ABSTRACT
Stem cells are a class of undifferentiated cells with great self-renewal and differentiation capabilities that can differentiate into mature cells in specific tissue types. Stem cell differentiation plays critical roles in body homoeostasis, injury repair and tissue generation. The important functions of stem cell differentiation have resulted in numerous studies focusing on the complex molecular mechanisms and various signalling pathways controlling stem cell differentiation. Circular RNAs (circRNAs) are a novel class of noncoding RNAs with a covalently closed structure present in eukaryotes. Numerous studies have highlighted important biological functions of circRNAs, and they play multiple regulatory roles in various physiological and pathological processes. Importantly, multiple lines of evidence have shown the abnormal expression of numerous circRNAs during stem cell differentiation, and some play a role in regulating stem cell differentiation, highlighting the role of circRNAs as novel biomarkers of stem cell differentiation and novel targets for stem cell-based therapy. In this review, we systematically summarize and discuss recent advances in our understanding of the roles and underlying mechanisms of circRNAs in modulating stem cell differentiation, thus providing guidance for future studies to investigate stem cell differentiation and stem cell-based therapy.
Abbreviations: CircRNAs: circular RNAs; ESCs: embryonic stem cells; ADSCs: adipose-derived mesenchymal stem cells; ecircRNAs: exonic circRNAs; EIciRNAs: exon-intron circRNAs; eiRNAs: circular intronic RNAs; tricRNAs: tRNA intronic circRNAs; pol II: polymerase II; snRNP: small nuclear ribonucleoprotein; m6A: N6-methyladenosine; AGO2: Argonaute 2; RBPs: RNA-binding proteins; MBNL: muscleblind-like protein 1; MSCs: mesenchymal stem cells; hiPSCs: human induced pluripotent stem cells; hiPSC-CMs: hiPSC-derived cardiomyocytes; hBMSCs: human bone marrow mesenchymal stem cells; hADSCs: human adipose-derived mesenchymal stem cells; hDPSCs: human dental pulp stem cells; RNA-seq: high-throughput RNA sequencing; HSCs: haematopoietic stem cells; NSCs: neural stem cells; EpSCs: epidermal stem cells; hESCs: human embryonic stem cells; mESCs: murine embryonic stem cells; MNs: motor neurons; SSUP: small subunit processome; BMSCs: bone marrow-derived mesenchymal stem cells; OGN: osteoglycin; GIOP: glucocorticoid‑induced osteoporosis; CDR1as: cerebellar degeneration-related protein 1 transcript; SONFH: steroid-induced osteogenesis of the femoral head; rBMSCs: rat bone marrow-derived mesenchymal stem cells; QUE: quercetin; AcvR1b: activin A receptor type 1B; BSP: bone sialoprotein; mADSCs: mouse ADSCs; PTBP1: polypyrimidine tract-binding protein; ER: endoplasmic reticulum; hUCMSCs: MSCs derived from human umbilical cord; MSMSCs: maxillary sinus membrane stem cells; SCAPs: stem cells from the apical papilla; MyoD: myogenic differentiation protein 1; MSTN: myostatin; MEF2C: myocyte enhancer factor 2C; BCLAF1: BCL2-associated transcription factor 1; EpSCs: epidermal stem cells; ISCs: intestinal stem cells; NSCs: neural stem cells; Lgr5+ ISCs: crypt base columnar cells; ILCs: innate lymphoid cells.
1. Introduction
Stem cells are a group of cells with the capability to self-renew and differentiate that can evolve into specialized cellular populations through differentiation. Stem cell differentiation enables stem cells to change from a non-specific state to a morphologically and functionally specific state, thus forming different tissues and organs in the body [Citation1]. Stem cells are mainly classified into two groups: embryonic stem cells (ESCs) and somatic stem cells. Stem cells participate in various physiological and pathological processes, such as tissue development, wound healing and tumorigenesis [Citation1,Citation2]. Normal stem cell differentiation plays a pivotal function in homoeostasis in the body, and abnormal stem cell self-renewal and differentiation can lead to human malignancy. Currently, stem cells have been widely applied in regeneration medicine to treat conditions such as bone fracture, neurological disorders and haematopoietic defects. For example, several clinical trials have identified the clinical feasibility of adipose-derived mesenchymal stem cell (ADSC)-based therapy for bone regeneration or reconstruction [Citation3,Citation4]. To date, the regulatory mechanisms controlling stem cell differentiation have been extensively investigated by various studies and include various cellular components, signalling pathways and epigenetic modifications [Citation5,Citation6]. However, the detailed mechanisms and regulatory networks of stem cell differentiation have not been conclusively determined.
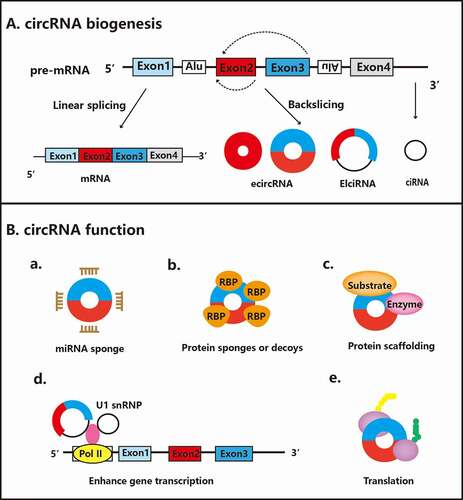
Circular RNAs (circRNAs) are a novel class of noncoding RNAs with a covalently closed-loop structure and tissue-specific and cell-specific expression patterns in eukaryotes [Citation7]. In 1976, circRNAs were first discovered in RNA viruses using electron microscopy [Citation8]. In contrast to the biogenesis of mRNAs, circRNAs are formed by a back-splicing process between the 5ʹ splice donor site and 3ʹ splice acceptor site. Most circRNAs are derived from protein-coding genes and contain one or more exons. CircRNAs are mainly classified into exonic circRNAs (ecircRNAs), exon-intron circRNAs (EIciRNAs), circular intronic RNAs (ciRNAs) and tRNA intronic circRNAs (tricRNAs) based on different splicing processes [Citation9,Citation10]. The biogenesis of circRNAs has been proposed to compete with the linear pre-mRNA splicing process and is regulated by multiple cis-elements and trans-factors [Citation11]. However, several studies report controversial points that both linear and circular forms of the same host gene have the same expression pattern and are regulated in similar manners in several cellular contexts. For example, both linear and circular CDKN2B-AS1 are downregulated in inflammatory bowel disease and play a role in regulating intestinal barrier formation together [Citation12]. In addition, some EIciRNAs and ciRNAs directly target RNA polymerase II (pol II) or the essential splicing factor U1 small nuclear ribonucleoprotein (snRNP), thereby promoting the linear pre-mRNA splicing and transcription of their host genes [Citation9,Citation13]. Following biogenesis, most circRNAs, except intron-containing circRNAs, are exported to the cytoplasm by ATP-dependent RNA helicase DDX39A and spliceosome RNA helicase DDX39B based on their different lengths [Citation14]. The mechanisms of circRNA turnover are still inconclusive, and only a few recent studies have proposed some hypotheses. For example, circRNAs containing the N6-methyladenosine (m6A) RNA modification may be degraded in an endonuclease-dependent manner [Citation15]. Additionally, argonaute protein-dependent cleavage may be another method of circRNA degradation. Notably, miR-671 binds to a highly complementary miRNA binding site in CDR1as, thus inducing its cleavage via Argonaute 2 (AGO2) [Citation16,Citation17].
Emerging evidence suggests that circRNAs have multiple biological functions (). The most extensively investigated biological function of circRNAs is to serve as sponges of miRNAs and thus attenuate their effects on downstream targets [Citation18]. CDR1as, which is perhaps the most representative circRNA, contains more than 70 target sites for miR-7 and has been found to sponge miR-7 in several cellular contexts [Citation19,Citation20]. According to a previous report, circITCH functions as a sponge for several different miRNAs, including miR-7, miR-17 and miR-214, thus increasing the expression of downstream targets [Citation21]. In addition, several circRNAs, such as circMbl, directly interact with different RNA-binding proteins (RBPs) to regulate RBP-dependent functions. For example, circMbl has many binding sites for muscleblind-like protein 1 (MBNL1), and MBNL1 affects the biogenesis of circMbl [Citation22]. Meanwhile, a few circRNAs, such as circAmotl1 and circFoxo3, function as protein scaffolds to promote substrate–enzyme colocalization, subsequently mediating various cellular processes [Citation23,Citation24]. Moreover, very recent studies have shown that some circRNAs, including circMbl, circFBXW7, circZNF609 and circSHPRH, are translated into proteins. However, the detailed mechanisms underlying the translation of these circRNAs are still unknown. Recently, m6A was shown to potentially promote this biological function of circRNAs [Citation25–27]. Because circRNAs have multiple biological functions, they likely play crucial roles in both normal physiological processes and diverse human diseases, such as human malignancy, neurological disorders and cardiovascular diseases [Citation28,Citation29]. Furthermore, circRNAs are characterized as extremely stable molecules, and thus multiple circRNAs may serve as biomarkers of both physiological and pathological processes. For example, the promising roles of circ_0000711, circPVT1 and circHIPK3 as prognostic and clinical biomarkers for various human cancers has been reported in several studies [Citation30-32].
Figure 1. Biogenesis and functions of circRNAs. A. The biogenesis of circRNAs: CircRNAs are formed by a noncanonical back-splicing process that competes with linear premRNA splicing. B. Biological functions of circRNAs: a. CircRNAs can function as miRNA sponges to regulate the expression of relevant target genes. b. CircRNAs containing RNA binding protein (RBP) binding motifs can interact with proteins. c. CircRNAs can function as protein scaffolds to facilitate the colocalization of enzymes and their substrates. d. CircRNAs can enhance gene transcription by interacting with RNA pol II or U1 small nuclear snRNP. e. CircRNAs can be translated into proteins
With the rapid development of bioinformatics analysis tools and high–throughput RNA sequencing in recent decades, thousands of circRNAs have been identified in different tissues and cell types. Notably, an increasing number of studies have reported circRNA profiles during the differentiation of diverse stem cell types, such as ESCs, mesenchymal stem cells (MSCs) and myoblasts () [Citation33-35]. For instance, Lei et al. identified 5602 circRNAs in human induced pluripotent stem cells (hiPSCs) and hiPSC-derived cardiomyocytes (hiPSC-CMs), with 1612 expressed in hiPSCs, 4260 expressed in hiPSC-CMs and 270 expressed in both, using high-throughput sequencing. Further investigations indicated that circSLC8A1, circCACNA1D and circSPHKAP may serve as novel biomarkers of the cardiogenesis of hiPSCs [Citation36]. With regard to bone marrow mesenchymal stem cells (BMSCs), upregulation of 21 circRNAs and downregulation of 21 circRNAs during osteogenic differentiation and upregulation of 130 circRNAs and downregulation of 97 circRNAs during chondrogenic differentiation have been identified in human bone marrow mesenchymal stem cells (hBMSCs). Furthermore, upregulation of circular FKBP5 is observed during both the osteogenesis and chondrogenesis of hBMSCs, and dexamethasone alters the expression of circular FKBP5 during differentiation [Citation37]. The microarray profile of human dental pulp stem cells (hDPSCs) revealed that 43 circRNAs are upregulated and 144 circRNAs are downregulated during the odontogenic differentiation process. The qRT-PCR analysis results confirmed that hsa_circRNA_104101, hsa_circRNA_406763, hsa_circRNA_002161, and hsa_circRNA_005044 are upregulated, while hsa_circRNA_079813 and hsa_circRNA_008336 are downregulated during odontogenic differentiation of hDPSCs [Citation38]. Five hundred eighty-one circRNAs were evidently differentially expressed in C2C12 myoblast cells, with 346 upregulated and 235 downregulated compared with myotubes, using a mouse circRNA microarray. Among these circRNAs, circRNA_34451 regulates the myogenesis-specific genes Mef2a, Myh7b, and Myog, and circRNA_19008 regulates Mef2a, Myh1, and Myog, indicating the important roles of these circRNAs in the differentiation of C2C12 myoblasts [Citation39]. In addition to the studies described above, many other studies focusing on dysregulated circRNAs during the differentiation of multiple stem cell types, such as haematopoietic stem cells (HSCs), neural stem cells (NSCs) and epidermal stem cells (EpSCs), are also listed in . In summary, these research results strongly indicate that a large number of circRNAs are dysregulated during the differentiation of diverse stem cell types. In particular, several circRNAs were identified to play a potential role in stem cell differentiation through complex mechanisms. Several circRNAs, such as CDR1as, circFOXP1 and circZNF91, have been reported to regulate stem cell differentiation in different lineages through circRNA-miRNA-mRNA interaction networks, protein sponges and various signalling pathways, such as the PI3K-Akt, MAPK and Wnt signalling pathways [Citation40,Citation41].
Table 1. CircRNAs profiles in stem cell differentiation
In this review, we systematically summarize the roles of circRNAs in regulating the differentiation of diverse stem cell types and the underlying mechanisms, thus highlighting the potential role of circRNAs as biomarkers of stem cell differentiation and circRNA-based stem cell therapy for regenerative medicine.
2. CircRNAs in stem cell differentiation
2.1 CircRNAs in embryonic stem cell (ESC) differentiation
Embryonic stem cells (ESCs), which are derived from the inner cell mass (ICM), possess an outstanding capability of self-renewal and pluripotency and can differentiate into diverse cell lineages [Citation50]. Numerous studies have investigated the regulation of the pluripotency and differentiation of ESCs, including the involvement of circRNAs. Notably, a number of circRNAs are enriched in undifferentiated human embryonic stem cells (hESCs), and the expression of these circRNAs may be correlated with the pluripotency and differentiation of hESCs. Further investigation indicates that circBIRC6 and circCORO1C are involved in regulating the pluripotency and differentiation of hESCs. Downregulation of circBIRC6 or circCORO1C has been shown to impair pluripotency and promote the differentiation of hESCs. Mechanistically, miR-34a and miR-145 are the direct targets of circBIRC6 involved in regulating hESC pluripotency. As shown in previous studies, miR-34a and miR-145 promote hESC differentiation by inhibiting the expression of several pluripotency-associated genes, such as OCT4, SOX2, and KLF4. Therefore, circBIRC6 maintains pluripotency and inhibits the differentiation of hESCs induced by miR-34a and miR-145. Moreover, NANOG and OCT4 have been shown to increase the expression of the splicing factor ESRP1, which induces the generation of circBIRC6 in hESCs, thus promoting hESC pluripotency [Citation51]. In murine embryonic stem cells (mESCs), the RNA-binding protein FUS controls the motor neuron (MN) differentiation of mESCs by regulating the expression of multiple circRNAs. Fourteen downregulated and 5 upregulated circRNAs were detected during the MN differentiation of mESCs. More investigations are needed to identify the role and molecular mechanisms of FUS and specific circRNAs in mESC MN differentiation [Citation52]. In addition, several RBPs have been identified to play a role in maintaining mESC pluripotency, and some RBPs, such as Krr1 and Ddx47, are components of the small subunit processome (SSUP) that modulates the biogenesis of 18S rRNA. SSUP components are expressed in stem cells and regulate stem cell pluripotency. Because circRNAs function as sponges or decoys of RBPs and modulate RBP-mediated functions, circRNAs may modulate mESC differentiation by regulating SSUP components [Citation53].
2.2 CircRNAs in mesenchymal stem cell (MSC) differentiation
Mesenchymal stem cells (MSCs), a type of pluripotent stem cell, reside in various adult tissues, such as bone marrow, adipose tissue and dental pulp. MSCs have the ability to differentiate into specific cell types, including osteoblasts, adipocytes and chondrocytes [Citation54]. Genetic and epigenetic regulation of the differentiation of MSCs has been investigated in numerous studies. Very recently, a number of circRNAs have been shown to be dysregulated in MSCs and may participate in MSC differentiation ().
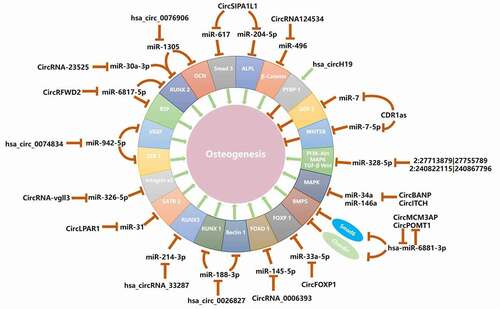
Figure 2. Roles and regulatory mechanisms of circRNAs in MSCs osteogenesis. Several circRNAs can promote or inhibit MSCs osteogenesis through sponging miRNAs and regulating expression of potential target genes and related signalling pathway
2.2.1 CircRNAs in bone marrow-derived mesenchymal stem cell (BMSC) differentiation
Bone marrow-derived mesenchymal stem cells (BMSCs) are a population of MSCs that reside in the bone marrow and differentiate into a limited number of cell types, such as osteoblasts. Due to their advantages in accessibility and the potential for differentiation and expansion, BMSCs have been widely used in the treatment of human disorders, such as bone defects [Citation55]. For example, after bone fracture, BMSCs migrate to the fracture site, secrete numerous extracellular matrix components and differentiate into osteoblasts, thus promoting bone fracture healing [Citation56]. The role of circRNAs in modulating BMSC differentiation has been explored in several studies (). For instance, circDAB1, which is derived from exon 8 of the DAB1 gene and is expressed at high levels during BMSC osteogenesis, promotes BMSC proliferation and osteogenesis. By upregulating RBPJ through sponging miR-1270 and miR-944, circDAB1 facilitates the osteogenic differentiation of BMSCs. Moreover, circDAB1 upregulates DAB1 expression by inducing RBPJ expression in BMSCs, indicating the regulatory role of the circDAB1/miR-1270/miR-944/RBPJ/DAB1 axis in BMSC proliferation and osteogenesis [Citation57]. A crucial role for hsa_irc_0074834, which was evidently downregulated in BMSCs from bone non-union samples, has been identified in BMSC differentiation. Notably, hsa_circ_0074834 serves as a sponge RNA for miR-942-5p and promotes BMSC osteogenic differentiation by attenuating the inhibitory effects of miR-942-5p on ZEB1 and VEGF. Furthermore, hsa_circ_0074834 has also been shown to enhance osteogenic differentiation of BMSCs and bone regeneration in vivo, indicating the potential role of hsa_circ_0074834 in the treatment of bone non-union [Citation58]. The osteogenic potential of hsa_circ_0076906 in osteoporosis has been reported recently. The expression of hsa_circ_0076906 is decreased in both bone tissue and serum in the osteoporosis group compared with the normal group, and it is upregulated during hBMSC osteogenesis. Additionally, knockdown of hsa_circ_0076906 decreases the expression of the osteogenesis-related genes OCN and RUNX2 to inhibit the osteogenic differentiation of hBMSCs. Furthermore, hsa_circ_0076906 promotes osteogenesis by sponging miR-1305 to induce the expression of the osteoglycin (OGN) gene, which enhances bone formation [Citation59]. Patients with glucocorticoid-induced osteoporosis (GIOP) exhibit the downregulation of circRNA_0006393. Mechanistically, circRNA_0006393 promotes osteogenesis by sponging miR-145-5p to induce FOXO1 expression in human BMSCs. Further in vivo studies indicated that hsa_circ_0006393 overexpression increases the bone mineral density (BMD) and RUNX2 and BMP2 expression in a GIOP model [Citation60]. Cerebellar degeneration-related protein 1 transcript (CDR1as), a well-investigated circRNA, has been found to play pivotal roles in diverse physiological and pathological processes. It potentially exerts its effects by sponging several miRNAs, mainly miR-7. The functional roles of CDR1as in stem cell differentiation have been widely investigated. CDR1as, which is upregulated in BMSCs from samples of steroid-induced osteonecrosis of the femoral head (SONFH), regulates the osteogenesis and adipogenesis of SONFH-BMSCs. By sponging miR-7-5p to induce WNT5B expression, CDR1as inhibits osteogenesis while improving the adipogenesis of SONFH-BMSCs in vitro [Citation41]. These functions of circRNAs under pathological conditions provide novel insights into the regulatory mechanisms of disorders related to BMSC differentiation and may serve as promising diagnostic biomarkers and therapeutic targets for these disorders. Several circRNAs have been shown to interact with oestrogen receptors to regulate the differentiation of rat BMSCs (rBMSCs). ERβ knockdown inhibits osteogenesis-related gene expression in rBMSCs. Mechanistically, ERβ may promote the osteogenesis of rBMSCs by regulating several circRNAs. Circ2:27,713,879|27,755,789 and 2:240,822,115|240,867,796 are further predicted to function as ceRNAs of miR-328-5p to modulate the PI3K-Akt, MAPK, TGF-β and Wnt signalling pathways, thereby regulating rBMSC osteogenesis [Citation61]. The hypothesis on the role of these circRNAs in regulating rBMSC osteogenesis has not been verified in experiments and in-depth explorations are needed. Similarly, quercetin (QUE), a member of the class of flavones, promotes the osteogenesis and adipogenesis of ERα-deficient rBMSCs by regulating the expression of some circRNAs. Further studies indicate that ERα and QUE may regulate rBMSC differentiation through the circRNA-miR-326-5p-mRNA axis, and miR-326-5p potentially serves as a regulator of ERα and QUE in turn [Citation62]. However, the evidence for the regulatory circRNA-miR-326-5p-mRNA axis in rBMSC differentiation is still weak, and more research is required to confirm the underlying mechanisms. In contrast, circIGSF11, which is downregulated during hBMSC osteogenesis, exerts a negative effect on hBMSC osteogenesis. Knockdown of circIGSF11 promotes the osteogenic differentiation of hBMSCs by sponging miR-199b-5p [Citation63]. Similarly, hsa_circ_0127781 may exert an analogous effect on hBMSC osteogenesis by inhibiting the expression of both miR-335-5p and miR-210-5p. Previous studies have shown that miR-335-5p and miR-210-5p induce osteogenesis by inhibiting DKK1 and activin A receptor type 1B (AcvR1b), respectively [Citation63]. Future studies are warranted to validate the exact role of hsa_circ_0127781 in regulating hBMSC osteogenesis.
Table 2. CircRNAs in bone marrow-derived mesenchymal stem cell differentiation
2.2.2 CircRNAs in adipose-derived mesenchymal stem cell (ADSC) differentiation
Adipose-derived mesenchymal stem cells (ADSCs) are a class of MSCs residing in the stromal-vascular section of adipose tissue. Compared with human BMSCs, human ADSCs have a stronger capacity for proliferation and maintain the potential to differentiate for a long time in culture [Citation64,Citation65]. Since the first identification in 2001, the promising role of ADSCs in tissue engineering and regeneration medicine has been investigated in numerous studies. However, the limited differentiation capability of ADSCs has become an obstacle to the clinical application of ADSCs. Several circRNAs have been shown to be associated with the differentiation of ADSCs (). For example, circFOXP1 has been reported to be expressed at high levels during hADSC osteogenesis in osteoporosis bone tissues. Mechanistically, circFOXP1 enhances hADSC osteogenic differentiation by sponging miR-33a-5p to upregulate FOXP1 expression both in vivo and in vitro. Therefore, circFOXP1 may be a promising candidate target for hADSC-based therapy in osteoporosis treatment [Citation66]. Another circRNA that is overexpressed during the osteoblastic differentiation of ADSCs is circRNA-23,524. It sponges miR-30a-3p to induce the expression of RUNX2, ALP and OCN, thus facilitating osteogenesis in ADSCs [Citation33]. Similarly, circRNA-vgll3 also increases the osteogenic differentiation ability of hADSCs, and miR-326-5p is the direct target of circRNA-vgll3. CircRNA-vgll3 has been found to serve as a ceRNA of miR-326-5p to stimulate the expression of integrin α5, a critical regulator of the ECM and osteoprogenitors, thus promoting osteogenesis in hADSCs [Citation67]. Both circRFWD2 and circINO80 have also been suggested to positively regulate NELL-1-induced osteogenesis in hADSCs. Mechanistically, circRFWD2 and circINO80 may function as sponges of miR-6817-5p, which inhibits the expression of RUNX2 and bone sialoprotein (BSP) in hADSCs [Citation68]. During the osteogenesis of mouse ADSCs (mADSCs), mmu_circRNA_013422 and mmu_circRNA_22566 are upregulated and are proposed to play a role in mADSC differentiation. The circRNA–miRNA network revealed miR-338-3p as a direct target of mmu_circRNA_013422 and mmu_circRNA_22566. Previous research has documented the inhibitory effect of miR-338-3p on mBMSC osteogenesis via the suppression of Runx2 and Fgfr2 [Citation69,Citation70]. Future studies should focus on the exact mechanisms by which mmu_circRNA_013422 and mmu_circRNA_22566 regulate mADSC osteogenesis mediated by miR-338-3p, which may provide novel mechanisms and targets for bone regeneration and formation treatment.
Table 3. CircRNAs in adipose-derived mesenchymal stem cell differentiation
In contrast, several circRNAs inhibit the differentiation of ADSCs. In hADSCs from patients with metabolic syndrome, knockdown of hsa_circH19 has been found to enhance adipogenesis and lipid accumulation. Mechanistically, hsa_circH19 may inhibit polypyrimidine tract-binding protein 1 (PTBP1) to suppress SREBP1 precursor cleavage [Citation71]. Similarly, decreased expression of circMCM3AP and circPOMT1 has been observed during the osteogenesis of hADSCs. Furthermore, hsa-miR-6881-3p facilitates hADSC osteogenesis, and Smad6 and Chordin, two negative regulators of BMPs, are considered direct targets of hsa-miR-6881-3p. Additionally, the expression of circMCM3AP and circPOMT1 is negatively associated with hsa-miR-6881-3p, thus indicating that circMCM3AP and circPOMT1 may play inhibitory roles in hADSC osteogenesis by upregulating Smad6 and Chordin expression through a mechanism mediated by hsa-miR-6881-3p [Citation72]. The roles of circMCM3AP and circPOMT1 in regulating hADSC osteogenesis are only predictions, and in-depth exploration is needed to reveal the underlying mechanisms by which these two circRNAs exert their effects.
2.2.3 CircRNA in dental pulp stem cell (DPSC) differentiation
Dental pulp stem cells (DPSCs) are a type of multipotent stem cell that develops from dental pulp tissues. DPSCs have strong self-renewal and multi-differentiation abilities with promising potential for use in regenerative medicine and bone disease treatment [Citation73]. Multiple circRNAs promote DPSC differentiation, such as circSIPA1L1, circLPAR1 and circRNA124534 (). During the osteogenic differentiation of DPSCs, upregulation of circSIPA1L1 and downregulation of miR-617 have been detected. CircSIPA1L1 promotes the osteogenesis of DPSCs by sponging miR-617 to induce Smad3 expression [Citation74]. Similarly, a high level of circLPAR1 was identified in osteogenic-induced DPSC-derived exosomes. Exosomal circLPAR1 promotes the osteogenic differentiation of hDPSCs by sponging hsa-miR-31 to activate SATB2 [Citation75]. Recent research also reported the involvement of the circRNA124534/miR-496/β-Catenin axis in regulating the osteogenic differentiation of hDPSCs. Exogenous expression of circRNA124534 promotes the osteogenic differentiation of hDPSCs by sponging miR-296 to increase β-Catenin expression both in vivo and in vitro [Citation76]. Ji et al. also identified significantly increased expression of hsa_circ_0026827 during the osteoblast differentiation of hDPSCs. By functioning as a sponge of miR-188-3p to upregulate Beclin1 and RUNX1 expression, hsa_circ_0026827 was found to enhance the osteogenic differentiation ability of hDPSCs in vitro. In addition, hsa_circ_0026827 promotes heterotopic bone formation in a heterotopic bone model in vivo, suggesting the promising potential of hsa_circ_0026827 in bone regeneration treatment [Citation77]. In addition, knockdown of hsa_circ_104101 inhibits the odontogenic differentiation of hDPSCs by suppressing the expression of various odontoblastic regulators, including DSPP, DMP1, ALP, and OCN [Citation38].
Table 4. CircRNAs in dental pulp stem cell differentiation
2.2.4 CircRNAs in periodontal ligament stem cell (PDLSC) differentiation
Periodontal ligament stem cells (PDLSCs) have the capacity for self-renewal and multilineage differentiation and are MSCs derived from the periodontal ligament. Inspiringly, PDLSCs are easily harvested and reported to possess a higher proliferative and self-renewal capacity than BMSCs and DPSCs, making PDLSCs a promising strategy for tissue regeneration therapy. PDLSCs play an important role in the formation of cementum/PDL-like structures, peripheral nerves, periodontal ligaments and blood vessels [Citation78]. Some circRNAs are involved in regulating PDLSC differentiation (). For instance, circBANP and circITCH are predicted to regulate PDLSC osteogenesis through the MAPK signalling pathway by sponging miR-34a and miR-146a [Citation79]. Further studies are required to elucidate the potential roles and underlying mechanisms of these two circRNAs in PDLSC differentiation. Significantly upregulated CDR1as expression and downregulated miR-17 expression have been detected during the osteogenic differentiation of PDLSCs. Mechanistically, CDR1as positively regulates PDLSC osteogenesis by downregulating miR-17, subsequently increasing the expression of the osteogenesis-related gene GDF5. Specifically, knockdown of GDF5 inhibits osteogenesis and partially represses the osteogenesis-promoting role of CDR1as in PDLSCs. Moreover, CDR1as/miR-7/GDF5 may exert osteogenic differentiation-promoting effects on PDLSCs by increasing the phosphorylation of Smad1/5/8 and p38 MAPK. Further in vivo experiments have shown that knockdown of CDR1as inhibits bone formation [Citation80]. These results reveal a novel CDR1as/miR-7/GDF5 regulatory axis in PDLSC osteogenesis, thus providing new therapeutic targets for periodontal tissue and bone formation. In contrast, circCDK8 has been found to serve as a negative regulator of PDLSC osteogenesis. Notably, circCDK8 was evidently upregulated in PDLSCs treated with CoCl2, an inducer of hypoxia. A hypoxic environment inhibits PDLSC osteogenesis and induces the expression of circCDK8, indicating the potential role of circCDK8 in PDLSC osteogenesis. Mechanistically, circCDK8 inhibits the osteogenesis of PDLSCs by inducing endoplasmic reticulum (ER) stress and autophagy, and knockdown of circCDK8 reverses the inhibitory effects of CoCl2 on PDLSC osteogenesis [Citation81].
Table 5. CircRNAs in periodontal ligament stem cell differentiation
2.2.5 CircRNAs in the differentiation of other MSC types
In addition to the MSC types mentioned above, several circRNAs have been reported to participate in the differentiation of other MSC types (). MSCs derived from the human umbilical cord (hUCMSCs) have remarkable self-renewal, differentiation and proliferation capacities. hUCMSCs have the ability to differentiate into various cell types of the three germ layers and secrete numerous molecules that interact with other cells, such as ESCs and immune cells, thus exhibiting promising therapeutic potential in diverse human diseases [Citation82]. Several studies indicate that some circRNAs are associated with hUCMSC differentiation. For instance, 127 circRNAs were upregulated and 99 circRNAs were downregulated during the differentiation of hUCMSCs into cardiomyocyte-like cells. Furthermore, three circRNAs, circRNA_05432, circRNA_08441 and circRNA_01536, are suggested to be critical regulators of the differentiation of hUCMSCs into cardiomyocyte-like cells, and hsa-miR-3620-5p may be the direct target of these circRNAs [Citation83]. The roles of these circRNAs in hUCMSC differentiation are only predicted and worthy of in-depth research. The involvement of CDR1as in hUCMSC differentiation has also been reported. CDR1as is overexpressed in hUCMSCs and enhances the adipogenic differentiation capacity in vitro. Moreover, knockdown of CDR1as inhibits differentiation and proliferation and induces the apoptosis of hUCMSCs [Citation20]. Maxillary sinus membrane stem cells (MSMSCs) are a group of MSCs isolated from the maxillary sinus membrane that are able to differentiate into osteoblasts. Importantly, hsa_circRNA_33287 promotes the osteogenic differentiation of MSMSCs by directly targeting Runx3 through miR-214-3p sponging. In vivo experiments have further shown that hsa_circRNA_33287 promotes and miR-214-3p inhibits ectopic bone formation by human MSMSCs [Citation84]. As a novel population of MSCs, stem cells from the apical papilla (SCAPs) reside in human immature impacted permanent teeth and play critical roles in pulp-dentin regeneration, periodontal tissue regeneration and bone regeneration. During the osteoblast differentiation of SCAPs, circSIPA1L1 which is markedly overexpressed, has been identified to positively regulate osteoblastic differentiation by blocking the miR-204-5p-dependent suppression of ALPL [Citation85]. Collectively, circRNAs are critically correlated with MSC differentiation, and we postulate that the functional characterization of more circRNAs is indispensable for understanding the molecular biology of MSC differentiation and developing novel MSC-based therapies.
Table 6. CircRNAs in the differentiation of other MSC types
hUCMSC: MSC derived from human umbilical cord, MSMSC: maxillary sinus membrane stem cell, SCAP: stem cell from the apical papilla.
2.3 CircRNAs in satellite cell differentiation
Myogenesis is a biological process in which satellite cells differentiate into myoblasts, followed by the subsequent differentiation and fusion of these cells into multinucleated myotubes [Citation86]. Satellite cells, which were first identified in 1961, are a specific muscle stem cell population that resides in skeletal muscle and plays important roles in skeletal muscle development and regeneration. Upon muscle injury or some pathological conditions, satellite cells in a quiescent state are activated and differentiate into myogenic progenitors to form multinucleated myotubes [Citation87]. Based on accumulating evidence, some circRNAs, including CDR1as and circTMTC1, are involved in satellite cell differentiation. For instance, CDR1as has been found to promote myogenesis via the induction of myofiber formation in skeletal muscle satellite cells (SMSCs). Mechanistically, CDR1as sponges miR-17 to activate IGF1R, a promoter of myogenesis. Specifically, myogenic differentiation protein 1 (MyoD1) has been identified to increase CDR1as expression by binding to the 5ʹ flanking region of CDR1as, thus promoting the myogenic differentiation of SMSCs [Citation88]. Additionally, circTMTC1 is expressed at high levels during chicken breast muscle development. CircTMTC1 inhibits chicken SMSC differentiation by sponging miR-128-3p, which binds to the 3ʹUTR of myostatin (MSTN) [Citation89]. Conversely, decreased expression of circFAM188B was detected during chicken SMSC differentiation. As shown in previous studies, circFAM188B encodes the novel protein circFAM188B-103aa that promotes proliferation and inhibits the differentiation of chicken SMSCs. This finding provides insights into a novel circRNA and protein that regulate chicken SMSC differentiation and reveals novel mechanisms for chicken muscle development [Citation90]. To date, little information is available on the role of circRNAs in satellite cell differentiation, and future studies should focus on their functions and mechanisms in satellite cell differentiation.
2.4 CircRNAs in myoblast differentiation
Myoblasts are also a group of proliferative muscle precursor cells that are indispensable for myogenesis. After satellite cells differentiate into myoblasts, myoblasts can differentiate into multinucleated myofibers with transcriptional and morphological alterations. Myoblast differentiation is a multistage and complex process that is controlled by multiple molecules and signalling pathways [Citation91,Citation92]. Several circRNAs have been identified to play critical roles in regulating myoblast differentiation (). For instance, circHIPK3-003 promotes the differentiation of chicken myoblasts by directly targeting myocyte enhancer factor 2 C (MEF2C) as a miR-30a-3p sponge [Citation93]. Similarly, circSVIL, which is upregulated during chicken embryonic leg muscle development, promotes chicken myoblast differentiation by upregulating the expression of both c-JUN and MEF2C as a miR-203 sponge in vitro [Citation94]. CircFGFR2, which is derived from exons 3–6 of the FGFR2 gene, is another circRNA that is overexpressed during embryonic chicken skeletal muscle development. As shown in a previous study, circFGFR2 promotes chicken myoblast differentiation by inhibiting the expression of both miR-133a-5p and miR-29b-1-5p. Both miR-133a-5p and miR-29b-1-5p suppress MYOD and MYOG expression to inhibit chicken myogenesis [Citation95]. Additionally, increased expression of circSamd4 has been observed in both human and mouse myoblasts. CircSamd4 suppresses the expression of the purine-rich binding proteins PURA and PURB, which are inhibitors of myosin heavy chain (MHC), thus inducing myoblast differentiation [Citation96]. Yao et al. detected high circHIPK3 expression in mouse skeletal muscle tissues. Further research indicated that circHIPK3 promotes C2C12 myoblast differentiation by sponging both miR-124-5p and miR-379-5p [Citation97]. Several circRNAs have been reported to be involved in promoting bovine myoblast myogenesis. Notably, miR-107, which is downregulated during myogenesis, negatively regulates bovine myoblast differentiation and apoptosis. CircFGFR4 promotes myogenesis and apoptosis by sponging miR-107 to attenuate its inhibition of Wnt3a in bovine myoblasts [Citation98]. CircFUT10 is also involved in regulating bovine myoblast differentiation. It functions as a miR-133a sponge to inhibit proliferation and facilitate the differentiation and apoptosis of bovine myoblasts in vitro [Citation34]. Similarly, the role of the circTTN/miR-432/IGF2/PI3K/AKT axis in bovine myoblast proliferation and differentiation has also been reported recently. Overexpressed circTTN sponges miR-432 to induce the IGF2/PI3K/AKT signalling pathway, thus positively regulating bovine myoblast differentiation [Citation99]. Another study also reported that circSNX29 functions as a sponge for miRNA-744 and promotes bovine myoblast differentiation by activating the Wnt5a/Ca2+/CaMKIId signalling pathway [Citation100]. Legnini et al. have found that several circRNAs, including circBNC2, circQKI and circZfp609, are regulated during myogenesis. The use of siRNAs targeting both circQKI and the QKI mRNA exerted anti–myogenic effects, indicating the positive role of circQKI and the QKI mRNA in regulating myogenesis. CircBNC2 is upregulated during myogenesis and may facilitate myoblast differentiation by competing with the production of the BNC2 mRNA [Citation28]. The regulatory mechanisms of circBNC2 and circQKI in myogenesis are still poorly understood, and further studies are needed to identify the exact roles and mechanisms of the two circRNAs in myogenesis.
Table 7. CircRNAs in myoblast differentiation
Conversely, several circRNAs serve as negative regulators of myoblast differentiation. CircHUWE1, which originates from exons 3-7 of the HUWE1 gene, is downregulated during myogenesis in bovine myoblasts. By stimulating AKT3 expression as a miR-29b sponge, circHUWE1 represses myoblast differentiation, while miR-29b reverses circHUWE1-induced myogenesis inhibition [Citation101]. The expression of circFoxO3 gradually increases with the differentiation of C2C12 myoblasts. After circFoxO3 silencing, the expression level of MyoG is upregulated in C2C12 myoblasts, indicating the inhibitory effect of circ-FoxO3 on myoblast differentiation. Further studies identified miR-138-5p as a direct target of circ-FoxO3 [Citation40]. Low expression of circLMO7 has been observed in bovine myoblasts. By downregulating the expression of the myogenesis-related genes MyoD and MyoG, exogenous expression of circLMO7 inhibits bovine myoblast differentiation by sponging miR-378a-3p [Citation102]. CircZfp609, which is also downregulated during myogenesis, encodes a protein that promotes myoblast proliferation [Citation28]. However, the detailed mechanisms underlying the translation of circZfp69 and the functions of this protein in myogenesis are still unknown. In addition, circZfp609 sponges miR-194-5p and attenuates its inhibitory effects on BCL2-associated transcription factor 1 (BCLAF1), subsequently inhibiting the myogenic differentiation of mouse myoblasts [Citation103]. Overall, the circRNA-miRNA-mRNA regulatory axis plays vital roles in myogenesis, and circRNAs may function as a potent tool to regulate muscle development. An understanding of the roles of circRNAs in regulating myogenesis may provide new insights into muscle development and reveal novel strategies for modulating circRNAs to treat muscular diseases.
2.5 CircRNAs in the differentiation of other stem cell types
In addition to ESCs, MSCs, satellite cells and myoblasts, other common types of stem cells, such as epidermal stem cells (EpSCs), intestinal stem cells (ISCs) and neural stem cells (NSCs), have been identified. Several circRNAs are associated with the differentiation of these stem cell types (). EpSCs are a specific class of multipotent cells located in the skin that are fundamental to epidermal regeneration [Citation104]. Multiple upregulated circRNAs have been observed during the differentiation of EpSCs into keratinocytes, including circZNF91, which contains 24 target sites for miR-23b-3p. Importantly, previous research has indicated that miR-23b-3p regulates human keratinocyte differentiation via the activation of the TGF-β-SMAD2 signalling pathway andrepression of TGIF1, indicating the potential role of circZNF91 in regulating EpSC differentiation [Citation35,Citation105]. Intestinal stem cells (ISCs) are a population of stem cells located at the base of intestinal crypts that are mainly divided into two groups: crypt base columnar cells (Lgr5+ ISCs) and Bmi+ label-retaining cells [Citation106]. The upregulation of circPan3 has been observed in mouse and human Lgr5-GFP+ ISCs. Knockdown of circPan3 contributes to the inhibition of immune cell-mediated self-renewal and epithelial regeneration in mouse ISCs. Mechanistically, circPan3 induces the expression of IL-13Rα1 by directly binding to the Il13ra1 mRNA in Lgr5-GFP+ ISCs. Moreover, IL-13 produced by innate lymphoid cells (ILCs) enhances self-renewal and epithelial regeneration by inducing IL-13Rα1 expression via circPan3 [Citation107]. Neural stem cells (NSCs) are a specific group of stem cells in the nervous system that differentiate into diverse cell lineages in the nervous system, such as neurons, astrocytes, and oligodendrocytes [Citation108]. CircRNAs are abundant in the brain, and their important roles in brain development have been reported recently. During neuronal differentiation and development, numerous circRNAs, such as CDR1as, circRTN4, circTULP4 and circRIMS2, are upregulated, and their expression changes at different stages of neuronal differentiation, suggesting the involvement of these circRNAs in neurogenesis [Citation109]. By sponging miR-561 and impairing its inhibitory effects on E2F transcription factor 8 (E2F8), hsa_circ_0002468 has been reported to repress cell proliferation and induce neuronal differentiation in SH-SY5Y cells [Citation110]. Overall, research on the regulatory roles of circRNAs in the differentiation of these stem cell types is at an early stage, and more efforts are needed to investigate the exact mechanisms.
Table 8. CircRNAs in the differentiation of other stem cell types
EpSC: epidermal stem cell, ISC: intestinal stem cell, NSC: neural stem cell.
3. Challenges and perspectives
An increasing number of circRNA transcriptional profiles have been reported, and several circRNAs have been shown to participate in regulating stem cell differentiation. In this review, we systematically summarized the roles of circRNAs and their regulatory mechanisms in the differentiation of diverse types of stem cells (). However, many limitations exist in exploring the functions and mechanisms of circRNAs in regulating stem cell differentiation, which must be further clarified.
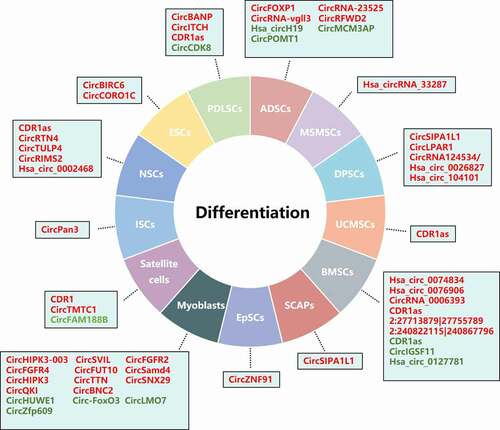
Figure 3. A summary diagram of circRNAs in stem cell differentiation. Several circRNAs have been found to be involved in the differentiation of diverse stem cell types through regulating expression of potential target genes and related signalling pathway
First, despite the recent advances that have been achieved in revealing the characteristics and biological functions of circRNAs, the biological processes and functional mechanisms of circRNAs remain largely unknown. For instance, the regulatory mechanisms underlying the biogenesis, nuclear export and decay of circRNAs are still largely unknown. The biological functions of circRNAs also require intensive research. Although researchers have widely hypothesized that circRNAs mainly exert their functions by sponging miRNAs, several studies have questioned ceRNA function. Notably, most studies regarding the ceRNA function of circRNAs are based on overexpression experiments, and only a few circRNAs have abundant binding sites for a specific miRNA, such as CDR1as and circZNF91, indicating that numerous circRNAs may not have miRNA sponge functions [Citation19,Citation35]. Meanwhile, more research is needed to understand other mechanisms by which circRNAs function, such as protein sponges, protein recruitment and translation.
Second, although multiple methodologies have been developed for the detection and functional characterization of circRNAs, many limitations and difficulties in circRNA research methodologies still exist. For instance, circRNAs usually present low abundance and overlap with their linear counterparts. The abundance of circRNAs is approximately less than 10% of their linear counterparts, and thus the estimation of circRNAs with low abundance may be biased [Citation111]. In addition, the unique structure of the ‘backspliced junction (BSJ) site’ of circRNAs is significantly different from that of their corresponding linear RNAs and is fundamental for their identification [Citation112]. Nonetheless, circRNAs consist of different components, including exons and/or introns, and hence using BSJs only to identify circRNAs may not be quite accurate and limit our detection and functional investigation of circRNAs. Hence, effective methodologies for circRNA detection and characterization must be developed to provide novel insights into the roles and regulatory mechanisms of circRNAs.
Third, why are a large number of circRNAs dysregulated during stem cell differentiation, and how do they regulate stem cell differentiation? The dysregulation of circRNAs is attributed to the imbalance among circRNA generation, localization, and degradation during stem cell differentiation. However, the underlying mechanisms of the dysregulation of circRNAs during stem cell differentiation have not been conclusively identified. Although diverse circRNA profiles have been detected during stem cell differentiation, the regulatory mechanisms underlying the expression of specific circRNAs remain largely unexplored. To date, the miRNA sponge mechanism is the most extensively investigated mechanism by which circRNAs exert their effects on regulating stem cell differentiation. Another mechanism underlying the regulatory functions of circRNAs in stem cell differentiation may be their ability to encode functional proteins. However, few studies have focused on the protein-encoding function of circRNAs or the role of circRNA-encoded proteins in stem cell differentiation. In addition, current research regarding the roles of circRNAs in stem cell differentiation is mostly based on in vitro experiments, and multiple studies have only predicted that several circRNAs might play a role in stem cell differentiation but lack sufficient evidence. Therefore, comprehensive and in-depth studies investigating the functions of circRNAs in stem cell differentiation are urgently needed.
Finally, future research efforts are needed to promote the effective application of circRNAs in clinical practice. Because circRNAs exhibit specific expression profiles during the differentiation of different stem cell types, multiple dysregulated circRNAs exhibit great potential as biomarkers for stem cell differentiation. Monitoring the expression of these circRNAs may help evaluate the stemness and differentiation capacity of stem cells. In addition, approaches targeting these dysregulated endogenous circRNAs may be a promising strategy to regulate stem cell differentiation, thus maintaining homoeostasis in the body. Most importantly, these circRNAs may serve as novel therapeutic targets in tissue engineering and regeneration medicine, and circRNA-based stem cell therapy has attracted the interest of numerous investigators. For example, exosomal circHIPK3 derived from hUCMSCs has been found to inhibit pyroptosis of skeletal muscle cells and promote ischaemic muscle injury repair in a mouse model in vivo, highlighting a promising role for circHIPK3 in stem cell-based therapy for tissue repair [Citation113]. However, the potential of most circRNAs in clinical practice has not been thoroughly identified, and although many circRNAs with regulatory roles in stem cell differentiation have been reported, the selection of specific circRNAs from these candidate circRNAs for stem cell-based therapy remains challenging. Future studies should focus on screening and validating target circRNAs for stem cell-based therapy in a large number of clinical samples. Comprehensive research is needed before circRNAs are incorporated into clinical practice.
4. Conclusions
In summary, emerging evidence has shown that the expression of numerous circRNAs is altered during stem cell differentiation and that several circRNAs play certain roles in regulating the differentiation of diverse stem cell types. These circRNAs may serve as biomarkers of stem cell differentiation and provide new avenues for stem cell-based therapy. However, the detailed mechanisms by which circRNAs regulate stem cell differentiation remain to be investigated, and additional in vivo research and clinical trials are urgently needed to confirm the potential clinical implications of circRNAs in stem cell therapy, which may significantly improve the effectiveness of stem cell-based therapy for the treatment of human disorders.
Authors’ contribution
Lin Zhengjun: Writing - Original Draft. Tang Xianzhe: Writing - Original Draft. Wan Jia: Writing - Review & Editing, Visualization. Zhang Xianghong: Writing - Original Draft, Visualization. Liu Chunfeng: Writing - Original Draft, Visualization. Liu Tang: Writing - Review & Editing, Supervision, Funding acquisition. All authors contributed to the writing and revision of the manuscript, knew the content of it, and approved its submission.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Acknowledgments
Not applicable.
Disclosure statement
The authors declare that they have no competing financial interests.
Availability of data and materials
Not applicable.
Additional information
Funding
References
- Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111.
- Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125.
- Castillo-Cardiel G, Lopez-Echaury AC, Saucedo-Ortiz JA, et al. Bone regeneration in mandibular fractures after the application of autologous mesenchymal stem cells, a randomized clinical trial. Dent Traumatol. 2017;33(1):38–44.
- Mesimaki K, Lindroos B, Tornwall J, et al. Novel maxillary reconstruction with ectopic bone formation by GMP adipose stem cells. Int J Oral Maxillofac Surg. 2009;38(3):201–209.
- Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: wnt signaling and stem cell control. Science. 2014;346(6205):1248012.
- Ermolaeva M, Neri F, Ori A, et al. Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol Cell Biol. 2018;19(9):594–610.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856.
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. .
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842.
- Zhang X-O, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–1287.
- Rankin CR, Lokhandwala ZA, Huang R, et al. Linear and circular CDKN2B-AS1 expression is associated with Inflammatory Bowel Disease and participates in intestinal barrier formation. Life Sci. 2019;231:116571.
- Zhang Y, Zhang X-O, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806.
- Huang C, Liang D, Tatomer DC, et al. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639–644.
- Park OH, Ha H, Lee Y, et al. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74(3):494–507. e498. .
- Kleaveland B, Shi CY, Stefano J, et al. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell. 2018;174(2):350–362. e317. .
- Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011;30(21):4414–4422.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. .
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Yang L, Bin Z, Hui S, et al. The Role of CDR1as in Proliferation and Differentiation of Human Umbilical Cord-Derived Mesenchymal Stem Cells. Stem Cells Int. 2019;2019:2316834.
- Li Y, Ge Y-Z, Xu L, et al. Circular RNA ITCH: a novel tumor suppressor in multiple cancers. Life Sci. 2020;254:117176.
- Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412.
- Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370.
- Zeng Y, Du WW, Wu Y, et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics. 2017;7(16):3842–3855. .
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21. e27. .
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. .
- Di Timoteo G, Dattilo D, Centron-Broco A, et al. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020;31(6):107641. .
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66(1):22–37. e29. .
- Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44.
- Feng XQ, Nie SM, Huang JX, et al. Circular RNA circHIPK3 serves as a prognostic marker to promote chronic myeloid leukemia progression. Neoplasma. 2020;67(1):171–177.
- Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321–330.
- Li J, Ni S, Zhou C, et al. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag Res. 2018;10:2777–2784.
- Guo Z, Zhao L, Ji S, et al. CircRNA-23525 regulates osteogenic differentiation of adipose-derived mesenchymal stem cells via miR-30a-3p. Cell Tissue Res. 2021;383(2):795–807.
- Li H, Yang J, Wei X, et al. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J Cell Physiol. 2018;233(6):4643–4651. .
- Kristensen LS, Okholm TLH, Veno MT, et al. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15(2):280–291.
- Lei W, Feng T, Fang X, et al. Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther. 2018;9(1):56.
- Della Bella E, Menzel U, Basoli V, et al. Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells. 2020;9(2):398.
- Chen M, Yang Y, Zeng J, et al. circRNA Expression Profile in Dental Pulp Stem Cells during Odontogenic Differentiation. Stem Cells Int. 2020;2020:1–19.
- Chen R, Jiang T, Lei S, et al. Expression of circular RNAs during C2C12 myoblast differentiation and prediction of coding potential based on the number of open reading frames and N6-methyladenosine motifs. Cell Cycle. 2018;17(14):1832–1845.
- Li X, Li C, Liu Z, et al. Circular RNA circ-FoxO3 Inhibits Myoblast Cells Differentiation. Cells. 2019;8(6):616.
- Chen G, Wang Q, Li Z, et al. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone. 2020;133:115258.
- Siede D, Rapti K, Gorska AA, et al. Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol. 2017;109:48–56.
- Qian D-Y, Yan G-B, Bai B, et al. Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed Pharmacother. 2017;90:492–499.
- Kang Y, Guo S, Sun Q, et al. Differential circular RNA expression profiling during osteogenic differentiation in human adipose-derived stem cells. Epigenomics. 2020;12(4):289–302.
- Li Z, Li N, Ge X, et al. Differential circular RNA expression profiling during osteogenic differentiation of stem cells from apical papilla. Epigenomics. 2019;11(9):1057–1073.
- Zhang P, Xu H, Li R, et al. Assessment of myoblast circular RNA dynamics and its correlation with miRNA during myogenic differentiation. Int J Biochem Cell Biol. 2018;99:211–218.
- Yang Q, Wu J, Zhao J, et al. Circular RNA expression profiles during the differentiation of mouse neural stem cells. BMC Syst Biol. 2018;12(Suppl 8):128.
- Xie F, Zhao Y, Wang S-D, et al. Identification, characterization, and functional investigation of circular RNAs in subventricular zone of adult rat brain. J Cell Biochem. 2019;120(3):3428–3437.
- Nicolet BP, Engels S, Aglialoro F, et al. Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res. 2018;46(16):8168–8180.
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954.
- Yu C-Y, Li T-C, Wu -Y-Y, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149.
- Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8(1):14741. .
- You KT, Park J, Kim VN. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev. 2015;29(19):2004–2009.
- Ding D-C, Shyu W-C, Lin S-Z. Mesenchymal stem cells. Cell Transplantation. 2011;20(1):5–14.
- Baker N, Boyette LB, Tuan RS. Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone. 2015;70:37–47.
- Chen C, Uludag H, Wang Z, et al. Noggin suppression decreases BMP-2-induced osteogenesis of human bone marrow-derived mesenchymal stem cells in vitro. J Cell Biochem. 2012;113(12):3672–3680.
- Chia W, Liu J, Huang Y-G, et al. A circular RNA derived from DAB1 promotes cell proliferation and osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death Dis. 2020;11(5):372.
- Ouyang Z, Tan T, Zhang X, et al. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 2019;10(12):932.
- Wen J, Guan Z, Yu B, et al. Circular RNA hsa_circ_0076906 competes with OGN for miR-1305 biding site to alleviate the progression of osteoporosis. Int J Biochem Cell Biol. 2020;122:105719.
- Wang X-B, Li P-B, Guo S-F, et al. circRNA_0006393 promotes osteogenesis in glucocorticoid‑induced osteoporosis by sponging miR‑145‑5p and upregulating FOXO1. Mol Med Rep. 2019;20(3):2851–2858.
- Li X, Peng B, Zhu X, et al. Changes in related circular RNAs following ERβ knockdown and the relationship to rBMSC osteogenesis. Biochem Biophys Res Commun. 2017;493(1):100–107.
- Li X, Chen R, Lei X, et al. Quercetin regulates ERalpha mediated differentiation of BMSCs through circular RNA. Gene. 2020;2020:145172.
- Zhang M, Jia L, Zheng Y. circRNA Expression Profiles in Human Bone Marrow Stem Cells Undergoing Osteoblast Differentiation. Stem Cell Rev Rep. 2019;15(1):126–138.
- Wankhade UD, Shen M, Kolhe R, et al. Advances in Adipose-Derived Stem Cells Isolation, Characterization, and Application in Regenerative Tissue Engineering. Stem Cells Int. 2016;2016:3206807.
- Tabatabaei Qomi R, Sheykhhasan M. Adipose-derived stromal cell in regenerative medicine: a review. World J Stem Cells. 2017;9(8):107–117.
- Shen W, Sun B, Zhou C, et al. CircFOXP1/FOXP1 promotes osteogenic differentiation in adipose-derived mesenchymal stem cells and bone regeneration in osteoporosis via miR-33a-5p. J Cell Mol Med. 2020;24(21):12513–12524.
- Zhang D, Ni N, Wang Y, et al. CircRNA-vgll3 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells via modulating miRNA-dependent integrin alpha5 expression. Cell Death Differ. 2020;28(1):283–302.
- Huang X, Cen X, Zhang B, et al. The roles of circRFWD2 and circINO80 during NELL-1-induced osteogenesis. J Cell Mol Med. 2019;23(12):8432–8441.
- Long T, Guo Z, Han L, et al. Differential Expression Profiles of Circular RNAs During Osteogenic Differentiation of Mouse Adipose-Derived Stromal Cells. Calcif Tissue Int. 2018;103(3):338–352.
- Liu H, Sun Q, Wan C, et al. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J Cell Physiol. 2014;229(10):1494–1502.
- Zhu Y, Gui W, Lin X, et al. Knock-down of circular RNA H19 induces human adipose-derived stem cells adipogenic differentiation via a mechanism involving the polypyrimidine tract-binding protein 1. Exp Cell Res. 2020;387(2):111753.
- Huang X-Q, Cen X, Sun W-T, et al. CircPOMT1 and circMCM3AP inhibit osteogenic differentiation of human adipose-derived stem cells by targeting miR-6881-3p. Am J Transl Res. 2019;11(8):4776–4788.
- Mortada I, Mortada R, Al Bazzal M. Dental Pulp Stem Cells and Neurogenesis. Adv Exp Med Biol. 2018;1083:63–75.
- Ge X, Li Z, Zhou Z, et al. Circular RNA SIPA1L1 promotes osteogenesis via regulating the miR-617/Smad3 axis in dental pulp stem cells. Stem Cell Res Ther. 2020;11(1):364.
- Xie L, Guan Z, Zhang M, et al. Exosomal circLPAR1 Promoted Osteogenic Differentiation of Homotypic Dental Pulp Stem Cells by Competitively Binding to hsa-miR-31. Biomed Res Int. 2020;2020:6319395.
- Ji F, Pan J, Shen Z, et al. The Circular RNA circRNA124534 Promotes Osteogenic Differentiation of Human Dental Pulp Stem Cells Through Modulation of the miR-496/β-Catenin Pathway. Front Cell Dev Biol. 2020;8:230.
- Ji F, Zhu L, Pan J, et al. hsa_circ_0026827 Promotes Osteoblast Differentiation of Human Dental Pulp Stem Cells Through the Beclin1 and RUNX1 Signaling Pathways by Sponging miR-188-3p. Front Cell Dev Biol. 2020;8:470.
- Park J-Y, Jeon SH, Choung P-H. Efficacy of periodontal stem cell transplantation in the treatment of advanced periodontitis. Cell Transplantation. 2011;20(2):271–285.
- Gu X, Li M, Jin Y, et al. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18(1):100.
- Li X, Zheng Y, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9(1):232.
- Zheng J, Zhu X, He Y, et al. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann N Y Acad Sci. 2021;1485(1):56–70.
- Fan C-G, Zhang Q-J, Zhou J-R. Therapeutic potentials of mesenchymal stem cells derived from human umbilical cord. Stem Cell Rev Rep. 2011;7(1):195–207.
- Ruan Z-B, Chen G-C, Zhang R, et al. Circular RNA expression profiles during the differentiation of human umbilical cord–derived mesenchymal stem cells into cardiomyocyte-like cells. J Cell Physiol. 2019;234(9):16412–16423.
- Peng W, Zhu S, Chen J, et al. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacother. 2019;109:1709–1717.
- Li Y, Bian M, Zhou Z, et al. Circular RNA SIPA1L1 regulates osteoblastic differentiation of stem cells from apical papilla via miR-204-5p/ALPL pathway. Stem Cell Res Ther. 2020;11(1):461.
- Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4(2):a008342–a008342.
- Giordani L, Parisi A, Le Grand F. Satellite Cell Self-Renewal. Curr Top Dev Biol. 2018;126:177–203.
- Li L, Chen Y, Nie L, et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim Biophys Acta Gene Regul Mech. 2019;1862(8):807–821.
- Shen X, Liu Z, Cao X, et al. Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int J Biol Sci. 2019;15(10):2265–2281.
- Yin H, Shen X, Zhao J, et al. Circular RNA CircFAM188B Encodes a Protein That Regulates Proliferation and Differentiation of Chicken Skeletal Muscle Satellite Cells. Front Cell Dev Biol. 2020;8:522588.
- Stockdale FE, Hager EJ, Fernyak SE, et al. Myoblasts, satellite cells, and myoblast transfer. Adv Exp Med Biol. 1990;280:7–11.
- Du GQ, Du WJ, Liu JJ, et al. Wnt1-overexpressing skeletal myoblasts as an improved cell therapy for cardiac repair following myocardial infarction. Panminerva Med. 2015;57(4):153–166.
- Chen B, Yu J, Guo L, et al. Circular RNA circHIPK3 Promotes the Proliferation and Differentiation of Chicken Myoblast Cells by Sponging miR-30a-3p. Cells. 2019;8(2):177.
- Ouyang H, Chen X, Li W, et al. Circular RNA circSVIL Promotes Myoblast Proliferation and Differentiation by Sponging miR-203 in Chicken. Front Genet. 2018;9:172.
- Chen X, Ouyang H, Wang Z, et al. A Novel Circular RNA Generated by FGFR2 Gene Promotes Myoblast Proliferation and Differentiation by Sponging miR-133a-5p and miR-29b-1-5p. Cells. 2018;7(11):199.
- Pandey PR, Yang J-H, Tsitsipatis D, et al. circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res. 2020;48(7):3789–3805. .
- Yao R, Yao Y, Li C, et al. Circ-HIPK3 plays an active role in regulating myoblast differentiation. Int J Biol Macromol. 2020;155:1432–1439.
- Li H, Wei X, Yang J, et al. circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve Its Inhibition of Wnt3a. Mol Ther Nucleic Acids. 2018;11:272–283.
- Wang X, Cao X, Dng D, et al. Circular RNA TTN Acts As a miR-432 Sponge to Facilitate Proliferation and Differentiation of Myoblasts via the IGF2/PI3K/AKT Signaling Pathway. Mol Ther Nucleic Acids. 2019;18:966–980.
- Peng S, Song C, Li H, et al. Circular RNA SNX29 Sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca2+ signaling pathway. Mol Ther Nucleic Acids. 2019;16:481–493.
- Yue B, Wang J, Ru W, et al. The circular RNA circHUWE1 sponges the miR-29b-AKT3 axis to regulate myoblast development. Mol Ther Nucleic Acids. 2020;19:1086–1097.
- Wei X, Li H, Yang J, et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8(10):e3153. .
- Wang Y, Li M, Wang Y, et al. A Zfp609 circular RNA regulates myoblast differentiation by sponging miR-194-5p. Int J Biol Macromol. 2019;121:1308–1313.
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22(1):339–373.
- Barbollat-Boutrand L, Joly-Tonetti N, Dos Santos M, et al. MicroRNA-23b-3p regulates human keratinocyte differentiation through repression of TGIF1 and activation of the TGF-ss-SMAD2 signalling pathway. Exp Dermatol. 2017;26(1):51–57.
- Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15(1):19–33.
- Zhu P, Zhu X, Wu J, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. 2019;20(2):183–194.
- McKay R. Stem cells in the central nervous system. Science. 1997;276(5309):66–71.
- Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. 2017;14(8):1028–1034.
- Yang M, Xiang G, Yu D, et al. Hsa_circ_0002468 regulates the neuronal differentiation of SH-SY5Y cells by modulating the MiR-561/E2F8 Axis. Med Sci Monit. 2019;25:2511–2519.
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777.
- Li X, Zhang B, Li F, et al. The mechanism and detection of alternative splicing events in circular RNAs. PeerJ. 2020;8:e10032.
- Yan B, Zhang Y, Liang C, et al. Stem cell-derived exosomes prevent pyroptosis and repair ischemic muscle injury through a novel exosome/circHIPK3/ FOXO3a pathway. Theranostics. 2020;10(15):6728–6742.
