ABSTRACT
Transposable elements have both detrimental and beneficial effects on their host genome. Tetrahymena is a unicellular eukaryote that deals with transposable elements in a unique way. It has a separate somatic and germline genome in two nuclei in a single cell. During sexual reproduction, a small RNA directed system compares the germline and somatic genome to identify transposable elements and related sequences. These are subsequently marked by heterochromatin and excised. In this Review, current knowledge of this system and the gaps therein are discussed. Additionally, the possibility to exploit the Tetrahymena machinery for genome editing and its advantages over the widely used CRISPR-Cas9 system will be explored. While the bacterial derived CRISPR-Cas9 has difficulty to access eukaryotic chromatin, Tetrahymena proteins are adept at acting in a chromatin context. Furthermore, Tetrahymena based gene therapy in humans might be a safer alternative to Cas9 because the latter can trigger an immune response.
Introduction
Transposable elements (TEs) pose a threat to their host as they are able to move from one genomic location to another. However, TEs are also a source of genetic diversity that can drive evolution of the host genome. Therefore, hosts have developed defence mechanisms to detect TEs in their genome and render them harmless while at the same time maintaining them as genetic reservoirs. Mammals for example tolerate TEs in their genome but silence them at the transcriptional level. The unicellular Tetrahymena on the other hand does not merely suppress the activity of TEs, but it employs the more rigorous measure of removing them from its somatic genome.
Tetrahymena is able to clear its somatic genome from TEs because it has two nuclei in a single cell: a micronucleus (MIC) containing the germline genome and a macronucleus (MAC) containing the somatic genome. During sexual reproduction, Tetrahymena compares the MIC and MAC genome to identify TEs and subsequently eliminate them from the new MAC. In total, approximately 35% of the somatic genome is reproducibly removed and the remaining sequences are rejoined [Citation1]. These radical DNA rearrangements are tightly coordinated by an intricate RNA framework [Citation2,Citation3].
This Review covers the later stages of programmed DNA elimination in Tetrahymena, starting from the search for a target by the Tetrahymena Piwi protein Twi1p (see for an overview of the involved proteins). For a detailed overview of all stages, the reader is referred to a previous Review by Mochizuki and Noto [Citation4]. The focus of this Review is on the open questions that remain regarding the Tetrahymena DNA elimination mechanism and how components of this system can potentially be repurposed as a gene-editing technique. Although CRISPR-Cas9 has revolutionized gene-editing and has become indispensable for life science, it has several drawbacks, especially when applying it in humans. We discuss the potential of a gene-editing system based on Tetrahymena DNA elimination proteins or a combination of the two systems to overcome these drawbacks.
Table 1. Proteins involved in DNA elimination in Tetrahymena
Tetrahymena life cycle
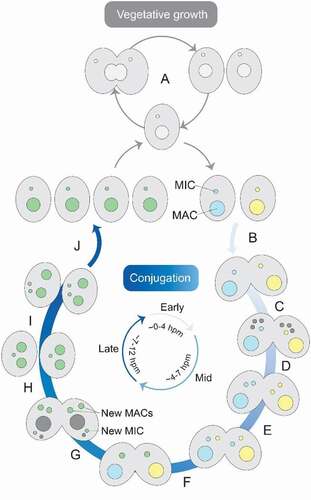
Tetrahymena is a unicellular eukaryote that lives in freshwater. When nutrients are plentiful, the MIC and MAC divide mitotically and amitotically, respectively, and binary fission follows to produce two daughter cells (). This changes under starvation conditions, when pairs of cells with different mating types reproduce sexually through conjugation (). During conjugation, both the new MAC and MIC initially contain the same genome. However, two types of programmed genome rearrangements occur in the developing MAC: chromosome breakage and DNA elimination. Additionally, the MAC genome undergoes multiple rounds of endoreplication.
Figure 1. Tetrahymena life cycle. Each cell contains a macronucleus (MAC) and a micronucleus (MIC). In the presence of sufficient nutrients, Tetrahymena reproduces asexually by binary fission (A). However, when there is a lack of nutrients, it reproduces sexually by conjugation (B-J). To start conjugation, two cells of complementary mating types fuse (B) and their MICs undergo meiosis (C). Three of the meiotic products are degraded, while the surviving nucleus undergoes mitosis (D). The fused cells then exchange a pronucleus (E) and the pronuclei fuse to create a zygotic nucleus (F), which undergoes two rounds of mitosis (G). Two products will develop into new MACs, one will form a new MIC and the fourth product is degraded. The parental MAC is also degraded and the fused cells separate (H). Finally, the MIC divides mitotically (I), which is followed by binary fission (J). The approximate time-scale of events is indicated in hours post-mixing (hpm)
The first type of programmed genome rearrangement, chromosome breakage, leads to the fragmentation of chromosomes at conserved chromosome breakage sequences [Citation5]. The second, DNA elimination, results in the removal of approximately 12,000 internal eliminated sequences (IESs), from the new MAC genome [Citation1]. The eliminated sequences mainly consist of TE-related sequences and elimination of these sequences is essential for cell viability, as indicated by the inability of mutants with defective DNA elimination to produce viable progeny [Citation6–8]. A small subset of 12 IESs have conserved inverted terminal repeats. These IESs are excised in a transposon-like fashion by the piggyBac transposases Tpb1p and Tpb6p. However, the majority of IESs do not share one common motif. Yet, they are excised from the genome with near base-pair accuracy by another domesticated piggyBac transposase: Tpb2p [Citation1,Citation9,Citation10]. Unlike the piggyBac transposases in other organisms such as Paramecium or Trichoplusia ni, Tpb2p does not appear to have strict specificity [Citation11–13].
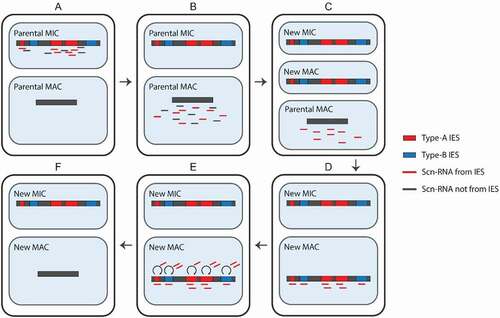
Excision of IESs by Tpb2p is orchestrated by small RNAs, termed scan RNAs (scnRNAs) [Citation2]. In the current model, early-scnRNAs are produced from certain IESs (referred to as type-A IESs) and their surrounding regions in the MIC genome during the early-conjugation stage () [Citation3].
Figure 2. Programmed DNA elimination. Early scan-RNAs (scnRNAs) are produced from type-A internal eliminated sequences (IESs) and surrounding regions (A). Next, they are transported to the MAC (B) and scnRNAs complementary to the MAC genome are degraded. Meanwhile, a new MIC and MAC have been formed (C). The remaining scnRNAs are transported to the new MAC, where they recognize both type-A and – B IESs (D). The scnRNAs induce heterochromatin formation and the production of late-scnRNAs, which further spread the heterochromatin (E). Finally, the parts of the genome marked by heterochromatin are excised and the ends are ligated (F)
Early-scnRNA induced heterochromatin formation
One of the crucial components of the programmed DNA elimination machinery is the Piwi protein Twi1p [Citation2]. In total Tetrahymena has 12 Twi proteins with varying functions, of which Twi1 and Twi11 are the only ones known to be essential for small RNA-directed DNA elimination [Citation17]. Shortly after scnRNA biogenesis and the subsequent transportation to the cytoplasm, Twi1p stabilizes the double-stranded scnRNAs and removes one of their strands. Once mature, scnRNA-Twi1p complexes are bound by Giw1p, enabling translocation to the new MAC [Citation18]. There the search for a sequence homologous to the scnRNA starts ().
Figure 3. From Twi1p-scnRNA target search to IES elimination. Together with Ema1p, the Twi1p-scnRNA complex searches for targets in nascent transcripts (A). At matching sequences, histone H3 is methylated at lysine 9 (H3K9me3) and 27 (H3K27me3) by Ezl1p (B). Pdd1p is recruited to the heterochromatin (C). This is a requirement for the production of late scnRNAs that further spread the heterochromatin. At the same time, boundary-protecting factors confine the heterochromatin to IESs (D). Finally, Tpb2p excises IESs marked by heterochromatin (E)
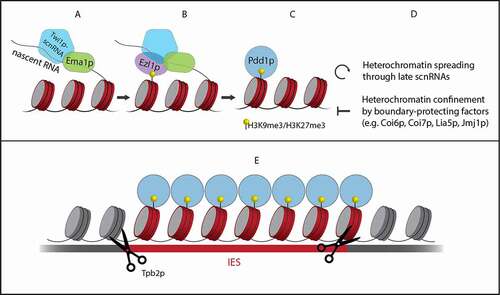
In the search for matching targets, the early-scnRNA-Twi1p complex identifies type-A IESs from which the early-scnRNAs were originally derived. Additionally, type-B IESs are found because they share repetitive sequences with type-A IESs [Citation3]. Once a sequence is identified that matches the scnRNA, the histone methyltransferase Ezl1p is recruited by the scnRNA-Twi1p complex through a mechanism that remains elusive (). A study by Liu and colleagues showed that Ezl1p catalyses methylation of histone H3 at lysine 27 (H3K27me3), which is a histone mark characteristic of heterochromatin [Citation16]. Additionally, they showed that Ezl1p-catalysed H3K27me3 regulates the methylation of histone H3 at lysine 9 (H3K9me3). In Tetrahymena, this second hallmark of heterochromatin is exclusively found at IESs in the developing MAC and the methylated histones attract additional proteins necessary for DNA elimination [Citation20].
Late-scnRNAs and IES boundary determination
The role for small RNAs in DNA elimination does not stop here. Noto and colleagues found that a second group of scnRNAs is produced during late conjugation [Citation3]. They showed that spreading of the heterochromatin induced by early-scnRNAs triggers the production of late-scnRNAs. This occurs through an unknown mechanism that requires Pdd1p, a protein that recognizes both H3K27me3 and H3K9me3 () [Citation3]. Late-scnRNAs are loaded into zygotic Twi1p and Twi11p, but whether the two zygotic Piwi proteins have distinct functions is unknown. Guided by the late-scnRNAs, they further promote heterochromatin formation at IESs in an Ezl1p-dependent manner. This can trigger the production of more late-scnRNAs from other IESs in trans. Thus, a positive feedback loop is formed (), which increases the robustness of the DNA elimination system. According to simulations by Noto and colleagues, this mechanism ensures that almost no IESs fail to be eliminated, even in the extreme case where only 1% of the type-A IESs produce early-scnRNAs [Citation3].
To prevent the excision of DNA outside IESs, heterochromatin spreading must be stopped exactly at IES boundaries (). In a study by Suhren and colleagues, several proteins were identified that are crucial for defining IES boundaries [Citation21]. One of these boundary-protecting factors is the heterochromatin-binding protein Coi6p, which localizes to heterochromatinized IESs. In its absence, heterochromatin is not sharply confined to IESs and late-scnRNAs are also produced from regions outside IESs. Two proteins that interact with Coi6p, Coi7p and Lia5p were also found to negatively regulate heterochromatin formation. A second level of boundary control comes in the form of the histone demethylase Jmj1p. It reverses heterochromatin marks and is crucial for proper DNA elimination. However, the exact mechanism through which Jmj1p contributes to defining IES boundaries has not yet been revealed.
DNA elimination
IESs marked by heterochromatin are excised from the genome (). The endonuclease responsible for this is the domesticated piggyBac transposase Tpb2p [Citation14,Citation15]. At IES boundaries it creates double-strand breaks (DSBs) with 4-nt long 5ʹ overhangs. Although piggyBac transposases are known to recognize and cleave 5ʹ-TTAA-3ʹ sequences, Tpb2p does not seem to have a strict sequence preference [Citation13]. It does however have the ability to interact with histone marks specific for heterochromatin [Citation13]. Hitherto, the exact mechanism ensuring that Tpb2p cleaves specifically at IES boundaries has not been elucidated.
Upon excision of the first IESs, aggregates of heterochromatinized IESs form [Citation22]. These structures, referred to as heterochromatin bodies, also appear in response to DNA damage induced by UV [Citation22]. This suggests that the function of heterochromatin bodies may be to facilitate DNA repair. Supporting this hypothesis is the finding by Lin and colleagues that a Ku80 homologue, TKu80p, is essential for the assembly of heterochromatin bodies [Citation23]. They show that TKu80p binds to and protects the ends of the DSBs created by Tpb2p. This is followed by repair of the break through the non-homologous end joining (NHEJ) pathway. Another possible role for heterochromatin bodies, not excluding the former, could be to sequester freed IESs. This would prevent them from interacting with other genomic sequences before they are degraded.
Discussion
The general system through which Tetrahymena clears its somatic genome of TEs and TE-related sequences has been unravelled and important players have been identified (). However, several key questions concerning the molecular mechanism remain unanswered. A first question is how early-scnRNA induced heterochromatin leads to late-scnRNA production. It seems counter-intuitive that heterochromatin, which generally corresponds to a transcriptionally inactive state, triggers expression. However, transcription might be promoted by one of the proteins recruited to the heterochromatin. The role of many of these proteins remains elusive, further studies into their function might reveal the link between heterochromatin formation and late-scnRNA production.
Arguably the most intriguing open question is how Tetrahymena is able to excise thousands of sequences from its genome with high precision without any apparent sequence motif at the IES boundaries. The molecular machinery led by scnRNAs marks IESs with heterochromatin, which at best demarcates IESs with nucleosome resolution. What further narrows down the IES boundary? A recent study by Lin et al. suggests the existence of cis-acting boundary elements [Citation24]. They identified multiple inverted repeats each of which potentially regulate a subset of IESs. These sequences are not located exactly at the IES boundaries but in a flanking region of about a hundred base pairs. Remarkably the same element is found at near equal distance from the two opposite boundaries of an IES. Altogether, they found six major groups of inverted repeats that are present in the flanking regions of approximately 60% of all IESs. Further studies focussing on the remaining 40% will point out whether cis-acting boundary elements are the global regulatory mechanism of determining IES boundaries or whether the boundaries of these IESs are set by a different mechanism. Additionally, identifying and characterizing proteins that bind the cis-acting elements could provide further insights into how these elements govern IES boundary determination.
Another open question is whether Tpb2p is recruited specifically to IES boundaries or whether it localizes to all heterochromatin. Although Tpb2p does not cleave DNA within the IES body, it could still bind to all heterochromatin if it is only activated at IES boundaries. In that case, an accessory protein is needed that localizes to euchromatin-heterochromatin transition regions and provides the trigger for Tpb2p activation. Studying the localization of Tpb2p, for example by ChIP analysis or high-resolution microscopy, could provide a first clue as to which of the two scenarios is correct. Additionally, finding interaction partners of Tpb2p will help to further unravel the mechanism regulating IES boundary determination.
Besides accurately removing such a large number of sequences with high accuracy, Tetrahymena also adequately degrades the excised sequences and ligates the remaining parts of the new MAC genome. Repair of the DSBs occurs through the NHEJ pathway, but these DSBs are different from typical DNA damage. When DNA damage occurs, the ends of the break are held close together to facilitate repair. However, in the case of DNA elimination, the ends that are to be ligated are different from the ends of the original DSB. It is crucial that the IES is removed and that the ends adjacent to the IES boundaries are the once that are ligated. Otherwise, IESs could be reintegrated in the genome or the genome could become scrambled. Little is known about this final stage of programmed DNA elimination. The aforementioned cis-acting boundary elements could play a role in keeping neighbouring MAC-destined regions together.
Perspective: potential for genome editing
Although multiple open questions remain, one can envision the Tetrahymena DNA elimination machinery being repurposed as a genome editing tool. Programmed DNA elimination in Tetrahymena is one of a wide variety of systems that organisms have developed to eliminate or silence foreign nucleic acid sequences. Another defence system against foreign DNA, the bacterial CRISPR-Cas9 system, has already been successfully employed for genome editing. Although it has quickly turned into an essential tool for life science research, the technique has several drawbacks including off-target effects [Citation25]. Because it acts through a different mechanism than CRISPR-Cas9, the Tetrahymena DNA elimination machinery might provide valuable tools to complement the current genome editing toolbox.
One of the drawbacks of the CRISPR-Cas9 system is that it is not optimized to act in a chromatin context. The system is originally derived from prokaryotes, where it acts on naked DNA. In a recent study, Horlbeck and colleagues observed that nucleosomes hinder Cas9 in binding to the genome and cleaving it [Citation26]. They noted that efficient targeting was almost exclusively achieved in nucleosome poor regions of the genome. Tetrahymena on the other hand is a eukaryote. Therefore, its DNA elimination machinery is tailored for accessing the eukaryotic genome. Thus, proteins from the Tetrahymena DNA elimination system might provide a gateway to chromatinized DNA, thereby increasing genome editing efficiency in eukaryotes.
A second drawback of the CRISPR-Cas9 system is that it might trigger an immune response in humans [Citation27]. The most frequently used Cas9 variants are derived from two bacterial species: Staphylococcus aureus and Streptococcus pyogenes. Infection with these bacteria is common among humans and as a result the majority of the human population has an adaptive immune response to Cas9 [Citation27]. This raises concerns regarding the safety and effectiveness of Cas9-based therapies as it might lead to the elimination of Cas9-expressing cells. Because Tetrahymena are not known to infect humans, there is expected to be no pre-existing adaptive immunity to Tetrahymena derived proteins. They might therefore provide a safer alternative to CRISPR-Cas9 for gene-editing in humans.
While CRISPR-Cas9 only requires a guide RNA and the Cas9 protein, a genome editing system based on the Tetrahymena DNA elimination machinery would consist of more components. First, a scnRNA-Twi1p complex is required to locate a target. This also requires Ema1p or another RNA helicase to provide access to nascent transcripts. The next crucial component is an endonuclease, which has to be recruited to the target site. Because many questions about Tpb2p are still unanswered, further research is required before it becomes clear whether Tpb2p can be used for genome editing. Tpb2p has for example been shown to have some sequence specificity, albeit not very strict [Citation13]. Can it be directed to cleave at a specific site of choice? If so, it provides an advantage over Cas9 which is limited by the requirement of a PAM sequence adjacent to the target. Finally, Tpb2p interacts with the histone H3 tail, especially in the presence of H3K9me3 or H3K27me3 [Citation28]. In Tetrahymena these heterochromatin marks are restricted to IESs, however in other organisms they are widespread through the genome. This could lead to off-target effects or inefficient targeting of euchromatin regions.
As an alternative, Cas9 could be used as the endonuclease. It would be interesting to explore ways to implement a form of ‘two-factor authentication’ by using a combination of Tetrahymena machinery and CRISPR-Cas9. Here a scnRNA with Twi1p would perform the first step of locating the target. Next, a conditionally active form of Cas9 would be used which is activated by a moiety carried by Twi1p. Cas9 would thus be activated at the target site, where its guide RNA ensures that it induces a DSB exactly at the right site. By using such a ‘two-factor authentication’ system, off-target effects are expected to be greatly reduced.
Conclusion
In conclusion, programmed DNA elimination in Tetrahymena is an intricate process of which the overall mechanism has been revealed. Further studies focussing on amongst others the open questions presented above are required to complement the current knowledge and obtain a detailed picture of the molecular mechanisms involved in programmed DNA elimination. This will not only increase our understanding of Tetrahymena biology, but it will also open new doors for exploiting the Tetrahymena DNA elimination system as a genome editing technique.
Acknowledgments
C.J. was supported by an ERC Consolidator grant (819299) of the European Research Council.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hamilton EP, Kapusta A, Huvos PE, et al. Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife. 2016 Nov; 5: DOI:10.7554/eLife.19090
- Mochizuki K, Fine NA, Fujisawa T, et al. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002 Sep;110(6):689–699.
- Noto T, Kataoka K, Suhren JH, et al. Small-RNA-mediated genome-wide trans-recognition network in tetrahymena DNA elimination. Mol Cell. 2015;59(2):229–242.
- Mochizuki K, Noto T. Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open Biol. 2017;7(10):170–172.
- Yao MC, Yao CH, Monks B. The controlling sequence for site-specific chromosome breakage in tetrahymena. Cell. 1990 Nov;63(4):763–772.
- Coyne RS, Nikiforov MA, Smothers JF, et al. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol Cell. 1999 Nov;4(5):865–872.
- Malone CD, Anderson AM, Motl JA, et al. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Molecular and Cellular Biology. 2005 Oct;25(20):9151–9164.
- Mochizuki K, Gorovsky MA. A Dicer-like protein in tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005 Jan;19(1):77–89.
- Cheng CY, Young JM, Lin CYG, et al. The piggyBac transposon-derived genes TPB1 and TPB6 mediate essential transposon-like excision during the developmental rearrangement of key genes in Tetrahymena thermophila. Genes Dev. 2016 Dec;30(24):2724–2736.
- Chao J-L, Lin C-YG, Yao M-C, et al. Setting boundaries for genome-wide heterochromatic DNA deletions through flanking inverted repeats in tetrahymena thermophila. Nucleic Acids Res. 2019 Mar;47(10):5181–5192.
- Bischerour J, Bhullar S, Wilkes CD, et al. Six domesticated piggybac transposases together carry out programmed DNA elimination in paramecium. Elife. 2018 Sep; 7: DOI:10.7554/eLife.37927
- Cary LC, Goebel M, Corsaro BG, et al. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989 Sep;172(1):156–169.
- Cheng C-Y, Vogt A, Mochizuki K, et al. A domesticated piggybac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in tetrahymena thermophila. Mol Biol Cell. 2010 Mar;21(10):1753–1762.
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004 Sep;18(17):2068–2073.
- Schoeberl UE, Kurth HM, Noto T, et al. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev. 2012 Aug;26(15):1729–1742.
- Liu Y, Taverna SD, Muratore TL, et al. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007 Jun;21(12):1530–1545.
- Couvillion MT, Lee SR, Hogstad B, et al. Sequence, biogenesis, and function of diverse small RNA classes bound to the piwi family proteins of tetrahymena thermophila. Genes Dev. 2009 Sep;23(17):2016–2032.
- Noto T, Kurth HM, Kataoka K, et al. The tetrahymena argonaute-binding protein giw1p directs a mature argonaute-siRNA complex to the nucleus. Cell. 2010 Mar;140(5):692–703.
- Aronica L, Bednenko J, Noto T, et al. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008 Aug;22(16):2228–2241.
- Wiley EA, Horrell S, Yoshino A, et al. Diversification of HP1-like chromo domain proteins in tetrahymena thermophila. J Eukaryot Microbiol. 2018 Jan;65(1):104–116.
- Suhren JH, Noto T, Kataoka K, et al. Negative regulators of an RNAi-heterochromatin positive feedback loop safeguard somatic genome integrity in tetrahymena. Cell Rep. 2017 Mar;18(10):2494–2507.
- Shieh AWY, Chalker DL. LIA5 is required for nuclear reorganization and programmed DNA rearrangements occurring during tetrahymena macronuclear differentiation. PLoS One. 2013 Sep;8(9):e75337–e75337.
- Lin I-T, Chao J-L, Yao M-C. An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in tetrahymena thermophila. Mol Biol Cell. 2012 Apr;23(11):2213–2225.
- Lin CYG, Chao JL, Tsai HK, et al. Setting boundaries for genome-wide heterochromatic DNA deletions through flanking inverted repeats in tetrahymena thermophila. Nucleic Acids Res. 2019;47(10):5181–5192.
- Zhang X-H, Tee LY, Wang X-G, et al. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015 Jan;4:e264.
- Horlbeck MA, Witkowsky LB, Guglielmi B, et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016 Mar;5:e12677.
- Charlesworth CT, Deshpande PS, Dever DP, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25(2):249–254.
- Vogt A, Mochizuki K. A domesticated piggyBac transposase interacts with heterochromatin and catalyzes reproducible DNA elimination in tetrahymena. PLoS Genet. 2013 Dec;9(12):e1004032.
