ABSTRACT
In all eukaryotic cells, the most abundant modification of ribosomal RNA (rRNA) is methylation at the ribose moiety (2ʹ-O-methylation). Ribose methylation at specific rRNA sites is guided by small nucleolar RNAs (snoRNAs) of C/D-box type (C/D snoRNA) and achieved by the methyltransferase Fibrillarin (FIB). Here we used the Illumina-based RiboMethSeq approach for mapping rRNA 2ʹ-O-methylation sites in A. thaliana Col-0 (WT) plants. This analysis detected novel C/D snoRNA-guided rRNA 2ʹ-O-methylation positions and also some orphan sites without a matching C/D snoRNA. Furthermore, immunoprecipitation of Arabidopsis FIB2 identified and demonstrated expression of C/D snoRNAs corresponding to majority of mapped rRNA sites. On the other hand, we show that disruption of Arabidopsis Nucleolin 1 gene (NUC1), encoding a major nucleolar protein, decreases 2ʹ-O-methylation at specific rRNA sites suggesting functional/structural interconnections of 2ʹ-O-methylation with nucleolus organization and plant development. Finally, based on our findings and existent database sets, we introduce a new nomenclature system for C/D snoRNA in Arabidopsis plants.
Introduction
In all eukaryotic cells, ribosomal RNAs (rRNAs) precursors are substrate of two major types of nucleotide modifications: methylation of sugars (2ʹ-O-ribose methylation) and isomerization of uridine to pseudouridine. Purine and pyrimidine rings in rRNAs can be also be methylated (m1N, m6N and m7N), aminocarboxypropilated (acp3N) and/or acetylated (ac4N) [Citation1–4].
2ʹ-O-methylation of rRNA might stabilize rRNA-mRNA, rRNA-tRNA or rRNA-protein interactions [Citation5,Citation6]. Furthermore, the significance of 2ʹ-O-methylation of rRNAs is highlighted through studies in animal cells showing that alterations of rRNA 2ʹ-O-methylation can be associated to diseases, mainly cancer and autoimmune syndromes. Indeed, ribosomes with altered rRNA 2ʹ-O-methylation profile translate mRNA with lower fidelity and increase internal ribosome entry site (IRES)-dependent translation initiation of key cancer genes [Citation4–9].
2ʹ-O-ribose methylations are guided by small nucleolar RNAs (snoRNAs), referred as C/D-box snoRNA (C/D snoRNA). The box C (5ʹPuUGAUGA3ʹ) and D (5ʹCUGA3ʹ) of C/D snoRNAs are short consensus sequences that localize a few nucleotides away from the 5ʹ- and 3ʹ-ends, respectively. In the central part, the C/D snoRNA might contain also less conserved C’ and D’ motifs. One or two of so-called antisense elements are located upstream of the D and/or D’ box. These antisense sequences are about 10–21 nucleotides long and are complementary to the region surrounding the site of 2ʹ-O-ribose methylation. The rRNA nucleotide to be methylated is located precisely at the fifth position upstream from the D or D’ box. The C/D snoRNA associates to four nucleolar proteins called the C/D-box core proteins: Fibrillarin/Nop1p, Nop56p/NOP56, Nop58p/NOP58 and snu13/L7Ae. Fibrillarin contains a characteristic SAM-binding methyltransferase motif required for the 2ʹ-O-ribose methylation activity Reviewed in [Citation10–13].
In plants, 2ʹ-O-methylation of rRNAs has been demonstrated at specific rRNA sites and/or predicted throughout in silico and functional characterization of C/D snoRNAs. On one hand, in Arabidopsis thaliana plants, over one hundred of C/D snoRNAs were first identified by computational screening of genomic sequences [Citation14,Citation15] and a plant snoRNA database was created [Citation16]. Later, sequencing of plant small RNA reported the expression of 151 C/D snoRNAs [Citation17] and sequencing of nucleolar RNA fraction identified 9 additional C/D snoRNA candidates [Citation18]. These results expanded the number of known C/D snoRNAs in Arabidopsis plants to over 200 (including variants). Similarly, hundreds of C/D snoRNAs predicted to guide 2ʹ-O-methylation of rRNA, were reported in O. sativa [Citation19,Citation20] and also other plant species [Citation21]. Notably, studies in Arabidopsis demonstrate that knockout of a single snoRNA (HID2) triggers strong developmental and growing defects [Citation22], while gene disruption of the C/D-box snoRNP assembly factor NUFIP inhibits 2ʹ-O-methylation at specific rRNA sites and leads to severe developmental phenotypes [Citation23]. On the other hand, over 125 2ʹ-O-methylated rRNA sites have been predicted in Arabidopsis [Citation24]. However, only about half of these potential rRNA modification sites has been mapped and/or validated [Citation14,Citation15,Citation22,Citation23,Citation25]. This is essentially due to the limits of current experimental approaches, like RT primer extension at low [dNTP] used to determine the methylation state of a single target site at a time.
Here, we used RiboMethSeq approach for global mapping all rRNA 2ʹ-O-methylation sites in leaves from Arabidopsis thaliana Col-0 (WT) and nucleolin 1 (nuc1) mutant plants. RiboMethSeq is based on the resistance of RNA at 2ʹ-O-methylated sites to alkaline fragmentation and employs Illumina sequencing of cloned RNA fragments for simultaneous mapping and quantification of hundreds ribose methylated sites in RNA [Citation26]. Furthermore, to identify C/D snoRNAs linked to the mapped 2ʹ-O-methylated rRNA candidate sites, we performed bioinformatics screening of the Arabidopis genome as well as experimental identification of expressed snoRNAs in a Fibrillarin immunoprecipitated fraction by small RNA-seq.
Results
rRNA 2ʹ-O-methylation in A. thaliana leaves
To map rRNA 2ʹ-O-methylated sites in A. thaliana, we extracted total RNA from Arabidopsis leaves (3 biological replicates, Figure S1) and performed RiboMethSeq analysis as previously reported [Citation10,Citation26].
After trimming and alignment of reads to the reference A. thaliana rRNA sequence [Citation25–29], we found substantial differences between the reference sequences for 18S (GenBank X16077.1) and 25S rRNA (GenBank X52320.1) and the observed rRNA reads. These rRNA sequences were thus corrected to fit to the observed sequencing data (Supplementary Information and Methods). Then, in order to map all possible candidate sites, we performed the same approach as reported in [Citation30]. We used threshold values for ScoreMEAN and Score A2, respectively 0.93 and 0.5; although in some cases, less strict ScoreMEAN limit (0.92 or lower) was applied. Combination of these parameters was found to give the best results for human rRNA having now well-established 2ʹ-O-methylation profile and thus to limit the number of false-positive/false-negative hits. We also compared the obtained RiboMethSeq hits with previously known [Citation14,Citation25] or tentatively assigned locations and rRNA 2ʹ-O-methylation list [Citation24]. See Supplemental Information and Methods and Table S1 for details.
RiboMethSeq mapped 117 potential ribose methylated sites: 38 in the 18S rRNA, 2 in the 5.8S rRNA and 77 in the 25S rRNA sequences ( and S1 for details): Among the 117 RiboMethSeq mapped sites, 52 were previously mapped [Citation14,Citation15,Citation22,Citation23,Citation25] while 25 others were only predicted from the sequences of C/D snoRNA guides [Citation24] but not experimentally validated in the previous studies. Notably, RiboMethSeq also mapped 40 potential rRNA methylated sites not reported in [Citation14,Citation13,Citation25] or predicted previously (Table S2). Nine (18S Um123, Cm1219 and 25S Cm2198, Am2257, Am2362, Gm2396, Um2494, Um2922 and Gm2923) of these 40 mapped candidates have an equivalent position in human, and for 31 of newly mapped positions we assigned the corresponding snoRNA. However, 7 candidate sites (18S Am812, Am1188 and Um1554 and 25S Um378, Cm2294, Gm2410 and Am2561) showing high RiboMethSeq signal still have no assigned snoRNA guide ().
Table 1. List of RiboMethSeq detected 2ʹ-O-methylation sites in A. thaliana 5.8S, 18S and 25S rRNAs. Mapped rRNA Nm sites (‘Mapped site’) are compared with previously known locations (‘Position 3D rRNAdb’ and associated C/D snoRNA [Citation22]). Conservation of rRNA modification sites with human (‘Position in human’) and yeast (‘Position in yeast’) profile was determined based on 2D rRNA structure and sequence [Citation8]. Human rRNA sites located in the same structural context, but not strictly conserved, are shown in grey. Newly detected snoRNA guides corresponding to the modified positions are shown (‘Assigned snoRNA’). The absence of identified snoRNA guide is shown in grey. Reduction of 2ʹ-O-methylation in nuc1-2 plants is shown in columns ‘ScoreC nuc1’ with corresponding p-value. Asterisks ****(≤0.0001), ***(≤0.001), **(≤ 0.01), *(≤0.05) represent significance level and ns, non-significant value ≥ 0.05
To validate 2ʹ-O-methylated candidates mapped by RiboMethSeq we performed additional orthogonal mapping of A. thaliana rRNA residues by high-throughput version of primer extension at low [dNTP]/low [Mg2+] conditions. The protocol was derived from published 2OMe-Seq [Citation31], with minor modifications at the adapter ligation step and uses RT enzymes AMV and MMLV (Supplemental Information and methods). Using this technique we validated 97 sites over the 117 mapped by RiboMethSeq, including 5 rRNA sites without a corresponding C/D snoRNA guide (Figure S2 and Table S2).
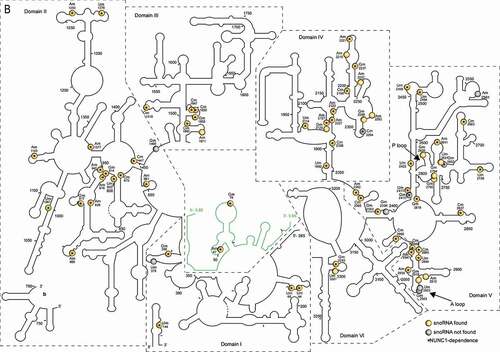
All sites mapped by RiboMethSeq are shown in the predicted 18S and 25S/5.8S RNA secondary structures (). All domains in the 18S, 25S and 5.8 rRNA sequences are 2ʹ-O-methylated at different extents. Noteworthy, the 5ʹ domain in the 18S rRNA and the domain V in the 25S rRNA contain the highest number of 2ʹ-O-methylation sites under our plant growth conditions.
Figure 1. Predicted 18S, 5.8S and 25S rRNA structures with mapped 2ʹ-O-methylations sites (A) 18S and (B) 5.8S/25S rRNA structures were generated on the basis of 2D structural predictions [Citation24]. 2ʹ-O-methylated sites in yellow circles or grey circles show rRNA sites with or without matching C/D snoRNAs (respectively). Star inside the circles indicates observed dependency of rRNA 2ʹ-O-methylation on NUC1 gene expression. In B, the 5.8S rRNA structure is coloured in green and two 25S fragments (5ʹ-fragment and 3ʹ-fragment, a and b, respectively) are split in the main structure for clarity. The 18S (5ʹ, central, 3ʹMajor and 3ʹMinor) and 25S (I–VI) rRNA domains are designated according to [Citation1]. P and A loop are indicated by arrows
![Figure 1. Predicted 18S, 5.8S and 25S rRNA structures with mapped 2ʹ-O-methylations sites (A) 18S and (B) 5.8S/25S rRNA structures were generated on the basis of 2D structural predictions [Citation24]. 2ʹ-O-methylated sites in yellow circles or grey circles show rRNA sites with or without matching C/D snoRNAs (respectively). Star inside the circles indicates observed dependency of rRNA 2ʹ-O-methylation on NUC1 gene expression. In B, the 5.8S rRNA structure is coloured in green and two 25S fragments (5ʹ-fragment and 3ʹ-fragment, a and b, respectively) are split in the main structure for clarity. The 18S (5ʹ, central, 3ʹMajor and 3ʹMinor) and 25S (I–VI) rRNA domains are designated according to [Citation1]. P and A loop are indicated by arrows](/cms/asset/adb173af-9895-4f41-8746-56646fef13d5/krnb_a_1869892_f0001a_oc.jpg)
Identification and characterization of C/D snoRNAs associated to Arabidopsis FIB2
To identify C/D snoRNAs that might guide 2ʹ-O-methylation of mapped rRNA sites, we characterized C/D snoRNAs that co-purify with Arabidopsis Fibrillarin 2 (FIB2). We performed immunoprecipitation (IP) experiments in triplicate using Arabidopsis plants expressing the 35S:FIB2-YFP (Yellow Fluorescent Protein) construct. The FIB2-YFP is a 61kDa protein and localizes in the nucleolus (Figure S3(A-B) and [Citation32,Citation33]).
Following IP with antibodies against GFP, we analysed by Western blot whole cell extract (WCE-input, lanes 1 and 4), unbound (lanes 2, 5, 7 and 9) and co-immunoprecipitated (CoIP, lanes 3, 6, 8 and 10) protein fractions. We observed that anti-GFP antibodies immunoprecipitate FIB2-YFP protein in all three FIB2-YFP samples (CoIP_1 to 3), but not from Col-0 (CoIP_1 mock) protein extract (Figure S3C).
Firstly, we performed LC-MS/MS and differential analysis on FIB2-YFP and Col-0 IP fractions to verify that C/D snoRNP proteins co-immunoprecipitate with FIB2-YFP. A total of 197 proteins were specifically identified in fractions FIB2-YFP compared to the Col-0 CoIP fractions (Table S3). Arabidopsis orthologues of Fibrillarin, NOP56, NOP58 and L7Ae proteins were identified and the first three were among the most enriched (top 10) proteins. The genome of Arabidopsis contains two Fibrillarin (FIB1 and FIB2), NOP56 and NOP58 and at least five L7Ae protein genes (). LC-MS/MS data did not allow precise discrimination of orthologue(s) co-immuno-precipitating with FIB2-YFP, but clearly detected at least one orthologue of each protein factor. Altogether, these data indicate that FIB2-YFP interacts with C/D-box snoRNP core proteins and likely forms a functional C/D-box snoRNP complex.
Figure 2. Characterization of FIB2-YFP snoRNP (A) Arabidopsis C/D-box protein orthologues (left) and C/D snoRNA (right) detected in the FIB2-YFP IP fractions using LC-MS/MS and RNA-seq approaches. The number of peptides and % of protein coverage for each protein is shown and the number of predicted 25S, 18S and 5.8S rRNA targets identified in the 5ʹ and 3ʹ antisense sequences of 141 C/D snoRNA. (B) Representation of 6 novel C/D snoRNA detected in the FIB2-IP fraction. C and D boxes are boxed. These C/D snoRNA do not contain a matching rRNA target sequence
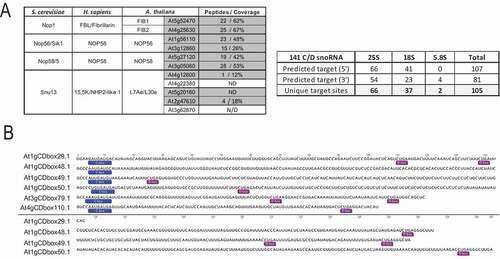
Next, to identify C/D snoRNAs in the FIB2-YFP fractions (CoIP_1 to 3), we performed RNA-seq analysis. Total RNAs were extracted from FIB2-YFP CoIP fractions, converted to library and sequenced. Between 7–8.7 × 106 C/D snoRNA reads are detected in each RNA samples (Figure S3D) and count for a total of 193 C/D snoRNAs (with at least 10 reads in one of the CoIP fractions; ). Among these, 187 were known C/D snoRNAs while the other 6 were novel species. Noticeable, 141 C/D snoRNAs target respectively 66, 37 and 2 sites in the 25S, 18S and 5.8S rRNA sequences while the other 52 C/D snoRNAs do not target rRNAs sites identified by RiboMethSeq ( and and S4).
Table 2. List of C/D box snoRNAs identified from RNA-seq data. The number of C/D snoRNA reads are provided for each Co_IP fraction. Are indicated, rRNA positions mapped as 2ʹ-O-methylated in this study, when predicted to be guided by a snoRNA and the names of known C/D snoRNA in databases Are highlighted in red snoRNAs with predicted rRNA methylation sites but not detected by RiboMethSeq. ID with an asterisk identifies a snoRNA which is dicistronic with a tRNA. ID with two asterisks represents a snoRNA which is polycistronic with a miRNA
Up to four C/D snoRNA might target each of the 105 ribose methylated sites detected by RiboMetSeq ( and ). However, based on the number of mapped reads in the same replicate, some C/D snoRNAs targeting the same rRNA methylation site seem to be differentially expressed. For instance ~80 thousands and ~3 thousands reads are respectively counted for At3gCDbox92.2 and At4gCDbox92.3 targeting 18S-Am28 while ~14 thousands reads are counted for At4gCDbox113.1 and none for At5gCDbox113.2 (not detected), both targeting 25S-Am1459. In contrast, for 14 rRNA 2ʹ-O-methylated sites (all with at least one annotated C/D snoRNA) we have not detected a corresponding C/D snoRNA in the FIB2-IP fraction, including 25S-Gm2794 and 25S-Cm1847 sites for which C/D snoRNAs were identified in silico ( and S4).
In conclusion, this analysis demonstrated expression for most of C/D snoRNA driving 2ʹ-O-methylation of rRNA sites detected by RiboMethSeq and also identified novel FIB2 interacting C/D snoRNAs without rRNA target site(s).
C/D snoRNA bioinformatics search
To identify RNA guides of ribose methylated rRNA sites () without known and/or immunoprecipitated C/D box snoRNAs, we performed a bioinformatics analysis of the Arabidopsis genome ().
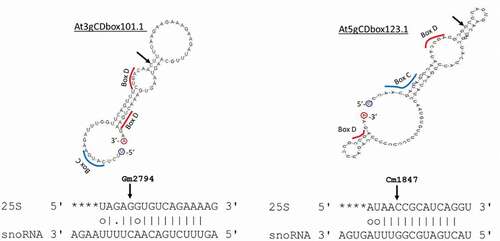
A first catalogue of 217 C/D box snoRNA sequences was established by collecting data from existing resources including SNOOPY [Citation34], ARAPORT [Citation35] and TAIR [Citation36]. This first catalogue was enriched by 3 novel C/D snoRNAs (At3gCDbox101.1, At5gCDbox123.1, At5gCDbox129.1) identified by searching the genome sequence for C/D snoRNA targeting orphan rRNA 2ʹ-O-methylated sites ( and and S4) . FIB2-YFP CoIP sequencing gave experimental evidence for 6 novel species mentioned before () resulting in an extended catalogue of 226 C/D snoRNAs. Altogether, 193 of the 226 C/D snoRNAs show expression evidence and represent 144 families. Most of them are organized into clusters of C/D snoRNAs. Notably, as previously described, 12 C/D box snoRNAs share a dicistronic organization with tRNAs [Citation14] and 2 snoRNA clusters share a polycistronic organization with miRNAs [Citation37].
Figure 3. Mfold structure of At3gCDbox1011 and At5gDbox123.1 identified in silico and alignment with 25S rRNA sequences. Arrows show mapped rRNA sites Gm2794 and Cm1847 in the 25S and their counterpart in the snoRNA. Blue and red bars underline potential C and D boxes sequences
All C/D box snoRNA genes were (re)named as AtchrgCDboxnumber.isoform and consecutively numbered. For example, At4gCDbox94.3 is the name of the C/D box (CDbox) snoRNA which is located on chromosome 4 of the genome (At4g) and is the third isoform of a previously annotated C/D box snoRNA which is numbered 94 (94.3). This nomenclature simplifies current annotation and provide useful information concerning the genomic organization of C/D snoRNA variants.
All identified C/D box snoRNAs were searched for their ability to guide known predicted and/or mapped rRNA 2ʹ-O-methylations (Table S4). We found 181 C/D box snoRNAs that contain predicted guide sequences upstream of their D’ or D box. Among them, 111 have predicted guide sequences upstream of each box, 155 have a predicted guide sequence upstream the D’ box and 136 have a predicted guide sequence upstream the D box (predicted target 5ʹ and 3ʹ respectively in Table S4). Several box C/D snoRNAs may have close homologs that are able to guide methylation at similar rRNA sites. Members of different families may also guide a modification at the same rRNA site. For instance, 3 different C/D snoRNAs potentially guide methylation of 18S-Am545 and 4 others guide methylation of 25S-Cm2338 (, and S4). Altogether, 4 snoRNAs are able to guide 2ʹ-O-methylation at the 2 positions of 5.8S rRNA, 101 at the 65 mapped/predicted positions of 18S rRNA and 183 at the 100 mapped/predicted positions of 25S rRNA. Exploring the updated catalogue of Arabidopsis thaliana C/D box snoRNAs with pairing constraints as defined in Materials and Methods, we could not find C/D box snoRNA guides for 12 of the mapped rRNA modifications ().
Differential 2ʹ-O-Methylation in nuc1-2 mutant plants
To determine if variations of rRNA 2ʹ-O-methylation might occur in Arabidopsis plants, we performed RiboMethSeq analysis of Nucleolin 1 (nuc1-2) mutants. Nucleolin is a phylogenetically conserved multifunctional protein required for transcription and processing of 45S pre-rRNA and assembly of ribosomes [Citation38,Citation39]. In Arabidopsis, NUC1 gene disruption affects rRNA expression and functional nucleolar structure and provokes major growth and development defects [Citation40–42]. RiboMethSeq analysis was performed in 3 biological replicates from 21-days-old Arabidopsis nuc1-2 mutant plants and compared with results from Col-0 (WT) control plants.
Quantitative 2ʹ-O-methylation score was calculated for each mapped position in 18S, 5.8S and 25S rRNAs both in Col-0 and nuc1-2 plants (Table S1) and compiled in a heat map (). All methylation scores were clustered with the software ‘R’ (hclust). This analysis revealed 65 sites that were significantly (p-value <0.01) hypo-methylated in the nuc1-2 compared to WT plants (). Remarkably, the RiboMethSeq analysis has not detected any hypermethylation of mapped sites in nuc1-2 mutant compared to Col-0 plants.
Figure 4. Heat map representation of rRNA 2ʹ-O-methylation in Arabidopsis. Differential methscore levels for 115 rRNA 2ʹ-O-methylated sites observed in three Col-0 and three nuc1-2 biological replicates (R1-R3) are shown. The rRNA sites are clustered according to hclust/ward.D2 method. Arrow heads show rRNA sites without associated guiding snoRNA. The colour key, histogram and values are shown
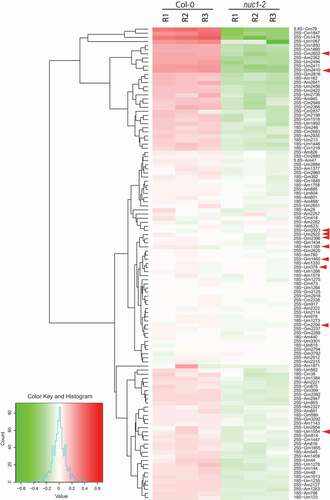
65 rRNA 2ʹ-O-methylation sites were differentially down regulated in the nuc1-2 plants. 61 are guided by C/D snoRNA, while the other 4 (18S Um1554 and 25S Cm2294, Gm2410, Gm2652) have no matching C/D snoRNA ( and yellow circles and grey circles with star in ). Noteworthy, the rRNA site 25S-Gm2652, is located in the domain V and close to the P loop (). We also noticed that out of 65 sites undermethylated in nuc1-2 plants, 30 and 16 are conserved in human and yeast rRNA respectively, including the 25S-Gm2652, equivalent to the 28S-Gm4228 in human ().
Altogether, this analysis supports the idea that 2ʹ-O-methylation of conserved and non-conserved rRNA sites could be modulated in Arabidopsis plants.
Discussion
In this work we used RiboMethSeq and mapped 117 rRNA 2ʹ-O-methylation sites in leaves from 21-days-old Arabidopsis thaliana plants; among these sites, 65 are mapped for the first time. Notably, 38 of the 65 sites mapped in leaves were recently reported as well in plant stems using a nuclease-base detection protocol [Citation43], ratifying that they are truthfully rRNA methylation targets in Arabidopsis thaliana rRNA (Table S2). Only 6 rRNA sites mapped by RiboMethSeq were not supported by at least two independent protocols or reported elsewhere (Table S2). Sites without such orthogonal validation were thus considered as potential false-positive hits. Thus, together with 66 previously [Citation14,Citation15,Citation22,Citation23,Citation25] and the 20 newly mapped sites in stems reported in [Citation41] a total of 151 ribose methylated sites have been now mapped in Arabidopsis ().
Figure 5. Illustration shows the number of 2ʹ-O-methylated sites mapped by RiboMethSeq (117) and C/D snoRNA detected in the FIB2 immunoprecipitated fraction (193) and/or identified by bioinformatics search (9). From these analyses we account 65 first time mapped 2ʹ-O-methylated sites and 9 novel C/D snoRNAs, increasing the number of 2ʹ-O-methylated mapped sites and C/D snoRNA identified and listed in Arabidopsis databases to 151 and 226 respectively
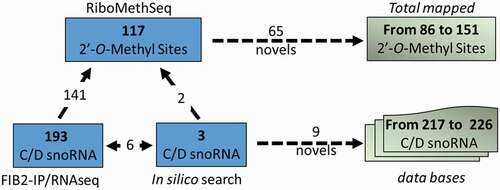
We noted that the majority of the mapped rRNA sites are guided by ~150 distinct C/D snoRNAs (). As expected, a number of these rRNA sites is targeted by different C/D snoRNA isoforms resulting from gene duplications of the Arabidopsis genome [Citation14,Citation15,Citation25].
Interestingly, 49 rRNA 2ʹ-O-methylation sites previously mapped or predicted were not detected by RiboMethSeq (Table S2). Among these, 4 sites were mapped in young seedlings [Citation15,Citation25], indicating that 2ʹ-O-methylation of these sites might occur specifically only at early plant growth and/or developmental stages. Then, it can be expected, that a fraction of rRNA methylated sites detected by RiboMetSeq could be specific to later stage of plant organs, including leaves.
The total number of rRNA mapped sites (117, ) in our plant growth conditions is relatively similar to human with 110 positions guided by 118 C/D snoRNA [Citation10,Citation44] but much higher compared to yeast with 55 positions guided by 43 C/D snoRNA [Citation45]. A subset of 63 and 36 rRNA 2ʹ-O-methylated sites in Arabidopsis are conserved in human and/or yeast, respectively (), indicating that these rRNA sites may be important for ribosome assembly and/or function in eukaryotic cells. On the other hand, most of the Arabidopsis specific sites are located in the 25S rRNA and more precisely in the domain V (), responsible for peptidyl-transferase activity and tRNA binding [Citation4,Citation46]. This might suggest that rRNA methylation at these specific sites in domain V could be required for tuning translation in plants, and likely linked to the usage of synonymous codons and cognate tRNAs to optimize protein translation at particular conditions, including tissue-specific translation in Arabidopsis [Citation47].
Plants are constantly subjected to different cellular and environmental stress conditions and might require additional RNA modifications to protect ribosome integrity or activity. We do not know yet if the Arabidopsis-specific rRNA sites are conserved in other plant species. However, specific rRNA methylation sites can be anticipated in other plant species, as reported in rice [Citation19].
Among the 12 rRNA sites without predictable guiding C/D snoRNAs, 6 were also mapped in Arabidopsis stems [Citation42] including 18S Um1554 and 25S Gm1460, Cm2294, Gm2396, Gm2652 and Gm2923 (Table S2). However, it is possible that some snoRNAs might guide methylation of the rRNA site adjacent to the targeted site. Indeed as shown for U24 in yeast [Citation48] and hypothesized in snoRNAdb for U24 in human [Citation49], the snoRNA At4gCDbox113.1, proposed to guide the modification of 25S-Am1459, might also guide the conserved 2ʹ-O-methylation of 25S-Gm1460. Similarly, the snoRNA At1gCDbox5.1 proposed to guide methylation at 25S-Um2651, might also guide the conserved 2ʹ-O-methylation of 25S-Gm2652. Interestingly, we have not found any C/D snoRNA guide able to guide modification at positions Gm2396, Um2922 and Gm2923 in 25S rRNA, as it is the case for conserved 2ʹ-O-methylated positions in human.
While most of the eukaryotic rRNA ribose modifications are guided by snoRNA, four different modifying enzymes direct ribose methylation in E. coli: rsmH/rsmL methylates 16S-Cm1402 while rlmB, rlmM and rlmE/rrmJ methylate Gm2251, Cm2498 and Um2552 in the 23S, respectively [Citation50]. For Arabidopsis 18S-Cm1645 and 25S-Gm2620, equivalent to E. coli 16S-Cm1402 and 23S-Gm2251, respectively, we have identified at least 2 C/D snoRNAs for each. In contrast, for 25S-Um2922, the equivalent of E. coli 23S-Um2552, we have not found either a specific and/or adjacent snoRNA that might guide methylation. This is an significant observation since methylation of E. coli 23S-U2552 influences the interaction of aminoacyl-tRNA with the ribosomal A-site and lack of methylation affect accuracy of translation [Citation50]. An Arabidopsis protein gene phylogenetically related to yeast genes encoding close homologues of E. coli rlmE/rrmJ and able to 2ʹ-O-methylate tRNA was reported [Citation51]. However, potential 2ʹ-O-metyltransferase activity of this Arabidopsis protein has not been demonstrated and requires further investigation.
For most of the rRNA methylated sites we detected, guiding C/D snoRNAs co-precipitate with FIB2 (). However, C/D snoRNAs At3gCDbox101.1 and At5gCDbox123.1 directing ribose methylation of Gm2794 and Cm1847 in the 25S, were not detected in any of the FIB2-IP fractions, suggesting that they are probably low expressed in our conditions. On the other hand, in the FIB2-IP fraction we found C/D snoRNAs At1gCDbox5.1 and At4gCDbox113.1, which, as mentioned before, might methylate adjacent rRNA sites without matching C/D snoRNA (). Interestingly, 6 novel C/D snoRNA that co-precipitated with FIB2, do not have recognized rRNA targets () and then they might guide fibrillarin methylation activity to other coding and/or non-coding RNA. Indeed, in eukaryotes, ribose methylation has been found in snRNA, tRNA, snoRNA and also mRNA [Citation16,Citation52,Citation53]. Alternatively, specialized C/D snoRNPs might also guide RNA base acetylation [Citation54].
Further bioinformatics analysis revealed that 18 C/D snoRNAs detected in the FIB2-IP fraction target 2ʹ-O-methylation of major U1, U2, U4, U6, and minor U12 and U6atac snRNAs (Figure S4). Among these 18 C/D snoRNA we have found that 5 are ‘orphans’ since do not have predicted rRNA target sites (Table S4). Furthermore, 5 other C/D box snoRNAs found in the FIB2-IP fraction and having no rRNA target are isoforms of U3 snoRNAs (). Unlike other C/D box snoRNAs, U3 is not directing 2ʹ-O methylation of other RNAs. Rather, U3 snoRNA is required to guide site-specific cleavage of rRNA during pre-rRNA processing and contains a longer 5′ region downstream the C box that pairs with pre-rRNA rRNA [Citation55–57]. All orphan snoRNAs (Table S4) were submitted to RNA central [Citation58] and show high conservation of all sequences and C/D box motifs in Arabidopsis lyrata.
138 C/D snoRNA in the FIB2- IP fraction were also detected among 154 C/D snoRNA recently reported in a nuclear fraction from Arabidopsis cell suspension culture [Citation59]. However, 16 others C/D snoRNAs detected in the nuclear fraction were not found to be associated to FIB2 in our conditions (Table S2). Altogether, the data and observations indicate specific expression of snoRNA in 21 days-old plant compared to Arabidopsis cell suspension culture. Indeed differential expression of C/D snoRNA has been reported in plants [Citation22,Citation60,Citation61] and in animal cells [Citation62,Citation63]. However, we do not rule out the possibility that some C/D snoRNA not detected in the FIB2-IP fraction are in fact expressed, but not assembled into C/D snoRNP in our plant conditions. Indeed, proper assembly of C/D snoRNA with core proteins, including Fibrillarin, is essential for activity of C/D snoRNP [Citation64]. Finally, the comparative analysis of FIB2-IP and in silico search analysis demonstrated expression of nearly two hundred C/D snoRNA species and identified 9 novel ones from which at least two direct rRNA 2ʹ-O-methylation (), expanding the number of known C/D snoRNA to 226 ().
Differentially 2ʹ-O-ribose methylated ribosomes and their impact in cellular function have been reported in animal and yeast cells (reviewed in [Citation4,Citation9,Citation10,Citation65]). Our results revealed rRNA 2ʹ-O-methylation alterations in the nuc1-2 mutant plants (), which display strong developmental phenotypes [Citation40–42]. Surprisingly, in the nuc1-2 plants we have not detected hypomethylation of 25S-Gm2620. Indeed, lack of methylation of 25S-Gm2620, by inhibiting expression of the guiding C/D snoRNA HID2, provokes developmental defects, which are reminiscent of plant mutants for specific ribosomal proteins and ribosome biogenesis factors, including Nucleolin [Citation22]. In contrast, the Um2422 and Am2641, located structurally closed to 25S-Gm2620 are strongly hypomethylated (), proposing a functional and/or structural connection between these three rRNA sites.
rRNA ribose hypomethylation of specific sites and major growth and developmental phenotypes were also observed in nufip mutant plants [Citation23]. NUFIP (Rsa1p in yeast) is a conserved and a central protein factor directing assembly of C/D snoRNPs [Citation66,Citation67]. Though, a functional link between Nucleolin and C/D snoRNP biogenesis/assembly has not been demonstrated yet, the mouse Nucleolin directly interacts with the MBII-52 snoRNA that assembles into a non-canonical snoRNPs and might function as a chaperone or assist shuttling of MBII-52 RNPs between nucleoplasm and nucleoli [Citation68]. Assembly and maturation of C/D-box snoRNP occurs in the nucleoplasm in human cells while in yeast maturation initiates in the nucleoplasm and terminates in the nucleolus [Citation69]. To our knowledge, it is not known where precisely assembly of snoRNP occurs in plants. The nucleolus is disorganized in nuc1-2 mutant plants [Citation42] and it is tempting to speculate its direct impact on C/D snoRNP biogenesis in plants and/or more directly on 2ʹ-O-methylation activity. If the assembly and/or maturation of pre-mature snoRNPs in functional C/D snoRNP is affected or not in nuc1 mutant plants remains an open question. Finally, how precisely rRNA ribose modification is impacted at different developmental stages and in response to environmental conditions, is also a next challenging question to be addressed.
Material and methods
Plant materials and growth conditions
All lines were derived from Arabidopsis thaliana Columbia (Col-0) ecotype. Plants mutant nuc1-2 and plants expressing FIB2-YFP nucleolar marker constructs were previously described in [Citation32,Citation42,Citation70]. Seeds were sown on soil (1/5 vermiculite and 4/5 soil) and left for 2 days at 4°C to synchronize. Plants were then grown in controlled growth chambers under a 16 h light/8 h dark cycle at 20°C. Light 100 µE.m−2.s−1 and Relative Humidity 60% (CLF Plant Climatics GmbH, Wertingen Germany). Aerial parts from three-week-old plant seedlings were collected, shock-frozen in liquid nitrogen and grinded in fine powder and store at −80°C.
RNA isolation
For RiboMethSeq, about 800 µL of frozen powder were supplemented with 5 mL of TRI Reagent® (Molecular Research Center, Inc), then 1 mL of cold chloroform was added and incubated for 3 minutes. The mix was centrifuged at 8,000xg for 15 min at 4°C. The aqueous phase was recovered and precipitated with 3 mL of cold isopropanol. After 30 min incubation, the mix was centrifuged at 8,000xg for 30 min at 4°C. Isopropanol was removed and 1.25 mL of 75% ethanol was added to the pellet and incubated over-night at −20°C. After centrifugation, the ethanol was removed and the pellet air dried. The RNA was suspended in RNAse free water and purity verified using Agilent RNA 6000 Pico Kit, analysed in an Agilent 2100 Bioanalyzer, according to the manufacturer’s protocol (Figure S1).
2ʹ-O-methylation sites cartography by RiboMethSeq
RiboMethSeq analysis was performed as previously described in [Citation24]. Briefly, 100 ng of total RNA from WT plants was subjected to alkaline hydrolysis for 12 min at 96°C. RNA was precipitated and end-repaired before being converted to library using NEBNext©Small RNA Library kit (NEB, USA). Library quality and quantity were assessed using a High Sensitivity DNA chip on a Bioanalyzer 2100 and using Qubit 2.0 fluorometer, respectively. Libraries were multiplexed and subjected for high-throughput sequencing using an Illumina HiSeq 1000 instrument with a 50 bp single-end read mode.
Heat map
Heatmap to compare RiboMethSeq methylation score for Col-0 and nuc1-2 mutants was constructed using position-normalized variations of MethScore relative to average value observed for a given position. In this presentation overmethylated sites are in red, while undermethylation compared to average is represented by green colour. Highly constitutive and invariable sites are in white. Clustering was performed using heatmap.2/hclust R functions using ward.D2 method.
Immunoprecipitation: Protein and RNA extraction
Leaves from 3 weeks-old Col-0 (WT) and p35S:FIB2-YFP seedlings were grinded into fine powder in liquid nitrogen. The whole cell extracts (2.4 g of p35S-FIB-YFP and 0.8 g for Col-0) were prepared in 3 volumes of buffer EB150 (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, and EDTA free proteases inhibitor cocktail from Roche) supplemented with 0.1% NP-40. p35S:FIB2-YFP cell extracts (input) were divided into three reactions, and all samples were incubated with 20 µL of GFP-Trap_ MA beads (Chromotek) for 2 h at 4°C with gentle rotation. The unbound fractions were collected and the beads were then washed three times with EB150.
RNA and proteins in IP fraction were extracted using TRI-Reagent (MRC research). RNA separated in aqueous phase were precipitated using glycogen as a carrier (SIGMA, 20 mg) following supplier instructions. Proteins were precipitated from phenolic phase by adding three volumes of cold acetone. After washing, pellets were resuspended in 4X SDS-Laemmli buffer, denatured to perform western blot analysis.
Alternatively, proteins bound to the beads were eluted and recovered in 4X SDS-Laemmli buffer (250 mM Tris-HCl pH 6.7; 8% SDS; 40% glycerol; 0.2% bromophenol bleu, 0.4 M DDT) in the experiments dedicated to Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) analysis.
SDS-PAGE and Western blot analysis
For SDS-PAGE and Western blot, proteins extracts were diluted in SDS-Laemmli buffer, supplemented with β-Mercaptoethanol, heated at 95°C for 10 min and subjected to 10% SDS-polyacrylamide gel electrophoresis. After electrophoresis proteins were either visualized by coomassie-blue staining or transferred to PVDF (Millipore) or nitrocellulose membranes (Bio-Rad) according to manufacturer’s instructions. The membranes were then blotted with α-GFP 1:5,000 (Tebu-Bio) and goat anti-rabbit coupled HRP antibodies (Bio-rad). Immunoreactive proteins were detected using Immobilon western chemiluminescent substrates (Millipore) and the acquisition of images was performed using Fusion Solo S camera (Vilber Lourmat). Once immunodetections performed, the membranes used to control IP experiments intended to RNAseq analysis were stained with colloidal blue coomassie solution.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) analyses
Immuno-precipitated protein fractions were diluted to 1X SDS-Laemmli buffer and supplemented with 10 mM DTT before being loaded on an in-house poured 4% acrylamide stacking gel. Gel was stained with Coomassie Blue and the stacking bands were manually excised. Proteins were then reduced, alkylated and digested overnight at 37°C with modified trypsin in a 1:100 enzyme:protein ratio (Promega, Madison, USA). Peptides were extracted during 1 hour with 80 μL of 80% acetonitrile, 0.1% formic acid, before being dried and suspended in water acidified with 0.1% formic acid prior to nanoLC-MS/MS analysis.
LC-MS/MS analyses were performed on a NanoAcquity LC-system (Waters, Milford, MA, USA) coupled to a Q-Exactive plus Orbitrap (Thermo Fisher Scientific, Waltham, MA, USA) mass spectrometer operated in Data-Dependent Acquisition mode as previously described [Citation32]. Peptides/proteins were identified using the Mascot search engine (version 2.5.1, MatrixScience, London, UK) against an Arabidopsis thaliana protein sequences database downloaded from The Arabidopsis Information Resource TAIR site (TAIR10 version gene model) to which common contaminants and decoy sequences were added (total of 2 × 27 534 protein entries). Identifications were validated and label-free extracted ion chromatogram-based quantification was performed using the Proline software suite. False Discovery Rate was optimized to be below 1% at PSM level using Mascot Adjusted E-value and below 1% at Protein Level using Mascot Mudpit score. Statistical analysis was performed using the Prostar software suite (version 1.12.11). Pairwised Limma t-tests were performed. P-values calibration was corrected using adapted Benjamini-Hochsberg method, and FDR was set to <1-2%. (For more details see Supplementary Information)
RNAseq of RNA from FIB2:YFP fractions
RNA samples from CoIP_1 (150 ng), CoIP_2 (300 ng) and CoIP_3 (225 ng) samples were subjected to alkaline hydrolysis for 5 min at 96°C. RNA was precipitated and end-repaired before being converted to library using NEBNext©Small RNA Library kit (NEB, USA). Library quality and quantity were assessed using a High Sensitivity DNA chip on a Bioanalyzer 2100 and using Qubit 2.0 fluorimeter, respectively. Libraries were multiplexed and subjected for high-throughput sequencing using an Illumina HiSeq 1000 instrument with a 50 bp single-end read mode.
C/D snoRNA bioinformatic search and analysis
Data. The genomic sequence of Arabidopsis thaliana used in these analyses is available at ftp.ensemblgenomes.org/pub/plants/current/fasta/arabidopsis_thaliana/dna/Arabidopsis_thaliana.TAIR10.dna.toplevel.fa.gz. Annotations are from TAIR. Fasta sequences of used ribosomal RNAs and snRNA are given in Supplemental Information.
C/D box snoRNA identification. The catalogue of Arabidopsis thaliana C/D box snoRNA genes was established using three approaches. The initial dataset of C/D box snoRNA genes was built from sequences available in the snoRNA Orthological Gene Database (snOPY) [Citation34] and in Arabidopsis thaliana repositories such as ARAPORT portal [Citation35] and TAIR resource [Citation36]. SnoRNA genes from ARAPORT and TAIR were manually curated to distinguish between H/ACA box and C/D box snoRNA sequences. Only C/D box snoRNA sequences that mapped on the genomic sequence with 100% of identity were kept for further analysis. The initial dataset of C/D box snoRNA sequences was enriched by using PatScan [Citation71] to search the genomic sequence for patterns encoding new C/D box snoRNAs. Such patterns were defined to contain motifs corresponding to the C box (RUGAUGA allowing one mismatch), the C/D box snoRNA region of the snoRNA:rRNA interaction (defined from each rRNA position orphan of a C/D box snoRNA guide at the region around a mapped methylation site and with at most three mismatches in the first eleven base pairs) and the D’/D box motifs (CUGA allowing one mismatch). This updated catalogue was enriched with regions expressed in the sequenced FIB2-YFP coIP fractions and matching the C/D box snoRNA gene pattern not considering the snoRNA:rRNA interaction constraint. Reads from sequenced FIB2-YFP coIP fractions were trimmed for the Illumina 3ʹ adaptor sequence using Cutadapt [Citation72] and aligned using the bowtie2 aligner [Citation73]. Highly expressed intronic and intergenic regions without annotation were inspected in IGV [Citation74], assembled in transcripts and searched for the C/D box snoRNA pattern. This dataset was also used to modify boundaries of previously identified C/D box snoRNA genes. All C/D box snoRNA genes were named as AtchrgCDboxnumber.isoform and consecutively numbered with chr giving the chromosome number, number increasing for each new C/D box snoRNA seen for the first time (paralogs have the same number) and isoform giving the number+1 of times an isoform was already found in preceding chromosomes or before this C/D box snoRNA in the same chromosome.
RNA folding structure prediction of C/D snoRNAs was performed using the mfold Web server http://unafold.rna.albany.edu/?q=mfold
Author contributions
J. A-F designed and performed protein related and IP experiments for sRNA analysis; and supervised master degree student training for IP LC-MS/MS experiments. C.M. and E.J performed RNA related experiments. Y.M., L.A. and V.M. performed RiboMethSeq analysis. M.R. and C.C performed LC-MS/MS and analyzed the data. C.G performed bioinformatics analysis. J.S-V, C.G and Y.M supervised and analyzed the data. J.S-V wrote the manuscript with the assistance of J. A-F., C.C., C.G and Y.M.
Supplemental Material
Download Zip (4 MB)Acknowledgments
We thank our master student, Clarisse Mariez, who contributed to IP-LC MS/MS and Anne de Bures for technical assistance.
Disclosure and availability Statement
The authors report no conflict of interest. Raw data were generated at the Epitranscriptomics and RNA Sequencing (EpiRNA-Seq) Core Facility, Nancy, France. Derived data supporting the findings of this study are available from the corresponding author [JSV] on request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27(7):344–351.
- Polikanov YS, Melnikov SV, Soll D, et al. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol. 2015;22(4):342–344.
- Sharma S, Lafontaine DL. ‘View from a bridge’: a new perspective on eukaryotic rrna base modification. Trends Biochem Sci. 2015;40(10):560–575.
- Sloan KE, Warda AS, Sharma S, et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14(9):1138–1152.
- Blanchard SC, Puglisi JD. Solution structure of the A loop of 23S ribosomal RNA. Proc Natl Acad Sci USA. 2001;98:3720–3725.
- Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728.
- Erales J, Marchand V, Panthu B, et al. Evidence for rRNA 2ʹ-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci U S A. 2017;114(49):12934–12939.
- Marcel V, Ghayad SE, Belin S, et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013;24(3):318–330. .
- Monaco PL, Marcel V, Diaz JJ, et al. 2ʹ-O-Methylation of Ribosomal RNA: towards an Epitranscriptomic control of translation? Biomolecules. 2018;8(4):1–13.
- Ayadi L, Galvanin A, Pichot F, et al. RNA ribose methylation (2ʹ-O-methylation): occurrence, biosynthesis and biological functions. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):253–269.
- Massenet S, Bertrand E, Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017;14(6):680–692.
- Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3(3):397–414.
- Yu G, Zhao Y, Li H. The multistructural forms of box C/D ribonucleoprotein particles. RNA. 2018;24(12):1625–1633.
- Brown JW, Clark GP, Leader DJ, et al. Multiple snoRNA gene clusters from Arabidopsis. RNA. 2001;7(12):1817–1832.
- Qu LH, Meng Q, Zhou H, et al. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 2001;29(7):1623–1630.
- Brown JW, Echeverria M, Qu LH. Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci. 2003;8(1):42–49.
- Chen HM, Wu SH. Mining small RNA sequencing data: a new approach to identify small nucleolar RNAs in Arabidopsis. Nucleic Acids Res. 2009;37(9):e69.
- Kim SH, Spensley M, Choi SK, et al. Plant U13 orthologues and orphan snoRNAs identified by RNomics of RNA from Arabidopsis nucleoli. Nucleic Acids Res. 2010;38(9):3054–3067.
- Chen CL, Liang D, Zhou H, et al. The high diversity of snoRNAs in plants: identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 2003;31(10):2601–2613.
- Liu TT, Zhu D, Chen W, et al. A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Mol Plant. 2013;6(3):830–846.
- Patra Bhattacharya D, Canzler S, Kehr S, et al. Phylogenetic distribution of plant snoRNA families. BMC Genomics. 2016;17(1):1–12.
- Zhu P, Wang Y, Qin N, et al. Arabidopsis small nucleolar RNA monitors the efficient pre-rRNA processing during ribosome biogenesi. Proc Natl Acad Sci U S A. 2016;113(42):11967–11972.
- Rodor J, Jobet E, Bizarro J, et al. AtNUFIP, an essential protein for plant development, reveals the impact of snoRNA gene organisation on the assembly of snoRNPs and rRNA methylation in Arabidopsis thaliana. Plant J. 2011;65(5):807–819.
- Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36: D178–D183.
- Barneche F, Gaspin C, Guyot R, et al. Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2ʹ-O-methylation sites. J Mol Biol. 2001;311(1):57–73.
- Marchand V, Blanloeil-Oillo F, Helm M, et al. Illumina-based RiboMethSeq approach for mapping of 2ʹ-O-Me residues in RNA. Nucleic Acids Res. 2016;44(16):e135.
- Gruendler P, Unfried I, Pointner R, et al. Nucleotide sequence of the 25S-18S ribosomal gene spacer from Arabidopsis thaliana. Nucleic Acids Res. 1989;17(15):6395–6396.
- Unfried I, Gruendler P. Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 1990;18(13):4011.
- Unfried I, Stocker U, Gruendler P. Nucleotide sequence of the 18S rRNA gene from Arabidopsis thaliana Co10. Nucleic Acids Res. 1989;17(18):7513.
- Pichot F, Marchand V, Ayadi L, et al. Holistic optimization of bioinformatic analysis pipeline for detection and quantification of 2ʹ-O-Methylations in RNA by RiboMethSeq. Front Genet. 2020;11:38.
- Incarnato D, Anselmi F, Morandi E, et al. High-throughput single-base resolution mapping of RNA 2΄-O-methylated residues. Nucleic Acids Res. 2017;45(3):1433–1441.
- Montacie C, Durut N, Opsomer A, et al. Nucleolar proteome analysis and proteasomal activity assays reveal a link between nucleolus and 26S Proteasome in A. thaliana. Front Plant Sci. 2017;8:1–13.
- Pontvianne F, Carpentier MC, Durut N, et al. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3d organization of the a. thaliana genome. Cell Rep. 2016;16(6):1574–1587. .
- Yoshihama M, Nakao A, Kenmochi N. snOPY: a small nucleolar RNA orthological gene database. BMC Res Notes. 2013;6:6.
- Krishnakumar V, Hanlon MR, Contrino S, et al. Araport: the Arabidopsis information portal. Nucleic Acids Res. 2015;43(D1):D1003–1009.
- Berardini TZ, Reiser L, Li D, et al. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis. 2015;53:474–485.
- Qu G, Kruszka K, Plewka P, et al. Promoter-based identification of novel non-coding RNAs reveals the presence of dicistronic snoRNA-miRNA genes in Arabidopsis thaliana. BMC Genomics. 2015;16(1):1–15.
- Tajrishi MM, Tuteja R, Tuteja N. Nucleolin: the most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol. 2011;4(3):267–275.
- Ugrinova I, Petrova M, Chalabi-Dchar M, et al. Multifaceted nucleolin protein and its molecular partners in oncogenesis. Adv Protein Chem Struct Biol. 2018;111:133–164.
- Kojima H, Suzuki T, Kato T, et al. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49(6):1053–1063. .
- Petricka JJ, Nelson TM. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007;144(1):173–186.
- Pontvianne F, Matia I, Douet J, et al. Characterization of AtNUC l1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC L2 gene in Arabidopsis. Mol Biol Cell. 2007;18(2):369–379.
- Tang Y, Wu Y, Xu R, et al. Identification and exploration of 2ʹ-O-methylation sites in rRNA and mRNA with a novel RNase based platform. bioRxiv. 2020;2020(2003):2027.011759.
- Dupuis-Sandoval F, Poirier M, Scott M. The emerging landscape of small nucleolar RNAs in cell biology. WIREs RNA. 2015;6(4):381–397.
- Decatur WA, Liang X-H, Piekna-Przybylska D, et al. Identifying effects of snorna-guided modifications on the synthesis and function of the yeast Ribosome. Methods Enzymol. 2007;425:283–316.
- Wu CC, Zinshteyn B, Wehner KA, et al. High-resolution ribosome profiling defines discrete ribosome elongation states and translational regulation during cellular stress. Mol Cell. 2019;73(959–970):e955.
- Camiolo S, Farina L, Porceddu A. The Relation of Codon Bias to Tissue-Specific Gene Expression in Arabidopsis thaliana. Genetics. 2012;192(2):641–649.
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, et al. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85(7):1077–1088.
- Lestrade L, Weber MJ. snoRNA-LBME-db, a comprensive database of human H/ACA and C/D box snoRNA. Nucleic Acid Research. 2006;34(90001):158–162.
- Sergeeva OV, Bogdanov AA, Sergiev PV. What do we know about ribosomal RNA methylation in Escherichia coli. Biochimie. 2015;117:110–118.
- Pintard L, Lecointe F, Bijnicki JM, et al. Trm7p catalyses the formation of two 2ʹ-O-methylriboses in yeast tRNA anticodin loop. Embo J. 2002;21(7):1811–1820.
- Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84(8):775–790.
- Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other rNAs. J Biol Chem. 2003;278(2):695–698.
- Sharma S, Yang J, van Nues R, et al. Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet. 2017;13(5):e1006804.
- Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. Embo J. 1995;14(17):4350–4356.
- Borovjagin AV, Gerbi SA. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J Mol Biol. 1999;286(5):1347–1363.
- Saez-Vasquez J, Caparros-Ruiz D, Barneche F, et al. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol Cell Biol. 2004;24(16):7284–7297.
- Consortium TR. RNAcentral: a hub of information for non-coding RNA sequences. NAR. 2019;47(D1):D221–D229.
- Streit D, Shanmugam T, Garbelyanski A, et al. The existence and localization of nuclear snoRNAs in Arabidopsis thaliana revisited. Plants (Basel). 2020;9(8):9.
- Wang Y, Li H, Sun Q, et al. Characterization of small rnas derived from trnas, rrnas and snornas and their response to heat stress in wheat seedlings. PLoS One. 2016;11(3):e0150933.
- Zheng J, Zeng E, Du Y, et al. Temporal small rna expression profiling under drought reveals a potential regulatory role of small nucleolar rnas in the drought responses of maize. Plant Genome. 2019;12(1):1–15. .
- Falaleeva M, Welden JR, Duncan MJ, et al. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: old dogs show new tricks. Bioessays. 2017;39(6):1–10.
- Stepanov GA, Filippova JA, Komissarov AB, et al. Regulatory role of small nucleolar RNAs in human diseases. Biomed Res Int. 2015;2015:206849.
- McKeegan KS, Debieux CM, Boulon S, et al. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol Cell Biol. 2007;27(19):6782–6793.
- Dimitrova DG, Teysset L, Carre C. RNA 2ʹ-O-Methylation (nm) modification in human diseases. Genes (Basel). 2019;10(2):1–23.
- Bizarro J, Charron C, Boulon S, et al. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J Cell Biol. 2014;7:1–18.
- Rothe B, Manival X, Rolland N, et al. Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly. Nucleic Acids Res. 2017;45(12):7455–7473.
- Soeno Y, Taya Y, Stasyk T, et al. Identification of novel ribonucleo-protein complexes from the brain-specific snoRNA MBII-52. RNA. 2010;16(7):1293–1300.
- Verheggen C, Lafontaine DL, Samarsky D, et al. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. Embo J. 2002;21(11):2736–2745.
- Pontvianne F, Abou-Ellail M, Douet J, et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010;6(11):1–13. .
- Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13(12):497–498.
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359.
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol 2001;29:24–26.