ABSTRACT
Complex cascades of RNA-binding proteins regulate the mRNA metabolism and influence gene expression. Several distinct proteins act at different stages of mRNA life cycle. SR family proteins in yeast are implicated in mRNA processing and nuclear export. In this report, we uncover the role of an SR/RGG-motif containing mRNA export factor Gbp2 in mRNA translation regulation. We demonstrate that Gbp2 localizes to cytoplasmic granules upon heat shock and oxidative stress. Our pull-down assays demonstrate that Gbp2 directly binds to the conserved translation factor eIF4G1 via its RGG motif. We further mapped the region on eIF4G1 to which Gbp2 binds and observed that the binding region overlaps with another translation repressor Sbp1. We found that the RGG-motif deletion mutant is defective in localizing to polysome fractions. Upon tethering Gbp2 to a GFP reporter mRNA in vivo, translation of GFP reporter decreased significantly indicating that Gbp2 acts as a translation repressor. Consistent with these results, we show that Gbp2 can directly repress mRNA translation in the in vitro translation systems in an RGG-motif dependent manner. Taken together, our results establish that the mRNA export factor Gbp2 has a vital role in repressing translation of mRNA. We propose that Gbp2 is a multifaceted RGG-motif protein responsible for translational repression without affecting mRNA levels.
Introduction
Messenger RNA, the central player in gene expression, is associated with RNA-binding proteins (RBPs) at every stage of its life cycle. The mRNA fate is orchestrated by the dynamic remodelling of these mRNA-RBP complexes in both the nucleus and the cytoplasm [Citation1,Citation2]. The nucleocytoplasmic shuttling proteins help in the transition of the messages across the nuclear membrane [Citation3]. Certain conserved RBPs like SR family proteins have drawn the attention of researchers because of their ability to influence the fate of mRNAs throughout their life cycle [Citation4]. The multifaceted roles performed by metazoan SR family proteins in mRNA processing, nuclear export, mRNA decay and mRNA translation have paved the way for their popularity as ‘the master regulators of gene expression’ [Citation5]. SR family of proteins are characterized by the presence of arginine and serine dipeptide-rich domains. The series in the SR motifs are subjected to phosphorylation by several protein kinases and this post-translational modification modulates the cellular functions of several SR proteins [Citation6]. SR proteins generally have one or more RNA binding domains (RBDs). RRM domains are the most common RBDs found in SR proteins. Deregulation of SR protein-functions is implicated in several diseases like cancer, spinal muscular atrophy and cystic fibrosis [Citation5,Citation7].
There are three nucleocytoplasmic shuttling SR-like proteins in the budding yeast: Npl3, Gbp2, and Hrb1. All three proteins are modular proteins having an SR domain and RRM domains. Npl3 has two RRM domains whereas Gbp2 and Hrb1 have three RRM domains each. All three SR–like proteins have been implicated in mRNA processing and export, similar to their metazoan relatives [Citation8–10]. In addition to the SR-rich region, they possess RGG-motif that contains repetitive arginine and glycine residues. A class of RGG-motif proteins in yeast are demonstrated to be translation repressors and target the conserved translation initiation factor eIF4G1 to repress mRNA translation [Citation11]. Npl3, an essential mRNA splicing factor, is also reported to repress cytoplasmic mRNA translation by binding eIF4G1 akin to other RGG–motif proteins like Scd6, Sbp1, and Khd1 [Citation11,Citation12]. But the cytoplasmic role of Gbp2 and Hrb1 is not known. Many other shuttling proteins like Pab1, Xrn1, eIF4G, and eIF4E that are key regulators of cytoplasmic mRNA translation/decay, have been reported to dictate nuclear events such as transcription, splicing, and export of certain mRNAs [Citation13–17]. These reports highlight the exciting phenomenon of coupled mRNA transcription, processing/export, translation, and decay. But the factors that are important and can bridge the wide range of functions performed by shuttling proteins is an underexplored arena.
Gbp2 was identified as a telomeric G-strand binding protein and later was demonstrated to be important for quality control of spliced mRNAs and nuclear export [Citation6,Citation18]. Gbp2 is recruited onto the mRNAs co-transcriptionally via the TREX complex and interaction with the THO/TREX complex couples it to mRNA export [Citation19,Citation20]. If the mRNA splicing is aberrant, then Gbp2 remains associated with the TRAMP degradation complex and the mRNA is subjected to degradation in the nucleus. Active transcription by RNA polymerase II and export factors like Mex67 and Nup159/Rat7 are important for the shuttling of Gbp2 across the nuclear membrane [Citation9]. In Plasmodium berghei parasites, Gbp2 has been recently shown to be important for gametocyte production by an unknown mechanism [Citation21]. Interestingly, the Mex67/Mtr2 orthologs are absent in the Plasmodium parasite. Gbp2 is imported to the nucleus via Mtr10 upon phosphorylation by Sky1 (a conserved protein kinase) at the serine residues of the SR-rich region [Citation6]. The cytoplasmic retention of Gbp2 is elevated upon mutation of these serine residues. Interestingly, Gbp2 is associated with mRNAs in the cytoplasm until translation, but the functional significance is not understood [Citation12]. Several observations hint towards a possible cytoplasmic role of Gbp2. Gbp2 is reported to be present in polyribosomes in the cytoplasm in both yeast and Trypanosomes, but the function is unknown [Citation12,Citation22]. The polyribosomal association of Gbp2 was not affected by the deletion of Sky1 suggesting that the ribosome association could be independent of its shuttling role. The cytoplasmic localization of Gbp2 is enhanced in hypoxic stress and Gbp2 forms cytoplasmic foci under glucose deprivation stress [Citation23,Citation24]. The significance of these observations is unclear. Above observations compelled us to explore the possible role of RBPs in mRNA regulation across the steps of gene expression cascade, and we hypothesized that Gbp2, an mRNA export factor, might act as a regulator of mRNA translation in yeast.
Results
RGG motif of Gbp2 is required for overexpression-mediated growth defect
A common feature of many translation repressors like Dhh1, Scd6, Pat1, and Sbp1 is that they impart a growth defect phenotype to the yeast cells upon overexpression [Citation25,Citation26]. The RGG-motif of Scd6 and Sbp1, important for their repression activity, is also important for causing the growth defect [Citation11,Citation26,Citation27]. Gbp2 is a multidomain, RGG-motif containing protein. The N terminus of Gbp2 is rich in SR and RGG repeats and is followed by the three RRM domains (). Gbp2 is reported to cause a growth defect upon overexpression [Citation6]. We hypothesized that Gbp2 overexpression mediated growth defect could be similar to the one caused by the above-known translation repressors. To test if the RGG-motif of Gbp2 is necessary for causing an overexpression mediated growth defect, we created an RGG-motif deletion mutant (Δ51-100) which comprised three -RGx and four -RGG repeats. Wild-type BY4741 cells were transformed with the plasmids bearing wild-type Gbp2GFP and Gbp2GFPΔRGG under the control of galactose promoter. We performed a growth assay with the transformants and observed that overexpression of wild-type Gbp2 caused a growth defect on the galactose media. The deletion of RGG-motif led to almost a complete rescue of the growth defect phenotype (). An empty vector and a plasmid expressing GFP alone served as controls. Gbp2ΔRGG protein expression was not compromised as compared to the wild-type Gbp2 (). The above results suggest that the RGG motif of Gbp2 is important for causing growth defect, a feature reminiscent of other RGG-motif containing translation repressors.
Figure 1. RGG motif is important for Gbp2 overexpression-mediated growth defect. (A) Schematic depicting the domain organization of Gbp2 (left) and Gbp2ΔRGG (right). (B) Wild-type cells overexpressing Gbp2GFP, Gbp2GFPΔRGG and GFP were serially diluted and spotted on glucose and galactose containing agar media, incubated at 30℃ for 36 h and 48 h for glucose and galactose plates, respectively, and imaged. (C) The cells from the first two spots on the galactose plate were scraped, lysed and analysed by western blotting using anti-GFP antibody. Ponceau stained blot served as the loading control
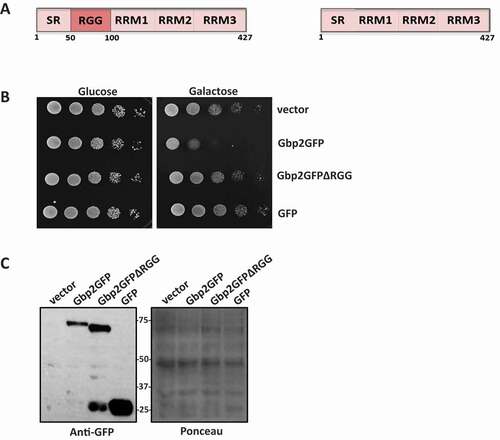
Wild type and nuclear import defective (NID) mutant of Gbp2 accumulate in cytoplasmic granules upon heat and sodium azide stress
Global translation repression is a key outcome of stress response mechanisms [Citation28–31]. Repressed mRNAs in complex with RNA-binding proteins often accumulate in cytoplasmic foci called P bodies and stress granules [Citation24,Citation32]. Poly(A)-tailed mRNAs accumulate in cytoplasmic granules upon heat shock. Gbp2 is a polyA-binding protein and has been reported to form nucleolar granules upon exposure of cells to heat shock for a short period (8 min) [Citation33]. We wanted to study the behaviour of Gbp2 upon prolonged exposure of cells to heat stress. We checked the localization of Gbp2GFP by live-cell imaging. We observed that some Gbp2 accumulated in cytoplasmic foci upon 45 min heat stress, whereas it remained nuclear with no cytoplasmic foci in cells grown at 30°C (). This is consistent with the report that Gbp2 forms cytoplasmic foci upon glucose deprivation stress [Citation24]. As described earlier [Citation6], a mutation in the SR region of Gbp2 causes an increased accumulation of Gbp2GFP in the cytoplasm ( bottom left). To assess if the cytoplasmic accumulation affects Gbp2 foci formation, we tested the localization of the Gbp2NID mutant. We observed that at 30°C the NID mutant shows significant cytoplasmic localization. Upon heat stress, this mutant formed a greater number of cytoplasmic foci (, bottom right), although the protein levels of the NID mutant were comparable to that of the wild type (). To check if Gbp2 localized to foci in response to other stresses that lead to global translation repression, we subjected the cells expressing Gbp2GFP to sodium azide stress. Sodium azide stress is widely used to study RNA granules [Citation34,Citation35]. We observed that Gbp2GFP formed foci under sodium azide stress and surprisingly, the nuclear localization of Gbp2 was much more reduced in sodium azide treated cells as compared to heat stress (). To assess if the Gbp2 foci formed under sodium azide stress are P bodies or stress granules, we checked for the colocalization of Gbp2 foci with Lsm1 and Scd6. Lsm1 is a P body component and Scd6 localizes to both P bodies and stress granules [Citation11,Citation36]. Under sodium azide stress, the Gbp2 foci largely did not colocalize with Lsm1 and Scd6 (). The significance of distinct behaviour of Gbp2 under different stress conditions needs to be explored in the future. Overall, the above results hint that Gbp2 might have a role in mRNA translation/metabolism in the cytoplasm.
Figure 2. Gbp2 forms cytoplasmic foci upon stress. (A) Gbp2GFP or Gbp2GFPNID expressing cells were grown until OD600 0.5 and subjected to heat stress at 460C for 45 min, followed by live cell imaging. (B) Quantitation of the GFP intensity of the heat-treated cells imaged in the experiment A. At least 75 cells from each cell type of two independent experiments were used for calculating the raw integrated intensity using ImageJ. The values were plotted as box plots. (C) Gbp2GFP expressing cells were grown as above and treated with water or 0.5% sodium azide for 30 min and live cell imaging was performed. (D) Live cell imaging was performed with cells expressing Gbp2GFP and either Lsm1mCherry or Scd6mCherry under sodium azide stress. The cells were grown and treated as described in (C)
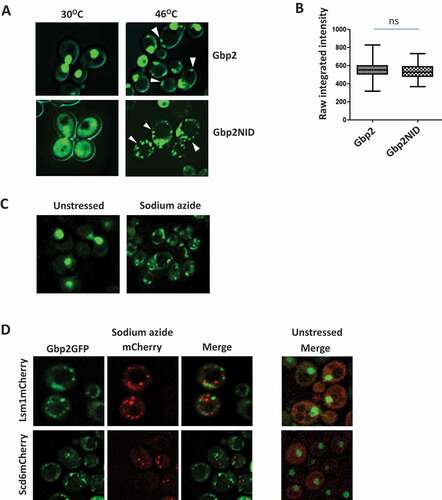
Gbp2 directly binds eIF4G1 via the RGG-motif
Translation repressors are known to target translation initiation factors to repress mRNA translation. We wanted to test if Gbp2 directly interacts with eIF4G1 and whether the RGG-motif is important for the binding. We incubated purified, recombinant Gbp2 and Gbp2ΔRGG with purified eIF4G1GST (Supplementary Fig. 1C) and performed a glutathione pull-down assay followed by western analysis to check their interaction. GST served as a negative control and did not show any interaction with Gbp2 and Gbp2ΔRGG. Wild-type Gbp2 was pulled down by eIF4G1GST indicating that Gbp2 directly binds eIF4G1. Strikingly, Gbp2ΔRGG mutant failed to interact with eIF4G1GST, thus highlighting the importance of the RGG-motif in eIF4G1-binding ().
Figure 3. Gbp2 directly binds eIF4G1. (A) Purified Gbp2 and Gbp2ΔRGG were incubated with either purified eIF4G1GST or GST. Glutathione pull-down was performed, followed by western analysis using anti-GST and anti-His antibodies. (B) Quantitation of three independent experiments performed as in (A). (C) Schematic depicting different fragments of eIF4G1 used in the pull-down assay shown in (D). (D) Purified Gbp2 was incubated with lysates containing overexpressed GST and different fragments of eIF4G1 as shown in (C), followed by glutathione pull-down and western blotting with anti-His antibody (*denotes the full-length band of each of the mutant fragments)
eIF4G1 is a modular protein and possesses multiple domains with distinct-binding regions for proteins like eIF4A, Pab1, and eIF4E (). Surprisingly, the binding regions of RGG-motif containing translation repressors like Scd6, Sbp1, and Npl3 on eIF4G1 are distinct from each other [Citation11]. Such a differential binding pattern might have distinct impacts on the mRNP function. We wanted to map the binding region of Gbp2 on eIF4G1 and hence tested the ability of wild-type Gbp2 to interact with different fragments of eIF4G1GST in vitro. Gbp2 did not bind the GST control and interacted with the full-length eIF4G1GST. We observed that Gbp2 bound eIF4G1 in the region containing the residues 490 to 656 (), overlapping with the Sbp1 binding region reported earlier [Citation11].
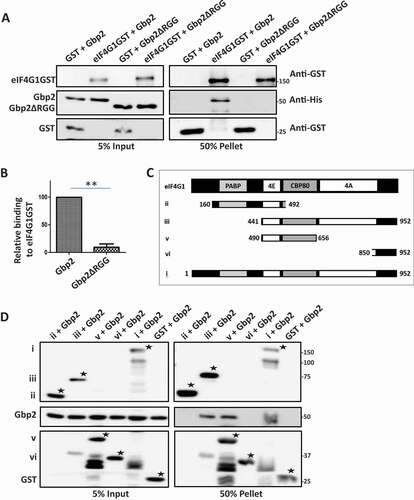
RGG motif of Gbp2 is important for localization to polysomes
Factors involved in mRNA translation repression and decay such as Scd6, Dhh1, Dcp2, and Xrn1 are reported to localize to polysome fractions [Citation37–39]. Gbp2 is reported to localize to polysome fractions in yeast and Trypanosomes [Citation12,Citation22], but the functional significance is not clear. We interpret this observation to indicate the possible role of Gbp2 in mRNA translation. eIF4G1 is known to be present in the polysome fractions [Citation40], and Gbp2 targets eIF4G1 and hence may localize to the polysome fractions to attenuate mRNA translation. As Gbp2 binds eIF4G1 in an RGG-motif dependent manner, we tested if RGG-motif of Gbp2 is required for the polysome localization. We performed sucrose gradient fractionation of cell lysates expressing wild-type Gbp2 or Gbp2ΔRGG and observed the pattern of Gbp2 protein distribution across the sucrose gradient fractions. Representative polysome profiles have been shown ( and Supplementary Fig. 1D). We isolated proteins from the sucrose gradient fractions, loaded them on SDS-PAGE (Supplementary Fig. 1A) and analysed them by western blotting. We observed that the wild-type Gbp2 localized to the polysome fractions as reported earlier, but Gbp2ΔRGG was enriched in the non-polysomal fractions (free mRNP and non-translating fractions) suggesting that the RGG motif of Gbp2 is crucial for polysome localization (, C).
Figure 4. RGG motif of Gbp2 is important for localization to polysomal fractions. (A) Gbp2His or Gbp2ΔRGGHis expressing cells were lysed and fractionated on a 10 to 50% sucrose gradient and 11 fractions of 1 ml each were collected using the BioComp polysome fractionator. The representative polysome profiles for Gbp2 (Black) and Gbp2ΔRGG (red) expressing cells are shown. (B) Proteins were isolated from the gradient fractions and were analysed by western blotting using anti-His antibody. (C) Quantitation of four independent experiments performed as in (B)
Gbp2 reduces GFP reporter mRNA translation in vivo without affecting mRNA levels
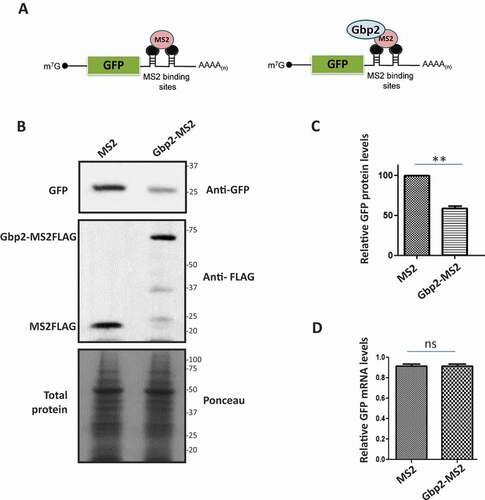
We next tested if Gbp2 indeed affects the translation of mRNA. We exploited the mRNA tethering assay used to demonstrate the direct effect of translation repressors on a reporter mRNA translation [Citation41,Citation42]. We used the GFP reporter mRNA containing MS2 binding sites in the 3ʹUTR (). A protein tagged to the MS2 coat protein will tether to the reporter mRNA with a high affinity through the MS2-binding site. We tagged Gbp2 with MS2 coat protein and co-expressed Gbp2-MS2FLAG with GFP reporter mRNA in wild-type cells. We analysed the protein levels of GFP reporter mRNA by western blotting using an anti-GFP antibody. We observed that the GFP protein levels were significantly reduced upon tethering of Gbp2-MS2FLAG compared to that of MS2FLAG alone that served as the control, indicating that Gbp2 reduces the GFP protein levels upon tethering (, C). It is reported that upon tethering of Scd6 to GFP reporter mRNA, the mRNA levels of GFP reporter are significantly reduced [Citation41]. To check if Gbp2 affects the mRNA levels of GFP reporter, we isolated RNA from the cells expressing Gbp2-MS2FLAG and MS2FLAG and performed qRT-PCR analysis. We observed that the GFP mRNA levels did not show a significant change upon tethering by Gbp2-MS2FLAG as compared to MS2FLAG alone (), suggesting that Gbp2 affects the translation of GFP reporter, but not mRNA levels.
Figure 5. Gbp2 decreases GFP protein levels upon tethering to GFP reporter mRNA. (A) Schematic showing the GFP reporter mRNA with MS2 binding sites in the 3ʹ UTR. Tethering of MS2 (left) or Gbp2-MS2 (right) is shown. (B) Cells co-expressing GFP reporter mRNA and MS2 or Gbp2-MS2 were lysed and analysed by western blotting with anti-GFP antibody. Anti-FLAG antibody was used to visualize MS2 and Gbp2-MS2 proteins. Ponceau staining served as the loading control. (C) Quantitation of three independent experiments performed as in (B). (D) GFP mRNA levels in the cells from three independent experiments as shown in B were determined by qRT-PCR analysis
Purified Gbp2 represses mRNA translation in vitro
We next tested if Gbp2 directly represses mRNA translation in vitro. We monitored the translation of luciferase reporter mRNA in the rabbit reticulocyte lysate (RRL) translation system in the presence of increasing concentrations of purified Gbp2 protein. The protein storage buffer served as the control. We observed a significant decrease in the luciferase activity upon the addition of increasing concentrations of Gbp2, suggesting that Gbp2 represses luciferase mRNA translation in vitro (). We confirmed that the luciferase mRNA levels in each reaction were comparable with the semi-quantitative RT-PCR analysis indicating that decreased luciferase levels were not due to decreased mRNA levels ().
Figure 6. Gbp2 represses luciferase mRNA translation in vitro. (A) Luciferase reporter mRNA was incubated in the rabbit reticulocyte lysate in the presence of increasing concentrations of Gbp2 (1 µM, 2 µM & 3 µM) and luciferase activity was measured after adding the substrate in a 96-well plate reader (Promega). The graph shows the quantitation of relative percentage of luciferase activity of three replicates. (B) Purified Gbp2 was loaded on SDS-PAGE and stained with CBB. (C) RNA was isolated from each of the reactions as shown in (A) and luciferase mRNA levels were analysed by semi-quantitative RT-PCR analysis. (D) Luciferase reporter mRNA was incubated in the rabbit reticulocyte lysate in the presence of Gbp2 or Gbp2ΔRGG and luciferase activity was measured after adding the substrate in a 96-well plate reader. The graph shows the quantitation of relative percentage of luciferase activity of three replicates. (E) Purified Gbp2 and Gbp2ΔRGG were loaded on SDS-PAGE and stained with CBB. (F) RNA was isolated from each of the reactions as shown in (D) and luciferase mRNA levels were analysed by semi-quantitative RT-PCR analysis
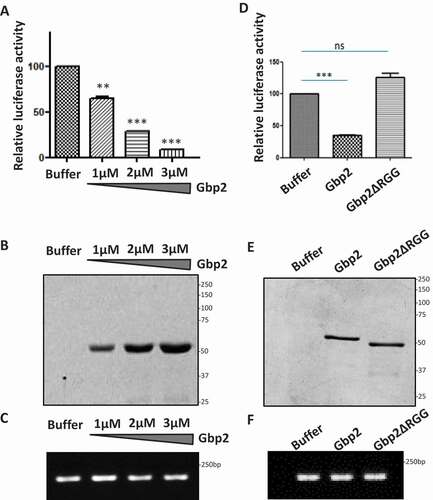
As our results highlight that RGG-motif of Gbp2 was important for polysome localization and eIF4G1 binding, we tested the effect of deletion of RGG motif on the Gbp2 mediated mRNA translation repression. We performed the in vitro translation assay in the presence of purified Gbp2 wild type and Gbp2ΔRGG mutant protein. We observed that the presence of Gbp2ΔRGG did not lead to a reduction in luciferase activity, suggesting that the RGG-motif of Gbp2 is important for its translation repression activity (). The luciferase mRNA levels were comparable in each of the reactions containing buffer, Gbp2, and Gbp2ΔRGG (), indicating that the lack of repression by the mutant was not due to altered mRNA levels. Overall, based on the above set of results, we identify a new role of Gbp2 as a translational repressor protein.
Discussion
Experimental results presented in this work establish that the RGG-motif containing mRNA export factor Gbp2 acts as a translation repressor of mRNAs without inducing degradation. Several results presented in this work point towards the role of Gbp2 as a translation repressor; i) Gbp2 accumulated in cytoplasmic granules upon heat and sodium azide stress (); ii) Gbp2 directly binds conserved translation initiation factor eIF4G1 (); iii) Tethering of Gbp2 to GFP reporter mRNA led to a significant reduction in GFP protein levels () and iv) Purified Gbp2 repressed mRNA translation in vitro (). Further, we demonstrate the importance of RGG-motif in the role of Gbp2 as a translation repressor. In addition to the rescue of growth defect phenotype, the deletion of RGG-motif led to a significant defect in eIF4G1 binding (), polysome association (), and translation repression activity of Gbp2 (), highlighting that RGG-motif of Gbp2 is important in the translation repression activity of Gbp2.
Gbp2 causes a growth defect upon overexpression like other translation repressors. The RGG-motifs of Scd6, Sbp1, and Npl3 are critical for the translation repression activity [Citation11,Citation43]. We have observed that upon deleting RGG-motif of Gbp2, the growth defect phenotype is rescued like in case of Scd6 and Sbp1, suggesting that RGG-motif of Gbp2 contributes significantly to its function (). Gbp2 overexpression mediated growth defect has been hypothesized to be due to the nuclear retention of mRNAs. But the RRM domain of Gbp2 is responsible for the interaction with the THO/TREX complex that couples it to the export [Citation19]. We observed that the deletion of RGG-motif alone rescues the growth defect phenotype in a manner comparable to the empty vector, suggesting that the growth defect could be due to the RGG-mediated function of Gbp2, like in the case of other RGG-motif translation repressors. Moreover, upon mutating the serine in the SR region of Gbp2 that renders it defective in nuclear import, we did not see significant rescue from the growth defect (data not shown), clearly suggesting that the growth defect phenotype is not entirely dependent on its nuclear function.
The formation of cytoplasmic foci under stress is a hallmark of proteins involved in translation repression/decay [Citation24,Citation44,Citation45]. The cellular redistribution of proteins is triggered due to a variety of stresses, and different sets of proteins respond in a distinct manner depending on the kind of stress and the cellular response that needs to be mounted. Under exponentially growing conditions, Gbp2 localization is predominantly nuclear. Heat stress and oxidative stress lead to extensive reprogramming of mRNA processing, export, and translation, with a concomitant formation of RNA granules in the cytoplasm [Citation32,Citation33,Citation46,Citation47]. We observed that, upon challenging the cells with heat stress or sodium azide stress, Gbp2 accumulated in cytoplasmic granules ( top panel & ). Further, the Nuclear Import Defective (NID) mutant of Gbp2 showed a robust increase in cytoplasmic foci formation upon heat stress ( bottom panel), indicating increased retention time in the cytoplasm leads to increased Gbp2 foci upon heat stress. This is consistent with the earlier observation that Gbp2 accumulates in foci under glucose deprivation stress indicating that cytoplasmic foci formation is a general feature of Gbp2. But it is noteworthy that the Gbp2 foci observed in sodium azide stress are distinct from that observed in heat and glucose deprivation stress (). Unlike in heat stress, the nuclear signal of Gbp2 is largely abrogated upon sodium azide stress suggesting a largely cytoplasmic role of Gbp2 in sodium azide stress. It is reported that Gbp2 foci in glucose deprivation condition colocalize with P bodies and stress granules [Citation24]. Interestingly, we observed that the Gbp2 foci under sodium azide stress largely did not colocalize with P body and stress granule components like Lsm1 and Scd6 (). The characterization of Gbp2 foci under sodium azide stress conditions and the identification of protein/mRNA components associated with Gbp2 foci will be an important future direction.
In this study, we add mRNA export factor Gbp2 to the repertoire of translation repressors that target the conserved translation initiation factor eIF4G1. We found that purified recombinant Gbp2 directly interacted with purified recombinant eIF4G1 (). Further, we elucidate that the RGG motif is important for eIF4G1 binding as we observed that Gbp2ΔRGG is highly defective in interacting with eIF4G1 (). For the first time, we have assigned a role for the RGG-motif of Gbp2. This observation is consistent with the observations that Scd6, Sbp1 and Npl3 interact with eIF4G1 via their RGG-motifs. This observation puts forth the interesting aspect that eIF4G1 acts as a nexus for the binding of many RGG-motif-containing proteins and could differentially regulate mRNAs in an mRNP dependent manner. RGG-motifs are generally disordered and are known to form higher order structures [Citation34,Citation48,Citation49]. Perhaps, the binding sockets on eIF4G1 favour such disordered regions. Interestingly, the RGG-motif of Gbp2 is predicted to be disordered (Supplementary Fig. 1B) and we have observed that purified Gbp2 exists as a multimer through gel filtration analysis (data not shown). Additionally, we mapped the binding region of Gbp2 on eIF4G1 and found that Gbp2 interacts with 490 to 656 residues of eIF4G1 (). Residues 492-539 of eIF4G1 are arginine and serine rich (RS-rich domain) and has been implicated in binding RNA. Binding of Gbp2 could therefore alter eIF4G1 interaction with RNA [Citation50]. Additionally, this binding region on eIF4G1 overlaps with the region reported to bind Sbp1 [Citation11]. Interestingly, genome-wide studies in yeast have shown that Gbp2 and Sbp1 interact [Citation51]. An exciting future task will be to determine if Gbp2 and Sbp1 can simultaneously bind to eIF4G1 or if they compete with each other to bind eIF4G1. It is possible that the binding of both the repressors to eIF4G1 is important to repress the translation of certain mRNAs effectively or replace one another on eIF4G1 depending on the context of the mRNA target and the physiological cues.
It was reported that Gbp2 localizes to polysome fractions in yeast and Trypanosomes, but the significance of this observation was not known. Several proteins whose cellular function pertains to translation repression or mRNA decay are reported to localize to polysome fractions, for example, Scd6, Npl3, and Dhh1 [Citation12,Citation37,Citation42]. But the individual domains responsible for such localization have not been demonstrated for any of those proteins. Our results indicate that Gbp2 RGG-motif is important for polysome localization because Gbp2ΔRGG was enriched in the non-polysomal fractions instead of polysome fractions (). To our knowledge, this is the first report elucidating that RGG-motif is involved in polyribosome association. The RGG-motif-mediated polysome association is likely important for the role of Gbp2 in translation regulation because Gbp2 targets eIF4G1, which is reported to be generally associated with the translating mRNPs [Citation40]. We hypothesize that Gbp2 localizes to the polysomes because it binds to eIF4G1 and inhibits translation via the RGG-motif.
An important future endeavour is to identify the endogenous mRNA translation targets of Gbp2 under normal conditions and under stress. Identifying mRNAs whose translation is affected by Gbp2 in the cytoplasm would pave the way to understand the impact of Gbp2 repression activity. mRNA export and translation repression functions of Gbp2 may be coupled, allowing repression of some mRNAs whose export is facilitated by Gbp2.
Finally, we ascertained the role of Gbp2 in translation repression by employing mRNA tethering and in vitro translation assays. We observed that Gbp2 can repress mRNA translation directly in rabbit reticulocyte lysates in a concentration-dependent manner (). But the RGG-motif deletion mutant was ineffective in repressing mRNA translation in vitro (). Further, a significant decrease in GFP protein levels was observed upon tethering of Gbp2 to GFP reporter mRNA (). Gbp2 did not affect the GFP reporter mRNA levels as indicated by the qRT-PCR results (), indicating that mRNA stability was unaffected. A very recent report implicated Gbp2 in repressing transcripts containing premature termination codons (PTCs) [Citation52]. The repression of these mRNAs by Gbp2 and Hrb1 aids in the degradation of these mRNAs by Upf1, indicating that Gbp2 primarily helps in repressing translation of defective mRNA en route to degradation. However, our work provides evidence that Gbp2 could play a role in directly repressing the translation of normal mRNAs without PTCs. Importantly, repression by Gbp2 does not lead to mRNA degradation (). We further provide a mechanism underlying repression activity, which is likely mediated by binding eIF4G1 through RGG-motif. Whether the RGG-motif is important for Gbp2-mediated translation repression on PTC-containing mRNAs will be an important future direction to understand mechanistic differences between the repression mechanisms on normal and PTC-containing mRNAs.
A future investigation of factors required for Gbp2 mediated translation repression is warranted. The SR-region of Gbp2 is subjected to phosphorylation by Sky1 and is important for the nuclear shuttling of Gbp2. The deletion of Sky1 led to a cytoplasmic accumulation of Gbp2 but did not affect its association with polysome fractions [Citation12]. Whether Sky1 mediated phosphorylation plays a role in mRNA binding/eIF4G1 interaction in the cytoplasm needs to be tested. RGG-motifs are the sites for protein arginine methylation, a post-translational modification that affects a wide range of cellular functions [Citation53]. Reports from our lab have demonstrated that arginine methylation augments the translation repression activity of Scd6 and Sbp1 [Citation27,Citation43]. Gbp2 is reported to be arginine methylated by Hmt1, and interestingly, Gbp2 shuttling is not affected by Hmt1, unlike Npl3 [Citation8]. Whether arginine methylation affects cytoplasmic foci formation, eIF4G1 binding, polysome association or translation repression activity of Gbp2 needs to be explored in the future. Arginine methylation could act as an interesting switch in tuning the function of the export factor Gbp2 into an mRNA translation repressor.
Overall, our current work establishes that the mRNA processing and export factor Gbp2 can act as a translation repressor of mRNAs. In response to physiological cues, Gbp2 perhaps represses translation of some mRNAs in the cytoplasm. These mRNAs could be the ones whose nuclear export is promoted by Gbp2. Our work suggests that Gbp2 has been co–opted to perform multiple functions in the nucleus and cytoplasm.
Materials and methods
Yeast cultures
BY4741 wild-type yeast strain and its derivatives were grown on standard Yeast extract Peptone media or Synthetic defined media (SD) with appropriate composition of amino acids, supplemented with 2% sucrose (SRL; 90701). Galactose (Sigma; G1750) was used in the medium for mRNA tethering experiments and growth assay. The list of the yeast strains used with their genotype and plasmids used in the study is given in Supplementary Tables 1 and 2, respectively.
Growth assay
Wild type BY4741 cells transformed with empty vector, GBP2GFP, GBP2GFPΔRGG, and GFP was serially diluted (starting from 10 OD600) and plated on agar plates containing SD media without uracil supplemented with either 2% glucose or 2% galactose. The plates were incubated at 30°C for 36 h and 48 h for glucose and galactose plates, respectively, and images were acquired.
Microscopy
Gbp2GFP and Gbp2GFPNID expressing cells were grown until OD600 0.5-0.55 in SD-Ura +2% sucrose at 30°C. For heat shock, half of the culture was incubated at 46°C for 45 min, and the rest half was incubated at 30°C, which served as the control. For sodium azide treatment, GBP2GFP cells were grown as above and treated with 0.5% sodium azide or water for 30 min. In , GBP2GFP strain transformed with plasmids containing Lsm1mCherry or Scd6mCherry, were grown in SD-Leu as above and treated with sodium azide. The cells were spotted on a 1 mm coverslip and immediately imaged using a GE Deltavision Elite microscope system with softWoRx 3.5.1 software (Applied Precision, LLC). Olympus 100x, oil-immersion 1.4 NA objective was used for imaging. For imaging, 1 s exposure with 100% GFP transmittance and 0.5 s exposure with 50% mCherry transmittance were used and images were collected as 512 × 512-pixel files with a CoolSnapHQ camera (Photometrics) with a 2 × 2 binning for yeast. SoftWoRx deconvolution algorithms were used to deconvolve all the images and all the images were set to equal contrast using ImageJ software. The GFP intensities in the images were quantified to assess protein levels of GFP tagged Gbp2 and Gbp2NID mutant. The images were converted to 8-bit format and background subtracted for Raw Integrated Intensity measurement was performed using ImageJ software. 75 cells from each of the cell types were analysed. The results were plotted in the form of boxplot with error bars indicating ± SEM.
Protein purification
HisGbp2 and HisGbp2ΔRGG were overexpressed in E. coli BL21 strain using 1 mM IPTG for 4 h at 37°C. The cells were pelleted and resuspended in cold Buffer A (50 mM NaH2PO4 pH8, 300 mM NaCl, 1 mM DTT, and 1 mg/ml Lysozyme) along with 10 mM Imidazole. This was followed by sonication, and the lysate was clarified by centrifugation (15000 rpm for 15 min). The supernatant was incubated with NiNTA beads (Thermo Fisher Scientific; 88222) in an end-to-end nutator at 4°C for an hour. The beads were washed thrice with Buffer A containing 20 to 50 mM Imidazole for 10 min each, and the proteins were eluted with Buffer A containing 500 mM Imidazole. During the lysis and binding steps, the extracts were treated with RNase A (10 µg/ml) to eliminate cellular RNA that might provide bridging interactions. All the proteins were concentrated and dialysed in dialysis bags (Sigma; Z371092). For pull-down reactions, the proteins were dialysed into Buffer B (10 mM Tris-Cl pH7, 100 mM NaCl, 10% glycerol, and 1 mM DTT) and that for in vitro translation assay in Buffer C (50 mM potassium acetate and 10 mM HEPES pH7).
GST and eIF4G1GST were similarly expressed in BL21 cells, lysed in Buffer D (20 mM Tris-HCl pH8, 300 mM NaCl, and 2 mM DTT), sonicated, and clarified as above. Lysates were incubated with 1 ml Glutathione sepharose (GE Healthcare; 17,075,605) for one hour and washed thrice with Buffer E (20 mM Tris-HCl pH8, 500 mM NaCl, and 2 mM DTT) and eluted with Buffer F (20 mM Tris-HCl pH8, 150 mM NaCl, 2 mM DTT, and 20 mM reduced glutathione pH8 [Amresco; 0399]). The proteins were dialysed into Buffer B.
Pull-down experiments
Glutathione Sepharose (30ul) was mixed with 200 pmol of each protein and diluted to 250ul in Buffer G (50 mM HEPES pH7, 100 mM NaCl, 1 mM DTT, 2 mM MnCl2, 2 mM MgCl2, 1% Triton-X 100, 10% glycerol, 10 mg/ml BSA and 0.25 mg RNase A). The tubes were nutated for 2 h at 4°C, and the beads were washed with Buffer G (without BSA and RNase A) three times (10 min each on the nutator). The beads were boiled after adding 10 μl of SDS-PAGE loading dye and analysed by SDS-PAGE followed by western blotting. For , the lysates with overexpressed eIF4G1GST fragments were incubated with HisGbp2, and Glutathione pull-down was performed as mentioned above.
Western blotting
Western analysis was performed after SDS-PAGE using standard protocols. The antibodies used were anti-GST (CST; 2624), anti-His (CST; 12698), anti-GFP (CST; 2555), and anti-FLAG (Sigma; F3165).
Polysome profiling
Wild type RPS6GFP cells expressing Gbp2 or Gbp2ΔRGG on plasmids were grown in 200 ml SD–Ura 2% sucrose until 0.55-0.6 OD600 and treated with cycloheximide (100 µg/ml) for 15 min at 30°C in a shaking incubator. Upon completion of cycloheximide treatment, the cells were immediately cooled on ice and pelleted at 4°C. The cells were washed with DEPC (SRL; 46791) treated water and 400ul Buffer H (10 mM Tris pH7.4, 100 mM NaCl, 30 mM MgCl2, 100ug/ml cycloheximide, 10 U RiboLock, 1x Complete mini-EDTA-free tablet [Roche; 04693132001] and 1 mM PMSF [SRL; 84375]) was added to the cell pellet, resuspended and broken open by adding glass beads followed by bead-beating at 4°C for 10 min. Lysates were spun at 5500rpm for 5 min and then at 15000 rpm for 2 min. The supernatant was taken and lysate corresponding to around 800 OD254 in total was loaded onto a 10 to 50% sucrose gradient. The sucrose gradient was prepared in Buffer I (140 mM KCl, 5 mM MgCl2, 5 mM Tris-HCl pH7.5, and 100ug/ml cycloheximide). The gradients were centrifuged at 41000 rpm at 4°C for 2 h in Beckman ultracentrifuge using the SW41Ti rotor. The gradient fractions (1 ml each) were collected with a BioComp Gradient profiler. Post fractionation, TCA was added to each fraction (to a final concentration of 10%), mixed, and stored at −20°C overnight for precipitation. Tubes were then spun at 15000 rpm for 15 min at 4°C. 1 ml of 100% cold acetone was used to wash the pellet twice. The tubes were dried in the fume hood for 30 min. The protein pellet was finally resuspended using 100ul of 1.5X of SDS-PAGE buffer. The tubes were vortexed for 1 min and the protein samples were boiled at 100°C for 5 min. The distribution of Gbp2 and Gbp2ΔRGG in the polysome fractions was analysed by western blotting.
mRNA tethering assay
The GFP reporter mRNA translation was assessed to check the in vivo translation repression activity of Gbp2. The GFP reporter mRNA has an MS2-binding site in the 3ʹ UTR (a kind gift from Dr. Alan Hinnebusch, NIH, USA). The Scd6 promoter and the ORF from the Scd6 Promoter-ORF-MS2-FLAG-3ʹUTR containing plasmid were replaced with Gbp2 promoter-ORF using the restriction enzymes Sph1 (Thermo Fisher Scientific; ER0601) and Xho1 (Thermo Fisher Scientific; ER0691). Gbp2-MS2FLAG or only MS2FLAG expressing plasmids were co-transformed into wild-type cells along with the plasmid containing GFP reporter (under the control of GAL10 UAS). The plasmid with MS2FLAG only served as the control. The transformants were grown in SD-Ura-Leu media with 1.9% sucrose and 0.1% galactose overnight at 30°C and harvested at around OD600 8. Cells were resuspended in 80ul Buffer J (50 mM Tris-Cl pH7.5, 50 mM NaCl, 2 mM MgCl2, 0.1% Triton-X100, 1 mM PMSF), and 20ul SDS-PAGE buffer was added, boiled, and analysed by western blotting with anti-GFP to visualize the change in GFP protein levels. MS2FLAG and Gbp2-MS2FLAG were visualized with anti-FLAG antibody.
RNA isolation and qRT-PCR
Total RNA was isolated using hot acidic phenol protocol and qRT-PCR analysis was performed as described earlier [Citation35]. For isolating RNA from the polysome fractions, the RNA was precipitated overnight in −20°C using 20μg glycogen (Thermo, R0551), 1/10th volume of sodium acetate and 2.5 volumes of ethanol. The pellet was washed with 70% ethanol, dried and resuspended in nuclease free water. The 18s and 25s rRNA were analysed using 1% formamide agarose gel electrophoresis. For qRT-PCR, briefly, 8µg of total RNA was treated with 4U of DNase1 (Thermo, 166 EN0525). Post DNase1 treatment, cDNA synthesis was performed using 1µg RNA with the RevertAid RT Reverse Transcription Kit (Thermo, K1691) as per the manufacturer’s protocol. Real-time PCR was carried out using TB Green™ Premix Ex Taq™ (TaKaRa) using 1:10 diluted cDNA. For qRT-PCR, triplicates were set up with 0.5µM primer and 2µl cDNA/reaction in BioRad CFX Connect Real-Time PCR Detection System. The PCR conditions were as follows: 950C 10 min-initial denaturation and 29 cycles of 95°C for 15 s, 48°C for 15 s and 72°C for 30 s. DNA was quantified in every cycle at the extension step. Melt curve acquisition was done at 64°C for 8 s. Ct values were extracted with an auto baseline and manual threshold. The Ct values obtained were normalized against the housekeeping gene PGK1 and plotted. The details of the primers used are given in supplementary Table 3.
In vitro translation
In vitro translation reactions were performed with the Rabbit Reticulocyte kit from Promega (Cat. L4960) according to the manufacturer’s protocol. Briefly, 100µg of luciferase reporter mRNA was incubated with the RRL mix and 20U of Ribolock (Thermo, EO0384) in the presence of purified Gbp2 and Gbp2ΔRGG. For the experiment in , 1µM, 2µM, and 3µM of purified Gbp2 were taken, and in , 1µM each of Gbp2 and Gbp2ΔRGG was taken. The dialysed elution buffer served as the control. The reaction mixture was incubated at 30°C for 90 min. 5µl of reaction mixtures were taken in triplicates and added with 5µl Luciferase assay reagent and luminescence was immediately measured using a Promega plate reader (GM3500). RNA was extracted from the reaction mixtures using the Zymogen Kit, and semi-quantitative RT-PCR analysis was performed to check the luciferase mRNA levels in each of the reactions.
Statistical analysis
For the quantified results in , the results were plotted as bar graphs showing the ‘Mean with SEM’. To find out the significance of the data, statistical analysis was performed for the plotted results using paired t-test.
Supplemental Material
Download Zip (218.3 KB)Acknowledgments
We thank Dr. Heike Krebber for the kind gift of Gbp2 plasmids. We also thank Dr. Alan Hinnebusch and Dr. Roy Parker for sharing the plasmids. We thank Dr. Sandeep M Eswarappa for the Promega plate reader, Dr. Dipankar Nandi, Dr. Sanmoy Pathak and Raju Roy for the help with BioRad CFX Connect machine.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;54:547–558.
- Rissland OS. The organization and regulation of mRNA-protein complexes. Wiley Interdisciplinary Reviews. RNA. 2016;8. DOI:https://doi.org/10.1002/wrna.1369
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205.
- Jeong S. SR proteins: binders, regulators, and connectors of RNA. Mol Cells. 2017;40:1–9.
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27.
- Windgassen M, Krebber H. Identification of Gbp2 as a novel poly(A)+ RNA-binding protein involved in the cytoplasmic delivery of messenger RNAs in yeast. EMBO Rep. 2003;4:278–283.
- Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242.
- Hacker S, Krebber H. Differential export requirements for shuttling serine/arginine-type mRNA-binding proteins. J Biol Chem. 2004;279:5049–5052.
- Hackmann A, Wu H, Schneider UM, et al. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun. 2014;5:3123.
- Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–734.
- Rajyaguru P, She M, Parker R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell. 2012;45:244–254.
- Windgassen M, Sturm D, Cajigas IJ, et al. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–10491.
- Volpon L, Osborne MJ, Borden KLB. Biochemical and structural insights into the eukaryotic translation initiation factor eIF4E. Curr Protein Pept Sci. 2019;20:525–535.
- Zahreddine HA, Culjkovic-Kraljacic B, Emond A, et al. The eukaryotic translation initiation factor eIF4E harnesses hyaluronan production to drive its malignant activity. Elife. 2017;6. DOI:https://doi.org/10.7554/eLife.29830
- Haimovich G, Medina DA, Causse SZ, et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011.
- Brune C, Munchel SE, Fischer N, et al. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. Rna. 2005;11:517–531.
- Das S, Sarkar D, Das B. The interplay between transcription and mRNA degradation in Saccharomyces cerevisiae. Microb Cell. 2017;4:212–228.
- Lin JJ, Zakian VA. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994;22:4906–4913.
- Martínez-Lumbreras S, Taverniti V, Zorrilla S, et al. Gbp2 interacts with THO/TREX through a novel type of RRM domain. Nucleic Acids Res. 2016;44:437–448.
- Hurt E, Luo MJ, Rother S, et al. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A. 2004;101:1858–1862.
- Niikura M, Fukutomi T, Fukui K, et al. G-strand binding protein 2 is involved in asexual and sexual development of Plasmodium berghei. Parasitol Int. 2020;76:102059.
- Wippel HH, Malgarin JS, Inoue AH, et al. Unveiling the partners of the DRBD2-mRNP complex, an RBP in Trypanosoma cruzi and ortholog to the yeast SR-protein Gbp2. BMC Microbiol. 2019;19:128.
- Dastidar RG, Hooda J, Shah A, et al. The nuclear localization of SWI/SNF proteins is subjected to oxygen regulation. Cell Biosci. 2012;2:30.
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455.
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886.
- Nissan T, Rajyaguru P, She M, et al. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783.
- Poornima G, Shah S, Vignesh V, et al. Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 2016;44:9358–9368.
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848.
- Liu B, Qian S-B. Translational reprogramming in cellular stress response. Wiley Interdiscip Rev RNA. 2014;5:301–315.
- Advani VM, Ivanov P. Translational control under stress: reshaping the translatome. BioEssays. 2019;41:e1900009–e1900009.
- Yamamoto Y, Izawa S. Adaptive response in stress granule formation and bulk translational repression upon a combined stress of mild heat shock and mild ethanol stress in yeast. Genes Cells. 2013;18:974–984.
- Buchan JR, Yoon J-H, Parker R. Stress-specific composition, assembly and kinetics of stress granules in <em>Saccharomyces cerevisiae</em>. J Cell Sci. 2011;124:228.
- Wallace EW, Kear-Scott JL, Pilipenko EV, et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 2015;162:1286–1298.
- Poornima G, Mythili R, Nag P, et al. RGG-motif self-association regulates eIF4G-binding translation repressor protein Scd6. RNA Biol. 2019;16:1215–1227.
- Garg M, Poornima G, Rajyaguru PI. Elucidation of the RNA-granule inducing sodium azide stress response through transcriptome analysis. Genomics. 2020;112:2978–2989.
- Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287.
- Weidner J, Wang C, Prescianotto-Baschong C, et al. The polysome-associated proteins Scp160 and Bfr1 prevent P body formation under normal growth conditions. J Cell Sci. 2014;127:1992–2004.
- Wang Z, Jiao X, Carr-Schmid A, et al. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A. 2002;99:12663–12668.
- Hu W, Sweet TJ, Chamnongpol S, et al. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229.
- Wang X, Xi W, Toomey S, et al. Stoichiometry and change of the mRNA closed-loop factors as translating ribosomes transit from initiation to elongation. Plos One. 2016;11:e0150616.
- Zeidan Q, He F, Zhang F, et al. Conserved mRNA-granule component Scd6 targets Dhh1 to repress translation initiation and activates Dcp2-mediated mRNA decay in vivo. PLoS Genet. 2018;14:e1007806.
- Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342.
- Bhatter N, Roy R, Shah S, et al. Arginine methylation augments Sbp1 function in translation repression and decapping. Febs J. 2019;286:4693–4708.
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646.
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489.
- Amesz WJC, Van Der Zeijst BAM. Azide as inhibitor of protein synthesis in yeast protoplasts. FEBS Lett. 1972;26:165–168.
- Zander G, Krebber H. Quick or quality? How mRNA escapes nuclear quality control during stress. RNA Biol. 2017;14:1642–1648.
- Ozdilek BA, Thompson VF, Ahmed NS, et al. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 2017;45:7984–7996.
- Järvelin AI, Noerenberg M, Davis I, et al. The new (dis)order in RNA regulation. Cell Commun Signal. 2016;14:9.
- Berset C, Zurbriggen A, Djafarzadeh S, et al. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. Rna. 2003;9:871–880.
- Gavin AC, Aloy P, Grandi P, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636.
- Grosse S, Lu YY, Coban I, et al. Nuclear SR-protein mediated mRNA quality control is continued in cytoplasmic nonsense-mediated decay. RNA Biol. 2021; Jan 7:1–18.
- Thandapani P, O’Connor TR, Bailey TL, et al. Defining the RGG/RG motif. Mol Cell. 2013;50:613–623.
