathways were upregulated while over 70 genes in aminoacyl-tRNA (aa-tRNA) biosynthesis were downregulated at the log phase. The upregulation of the metabolic pathways may implicate the burden of carrying plasmids. AminoacyltRNA is crucial for delivering amino acid to ribosome for polypeptide chain synthesis. Lower expression of aminoacyl tRNA biosynthesis genes suggested the fitness cost of plasmid to the bacterial host. Intriguingly, a significant number of genes which cluster in the pathways including flagella biosynthesis/assembly and bacterial chemotaxis family were downregulated at log phase (Supplementary Figure S2). The result was indeed the molecular explanation of our previous swimming motility experiment that showed lower motility in the transconjugants carrying the plasmid. In addition, we found that the ABC transporter genes (), especially the autoinducer 2 (AI-2) quorum-sensing (QS) operon (lsrABCDF), upregulated significantly in J53/pNDM-HN380 transconjugate at log phase. Previous studies have shown that both quantity and architecture of biofilms are influenced by the QS system [Citation32]. The presence of pNDM-HN380 may promote and facilitate the biofilm formation by moderating the motility and QS system of bacteria.
Figure 2. In silico analysis of E. coli J53/pNDM-HN380 at log and stationary phases. (a). Transcriptome analysis. The outermost circle (red) represents the genome coordinates (in Mbp) of J53. The first inner circle represents the names of genes of J53/pNDM-HN380 with fold-change greater than 2 folds (in red) and −2 folds (in blue) and the locations of genes in the genome. The second inner circle represents gene expression of J53 (J53) and J53/pNDM-HN380 (X3) in TPM (transcripts per million) in log2 scale. The third inner circle represents the change of gene expression in J53/pNDM-HN380 compared with J53, in a log2 scale. The red and blue dots represent upregulated and downregulated genes (>2-fold) respectively, and others are coloured in black. (b). KEGG enrichment pathway analysis. Columns in blue represent the number of genes with log2-fold change greater than 1 in the pathway. Area in orange represents the percentage of the number of genes with log2-fold change greater than 1 annotation under the pathway. Positive numbers represent the number of upregulated genes in the pathway whereas negative numbers represent the number of downregulated genes in the pathway
Identification and characterization of sRNAs of J53/pNDM-HN380
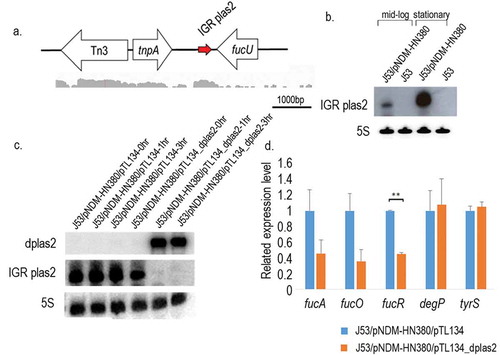
In addition to the global changes upon the carriage of pNDM-HN380, small RNA (sRNA) expressions influenced by pNDM-HN380 were profiled. Small RNAs transcribed from chromosome and their target genes were compared between J53 and J53/pNDM-HN380. We found that outer membrane proteins (OMP)-related sRNAs micF and motility moderator rprA downregulated in the presence of pNDM-HN380 at log phase, suggesting the regulatory roles of the plasmid on the sRNAs expression. In addition to those known sRNAs from bacteria, one novel plasmid-transcribed intergenic region (IGR) sRNA IGR plas2 (, SupplementaryTable 3) was identified from the sequencing result. The IGV software was employed to visualize sequencing reads and screen for the peak of reads that was not located on coding regions. We finally found a peak of aligned reads located in the intergenic region between tnpA and mpr on pNDM-HN380 (). We also used BLAST to further confirm that the sequence was not a coding mRNA or other sRNAs. The presence and corresponding size of IGR plas2 were validated by Northern blot analysis with 5 S rRNA as a control. As shown in , IGR plas2 is about 250 bp long and the expression level of IGR plas2 was found higher at stationary phase than at log phase ().
Table 2. Information of the novel sRNA
Figure 3. sRNA IGR plas2 and its decoy dplas2. (a). The genetic locus of IGR plas2. The arrow depicted in red is IGR plas2. RNA-sequencing reads showed below were captured using the Integrative Genomics Viewer (IGV) software. (b). Northern blot of IGR plas2. The experiment was performed at both log and stationary phases. The 5 S rRNA was used as control. (c). Northern blot validating the expression of dplas2 and the degradation of IGR plas2. The level of IGR plas2 was measured using the Northern blot assay at time points of 0, 1 and 3 hours after induction of dplas2. 5 S rRNA was used as control. (d). qRT-PCR of IGR plas2 target genes. Relative mRNA levels of predicted target genes of IGR plas2 were measured after the knockdown of IGR plas2. fucA, fucO, and fucR are functional genes in the fuc operon. J53/pNDM-HN380/pTL134 were used as controls. IGR: Intergenic region. Bars indicate the standard deviation. ** p-value <0.01
Construction of an IGR plas2-free system using sRNA decoy
Since the IGR plas2 is a plasmid-transcribed small RNA, the presence of pNDM-HN380 in bacteria host is indispensable for its functional characterization and study. Therefore, an IPTG-inducible pTL134_dplas2 (antisense sRNA of IGR plas2, dplas2) vector derived from pTL134 was constructed and transformed into J53/pNDM-HN380. As shown in Figure 3cand Supplementary Figure S3, the expression of dplas2 was detected after IPTG induction and stably presented in the host cell for 3 h. On the other hand, IGR plas2 was diminished once dplas2 was expressed, indicating that an IGR plas2-free system was established in J53/pNDM-HN380 using sRNA decoy.
Modulation of fucose metabolism by IGR plas2 sRNA
In order to study the function of IGR plas2, IntaRNA was used to predict targets of the sRNA. As shown in Supplementary Figure S4a and , IGR plas2 was predicted to interact withfucR which is a DNA-binding transcriptional activator that regulates genes involved in the fucose metabolic pathway including fucA and fucO [Citation33]. Quantitative RT-PCR was performed to validate the interaction between IGR plas2 and its target genes. Intriguingly, overexpression of IGR plas2 enhanced the expression of fucR (Supplementary Figure S4b). On the other hands, a 50% reduction in gene expressions of fucR, fucA and fucOwere observed when IGR plas2 was knockdown by the dplas2 decoy (). These results suggested the enhancer function of IGR plas2 in modulating the fucose metabolism which is vital for fitness to bacteria presented in gut microbiomes. To further investigate the functional consequence of plas2 sRNA in fucose metabolism, we measured the cell growth of bacteria (carrying pNDM-HN380 plasmid) overexpressing dplas2 decoy in M9 minimal medium with fucose as the only carbon source. As shown in and , reduced level of plas2 showed a defect in bacterial growth and increased doubling time whereas no difference was observed in bacteria without pNDM-HN380 plasmid under the same conditions (Supplementary Figure S5). This result suggested the functional relationship of plas2 sRNA in fucose metabolism of bacteria carrying pNDM-HN380 plasmid.
Table 3. Doubling time of E. coli strains in fucose medium
Figure 4. Growth curves in M9 minimal medium with fucose as solo carbon source. The growth of E. coli J53/pNDM-HN380/pTL134 and J53/pNDM-HN380/pTL134_dplas2 in M9 minimal medium supplemented with fucose was measured. Decoy plas2 (dplas2) was induced by IPTG at O.D.600nm = 0.5. J53/pNDM-HN380/pTL134 was used as control. Bacteria were cultured at 37°C with 250 rpm shaking. The experiments were conducted in biological triplicates. Bars indicate the standard deviation
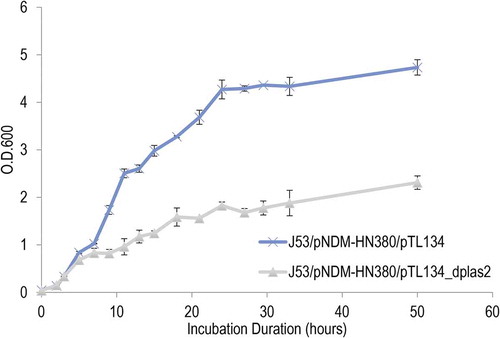
Reduced biofilm formation of J53/pNDM-HN380 with decoy of IGR plas2
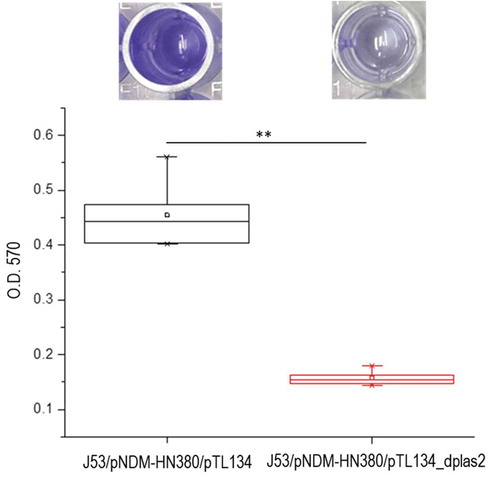
Biofilms are extracellular matrix formed by multicellular bacteria community. Fucose metabolism was reported to be closely related to the formation of gram-negative bacteria biofilm [Citation34]. To test if IGR plas2 also influenced bacterial biofilm formation, we compared the quantity of biofilm of J53/pNDM-HN380 in M9 medium using an inducible IGR plas2decoy system. Our result showed a significant inhibition of biofilm formation regardless of sugar supplement when IGR plas2 was suppressed (, Supplementary Figure S6a). No significant difference was observed between J53 and J53/pNDM-HN380 in M9 minimal medium (Data not shown). These evidences suggested sRNA IGR plas2 played a significant role in regulating biofilm formation.
Discussion
Dissemination of drug-resistant genes through the mobile element such as plasmids has been well studied in the last few decades. Nevertheless, recent evidences were found that the function of plasmids was not only restricted from disseminating drug-resistant gene [Citation35–38], but also mediating a multitude of adaptations of bacteria in stress environments [Citation39]. Previously, we identified and characterized an incompatibility group X3 plasmid (IncX3) carrying NDM-1 in Enterobacteriaceae isolates that epidemiologically links to multiple geographical areas in China [Citation11]. The blaNDM-1 gene was found in a transposon-like structure flanked by ISAba125 and IS26 which are identical to plasmids from Acinetobacter isolates carrying blaNDM-1–positive Tn125 [Citation12,Citation13]. Intriguingly, a recent report from United Arab Emirates also described three isolates of different species carrying identical, or very similar, IncX3 plasmids with an IS5 element upstream of blaNDM-1 gene and co-transferring with blaSHV-12 gene [Citation40]. These results indicated that IncX3 plasmids play a unique role in the inter-generic spread of the NDM-1 gene. In this work, we investigated the influence of IncX3 plasmid pNDM-HN380 to bacteria E. coliand found that this plasmid was able to reduce the motility of the host via attenuating the expression of flagella and chemotaxis-related genes at log phase, which was similar to other ESBL plasmids described previously [Citation41,Citation42]. Bacterial motility requires intensive energy and the synthesis of bacterial flagella system costs a lot of resources. Indeed, as shown in Figure S2b, flagella and chemotaxis-related genes were downregulated in both J53 and J53/pNDM-HN380 transconjugant in the stationary phase compared to the log phase, indicating bacteria reduced energy consumption of bacterial motility when nutrition was limited. However, the expressions of flagella and chemotaxis-related genes in J53/pNDM-HN380 at stationary phase were relatively higher compared to J53. The observed result suggested that the MDR plasmid may improve the ability of bacteria to avoid harmful materials by less reduced flagella and chemotaxis-related genes in stationary phase. These manipulations of genes expression could be the fitness cost of carrying ESBL plasmids. Indeed, Diel B et al. identified a novel plasmid-transcribed regulatory sRNA, QfsR, involved in bacterial quorum sensing and flagella synthesis [Citation43]. Besides bacterial motility, recent studies have also found plasmid-encoded genes that can regulate the expression of chromosomal virulence genes [Citation44] and bacterial secretion system [Citation45] suggesting the important roles of gene regulation of host bacteria are mediated by MDR plasmids.
In addition to profiling the transcriptomes of E. colicarrying pNDM-HN380, we also sequenced the small RNA repertiores that are influenced by the plasmid and found that IGR plas2, a novel sRNA can modulate fucose metabolism of host bacteria. It is well known that fucose is a major component of mucin glycoproteins and it is highly abundant in the intestine where bacteria colonize during infection [Citation46,Citation47]. The control of fucose consumption by IGR plas2 may facilitate the pathogens to modulate their virulence and metabolism in gut where fucose is mostly secreted. Over 80% of sRNAs in bacteria play role in suppressing their corresponding target genes by activating RNA degradation [Citation47]. However, IGR plas2 sRNA acts as an enhancer or modulator to control the expression of fucR genes. One of the possible explanations to this phenomenon is that IGR plas2 is able to protect and stabilize thefucR mRNA from degradation by either hybridization or inducing conformation change of fucR transcript. Indeed, the RNA-RNA gel shift experiment confirmed the direct binding of IGR plas2 tofucR mRNA (Supplementary Figure S7). Moreover, a higher level of IGR plas2 was observed at the stationary phase compared to the log phase, suggesting the sRNA was upregulated in a stressed conditon with limited nutritions.
Biofilm formation is important for bacterial survival and pathogenesis, for the reason that biofilm can also act as a barrier to protect the bacteria from antibiotic attack [Citation48]. Here, we managed to inhibit the formation of bacterial biofilm by the knockdown of IGR plas2 using an antisense decoy. Admittedly, we cannot exclude the possibility that the effect of IGR plas2 in biofilm formation is indirect: that is the deficiency in bacterial growth might be the main cause of the reduction of biofilm. However, it is known that nutrient in media could influence bacterial biofilm formation [Citation34]. Interestingly, the impair of biofilm formation was also observed in different sugar supplements including glucose and fucose medium (Supplementary Figure S6a). Fucose, as a major component of glycan, is closely related to biofilm formation and fucose sensing has been known to regulate bacterial intestinal colonization via alternating biofilm formation capability [Citation34]. Elimination of biofilm to remove pathogens and recolonization of probiotic are crucial for not only acute infection disease but also many chronic diseases. Several methods including ozonated water treatment [Citation49], phage therapy [Citation50], matrix targeting enzymes [Citation51], bacteriocins [Citation52], synthetic chemicals [Citation53] and natural origin compounds [Citation54] are currently applied for biofilm matrix disruption. Introduction of the dplas2 could be potentially used as a therapeutic approach to inhibit fucose metabolism and biofilm formation. To further confirm the function of IGR plas2, an IGR plas2 knock-out (KO) clone was constructed by replacing the IGR plas2 sequence on pNDM-HN380 with a chloramphenicol gene as a selection marker. However, the biofilm experiment using IGR plas2 KO showed no difference with the control (Supplementary Figure S8). We noticed that the knock-out of IGR plas2 also destroyed the promoter region and the 5ʹ of another plasmid-transcribed sRNA IGR plas1. This was inevitable due to the genic locations of the two sRNAs on pNDM-HN380 (Supplementary Figure S9). Notably, the predicted binding site of IGR plas1 shared the similar location (almost the same) of the binding site of IGR plas2 on fucRgene (Supplementary Figure S10), suggesting the possible interaction between the two sRNAs and fucR mRNA. In our biofilm experiment, therefore, it is possible that the plas2 KO strain affected the unknown sRNA regulatory pathways that involved IGR plas1 sRNA, and influenced the result. Indeed, further experiment is required to further identify the detailed mechanism. Moreover, we also explored the gene expression of three flagellar genes flhC, flhD and flgNwith IGR plas2 overexpression in J53 using qPCR (Supplementary Figure S11). We found that flhC and flhD, which are activators of the class 2 flagellar genes, were significantly upreguated (**, P < 0.01), suggesting IGR plas2 sRNA also plays a functional role in regulating flagella synthesis in Bacteria.
In conclusion, we profiled and compared the RNA repertoires of transconjugants E. coli J53/pNDM-HN380 with E. coliJ53 and showed that pNDM-HN380 affected the motility of the host in addition to conferring multiple antibiotic resistance. Moreover, we identified and characterized a novel sRNA IGR plas2 that regulates the fucR gene which modulates fucose metabolism and biofilm formation. Our finding provides new insights into plasmid fitness in bacterial host that facilitates bacterial survival in competitive environment, and the potential of utilizing plasmid-transcribed sRNA against pathogens.
Supplemental Material
Download MS Word (11.7 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054..
- Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602..
- Falagas ME, Karageorgopoulos DE. Extended-spectrum beta-lactamase-producing organisms. J Hosp Infect. 2009;73(4):345–354.
- Kong HK, Liu X, Lo WU, et al. Identification of plasmid-encoded sRNAs in a bla(NDM-1)-harboring multidrug-resistance plasmid pNDM-HK in enterobacteriaceae. Front Microbiol. 2018;9. DOI:10.3389/fmicb.2018.00532.
- Villa L, Poirel L, Nordmann P, et al. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J Antimicrob Chemother. 2012;67(7):1645–1650..
- Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011;55(9):4224–4229.
- Nordmann P, Poirel L, Walsh TR, et al. The emerging NDM carbapenemases. Trends Microbiol. 2011;19(12):588–595..
- Kim MN, Yong D, An D, et al. Nosocomial clustering of NDM-1-producing Klebsiella pneumoniae sequence type 340 strains in four patients at a South Korean tertiary care hospital. J Clin Microbiol. 2012;50(4):1433–1436..
- Ho PL, Li Z, Lai EL, et al. Emergence of NDM-1-producing Enterobacteriaceae in China. J Antimicrob Chemother. 2012;67(6):1553–1555..
- Ho PL, Cheung -Y-Y, Lo W-U, et al. Molecular characterization of an atypical incX3 plasmid pKPC-NY79 carrying bla KPC-2 in a klebsiella pneumoniae. Curr Microbiol. 2013;67(4):493–498..
- Ho PL, Li Z, Lo WU, et al. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emer Mic Infect. 2012;1:e39.
- Fu Y, Du X, Ji J, et al. Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J Antimicrob Chemother. 2012;67(9):2114–2122..
- Poirel L, Bonnin RA, Boulanger A, et al. Tn 125-related acquisition of bla NDM-like genes in acinetobacter baumannii. Antimicrob Agents Chemother. 2012;56(2):1087–1089..
- Abe T, Shimoyama T, Fukuda S, et al. Effects of Helicobacter pylori in the stomach on neutrophil chemiluminescence in patients with gastric cancer. Luminescence. 2000;15(5):267–271..
- Nikaido H, Pages J-M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. 2012;36(2):340–363.
- Miyakoshi M, Nishida H, Shintani M, et al. High-resolution mapping of plasmid transcriptomes in different host bacteria. BMC Genomics. 2009;10(1):1–15..
- Frost LS, Leplae R, Summers AO, et al. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–732..
- Miyakoshi M, Shintani M, Terabayashi T, et al. Transcriptome analysis of Pseudomonas putida KT2440 harboring the completely sequenced IncP-7 plasmid pCAR1. J Bacteriol. 2007;189(19):6849–6860..
- Cook LCC, Dunny GM. The influence of biofilms in the biology of plasmids. Microbiol Spectr. 2014;2(5):0012.
- Nakao R, Myint SL, Wai SN, et al. Enhanced biofilm formation and membrane vesicle release by Escherichia coli expressing a commonly occurring plasmid gene, kil. Front Microbiol. 2018;9:2605.
- Narenkumar J, Madhavan J, Nicoletti M, et al. Role of bacterial plasmid on biofilm formation and its influence on corrosion of engineering materials. J Bio Tribo Corro. 2016;2(4):24..
- Masse E, Majdalani N, Gottesman S. Regulatory roles for small RNAs in bacteria. Curr Opin Microbiol. 2003;6(2):120–124.
- Kim T, Bak G, Lee J, et al. Systematic analysis of the role of bacterial Hfq-interacting sRNAs in the response to antibiotics. J Antimicrob Chemother. 2015;70(6):1659–1668..
- Ramos CG, Grilo AM, Sousa SA, et al. Regulation of Hfq mRNA and protein levels in Escherichia coli and Pseudomonas aeruginosa by the Burkholderia cenocepacia MtvR sRNA. PLoS One. 2014;9(6):e98813..
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99(7):4620–4625.
- Thomas VC, Kinkead LC, Janssen A, et al. A dysfunctional tricarboxylic acid cycle enhances fitness of Staphylococcus epidermidis during β-lactam stress [published correction appears in MBio. 2014;5(3):e01307-14. Chittezham Thomas, Vinai [corrected to Thomas, Vinai C]]. mBio. 2013;4(4):e00437-13. Published 2013 Aug 20. DOI:10.1128/mBio.00437-13.
- Yi H, Cho Y-J, Yong D, et al. Genome sequence of Escherichia coli J53, a reference strain for genetic studies. J Bacteriol. 2012;194(14):3742–3743..
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–580.
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26..
- Krzywinski MI, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Research; 2009;19(9):1639–1645. DOI:10.1101/gr.092759.109.
- Bak G, Han K, Kim KS, Lee Y, et al. Electrophoretic mobility shift assay of RNA-RNA complexes. Methods Mol Biol. 2015;1240:153–163. DOI:10.1007/978-1-4939-1896-6_12.
- Li J, Attila C, Wang L, et al. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J Bacteriol. 2007;189(16):6011–6020..
- Chen YM, Lu Z, Lin EC. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on L-1,2-propanediol. J Bacteriol. 1989;171(11):6097–6105.
- Dwivedi R, Nothaft H, Garber J, et al. L-fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol Microbiol. 2016;101(4):575–589..
- Baquero F, Alvarez-Ortega C, Martinez JL. Ecology and evolution of antibiotic resistance. Environ Microbiol Rep. 2009;1(6):469–476.
- Falkow S, Citarella RV, Wohlhieter JA, et al. The molecular nature of R-factors. J Mol Biol. 1966;17(1):102–116..
- Sugino Y, Hirota Y. Conjugal fertility associated with resistance factor R in Escherichia coli. J Bacteriol. 1962;84(5):902–910.
- Livermore DM. Beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8(4):557–584.
- Top EM, Springael D. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol. 2003;14(3):262–269.
- Sonnevend A, Al Baloushi A, Ghazawi A, et al. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J Med Microbiol. 2013;62(Pt 7):1044–1050..
- Jiang X, Liu X, Law COK, et al. The CTX-M-14 plasmid pHK01 encodes novel small RNAs and influences host growth and motility. FEMS Microbiol Ecol. 2017;93(7). DOI:10.1093/femsec/fix090.
- Zhao K, Liu M, Burgess RR. Adaptation in bacterial flagellar and motility systems: from regulon members to ‘foraging’-like behavior in E. coli. Nucleic Acids Res. 2007;35(13):4441–4452.
- Diel B, Dequivre M, Wisniewski‐Dyé F, et al. A novel plasmid-transcribed regulatory sRNA, QfsR, controls chromosomal polycistronic gene expression in Agrobacterium fabrum. Environ Microbiol. 2019;21(8):3063–3075..
- Patton MJ, Chen C-Y, Yang C, et al. Plasmid negative regulation of CPAF expression is pgp4 independent and restricted to invasive chlamydia trachomatis biovars. mBio. 2018;9(1):e02164–17..
- Di Venanzio G, Moon KH, Weber BS, et al. Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc Nat Aca Sci USA. 2019;116(4):1378–1383.
- Pacheco AR, Curtis MM, Ritchie JM, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492(7427):113–117.
- Robbe C, CAPON C, CODDEVILLE B, et al. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384(Pt 2):307–316.
- Balcazar JL, Subirats J, Borrego CM. The role of biofilms as environmental reservoirs of antibiotic resistance. Front Microbiol. 2015;6:1216.
- Bialoszewski D, Pietruczuk-Padzik A, Kalicinska A, et al. Activity of ozonated water and ozone against Staphylococcus aureus and Pseudomonas aeruginosa biofilms. Med Sci Monit. 2011;17(11):Br339–344.
- Azeredo J, Sutherland IW. The use of phages for the removal of infectious biofilms. Curr Pharm Biotechnol. 2008;9(4):261–266.
- Torelli R, Cacaci M, Papi M, et al. Different effects of matrix degrading enzymes towards biofilms formed by E. faecalis and E. faecium clinical isolates. Colloids Surf B Biointerfaces. 2017;158:349–355.
- Chopra L, Singh G, Kumar Jena K, et al. Sonorensin: A new bacteriocin with potential of an anti-biofilm agent and a food biopreservative. Sci Rep. 2015;5:13412.
- de Lima Pimenta A, Chiaradia-Delatorre LD, Mascarello A, et al. Synthetic organic compounds with potential for bacterial biofilm inhibition, a path for the identification of compounds interfering with quorum sensing. Int J Antimicrob Agents. 2013;42(6):519–523.
- Magesh H, Kumar A, Alam A, et al. Identification of natural compounds which inhibit biofilm formation in clinical isolates of Klebsiella pneumoniae. Indian J Exp Biol. 2013;51(9):764–772.