ABSTRACT
As the only oxygenic phototrophs among prokaryotes, cyanobacteria employ intricate mechanisms to regulate common metabolic pathways. These mechanisms include small protein inhibitors exerting their function by protein–protein interaction with key metabolic enzymes and regulatory small RNAs (sRNAs). Here we show that the sRNA NsiR4, which is highly expressed under nitrogen limiting conditions, interacts with the mRNA of the recently described small protein PirA in the model strain Synechocystis sp. PCC 6803. In particular, NsiR4 targets the pirA 5ʹUTR close to the ribosome binding site. Heterologous reporter assays confirmed that this interaction interferes with pirA translation. PirA negatively impacts arginine synthesis under ammonium excess by competing with the central carbon/nitrogen regulator PII that binds to and thereby activates the key enzyme of arginine synthesis, N-acetyl-L-glutamate-kinase (NAGK). Consistently, ectopic nsiR4 expression in Synechocystis resulted in lowered PirA accumulation in response to ammonium upshifts, which also affected intracellular arginine pools. As NsiR4 and PirA are inversely regulated by the global nitrogen transcriptional regulator NtcA, this regulatory axis enables fine tuning of arginine synthesis and conveys additional metabolic flexibility under highly fluctuating nitrogen regimes. Pairs of small protein inhibitors and of sRNAs that control the abundance of these enzyme effectors at the post-transcriptional level appear as fundamental building blocks in the regulation of primary metabolism in cyanobacteria.
Introduction
Nitrogen (N) is an essential component of key biomolecules. Given that combined N is a valuable resource, its energy-intensive uptake and incorporation into metabolism is a highly regulated process in bacteria [Citation1,Citation2]. One of the most thoroughly investigated N assimilatory pathways is the glutamine synthetase – glutamine:2-oxoglutarate-aminotransferase (GS-GOGAT) cycle in enterobacteria and most Gram-negatives (for a review see [Citation3]). In this cycle, GS enzyme activity is fine-tuned by a bicyclic cascade of covalent modifications [Citation4–7] that sensitizes the enzyme for feedback inhibition by a suite of molecules involved in N and energy metabolism [Citation8,Citation9]. Moreover, N assimilation is controlled at transcriptional level by the NtrC-NtrB two component system, which regulates the transcription of genes involved in N assimilation, for example, glnA that encodes GS [Citation10,Citation11].
Cyanobacteria are major drivers of the global C and N cycles as they perform oxygenic photosynthesis and many representatives also N2 fixation [Citation12–17]. Albeit cyanobacteria possess a GS, which is structurally similar to enterobacterial GS [Citation18], its regulation is fundamentally different. For example, cyanobacterial GS is devoid of covalent modifications [Citation19–21]. Instead, most cyanobacteria tune specific GS activity via protein–protein interaction by means of small inhibitory proteins, so-called inactivating factors (IFs) that are deployed depending on the N status of the cell [Citation22,Citation23]. As the widespread NtrB-NtrC system is absent in cyanobacteria, transcriptional regulation of N assimilatory genes is achieved via the global N regulator NtcA, which can act both as activator (e.g. in case of glnA) and repressor (e.g. in case of the genes gifA/B which encode two different IFs, IF7 and IF17) [Citation24,Citation25].
The binding affinity of NtcA to the respective promoters and hence, its activity as transcriptional regulator is determined in response to 2-OG metabolite levels via the major signalling protein PII, in particular the PII-PipX-NtcA regulatory axis [Citation26,Citation27]. In bacteria, 2-OG represents a proxy for monitoring the C/N balance of the cell as it provides the carbon framework required as a substrate for N assimilation and its abundance negatively correlates with N availability [Citation28]. As soon as N limitation occurs, 2-OG levels increase, which leads to a release of PipX from PII and complex formation with NtcA, thus activating it for DNA binding. As a result, NtcA fosters N scavenging by activating transcription of N assimilatory genes and by preventing transcription of genes encoding inhibitory proteins.
Beyond N assimilation, regulation of storage and mobilization is achieved by the interaction of PII with N-acetyl-glutamate-kinase (NAGK), catalysing the rate-limiting step of arginine synthesis [Citation29,Citation30]. Again, 2-OG levels determine the interaction of PII with NAGK. As NAGK is feedback inhibited by arginine, PII interaction with the enzyme, which is only permitted during N sufficiency, relieves NAGK from feedback inhibition, and in turn accelerates arginine synthesis [Citation27].
Recently, additional players involved in regulating cyanobacterial N metabolism have been discovered in Synechocystis sp. PCC 6803 (from here: Synechocystis). In particular, the role of small proteins as effectors for the activity of key enzymes receives more and more attention (for a review see [Citation31]). One such small protein is PirA (PII-interacting regulator of arginine synthesis) which has recently been shown to modulate arginine synthesis via competition with NAGK for PII binding [Citation32]. As binding of NAGK to the PII protein reduces the effect of feedback inhibition, PirA interferes with arginine accumulation and acts as indirect inhibitor of NAGK. Similar to gifA and gifB, which encode the two known proteinaceous GS inhibitors IF7 and IF17, expression of the pirA gene (Gene ID: ssr0692) increases with rising N availability due to relief from NtcA-mediated transcriptional repression [Citation24,Citation32]. Moreover, novel RNA-based mechanisms regulating N assimilation have been described in Synechocystis and other cyanobacteria [Citation33–35].
Besides proteinaceous regulators, bacteria possess diverse regulatory small RNAs (sRNAs) [Citation36]. Many of these sRNAs are only expressed during specific environmental conditions, thus pointing towards specific biological functions. These sRNAs hold great potential for the discovery of novel regulatory mechanisms and metabolic engineering for biotechnological applications [Citation37,Citation38]. Several conditionally regulated sRNAs have been identified in Synechocystis [Citation39]. One of these, the nitrogen stress induced RNA 4 (NsiR4) is strongly up-regulated under N limitation. NsiR4 was found to partially control the synthesis of IF7 via direct interaction with the gifA mRNA, thereby also controlling glutamine synthesis [Citation40]. The regulons of bacterial sRNAs typically contain multiple target genes and these targets often reveal a distinct physiological role of the respective sRNA. For instance, the sRNA CrfA of Caulobacter crescentus, which accumulates under C starvation, promotes the expression of a multitude of genes involved in adaption to C starvation by preventing target mRNA degradation [Citation41]. In Pseudomonas aeruginosa O1, the sRNA NalA (nitrogen assimilation leader A) transcribed directly upstream of the nitrate assimilation nirBD-PA1779-cobA operon was shown to be essential for transcription of the nitrate assimilation operon by means of transcriptional antitermination [Citation42]. In Synechocystis, the sRNAs PsrR1 and IsaR1 were shown to control sets of genes associated with photosynthesis upon shifts in light intensity [Citation43] or iron availability [Citation44]. In the light of these studies, it is very likely that NsiR4 regulates additional genes of N assimilation beyond gifA that are yet to be identified.
Here, we show that the pirA gene of Synechocystis is post-transcriptionally regulated by the small RNA NsiR4, and thereby clearly interferes with arginine metabolism. This extends the role of NsiR4 as a key regulator of N metabolism in cyanobacteria. NsiR4 coordinates N assimilation, storage and remobilization at the post-transcriptional level. Moreover, the NsiR4-pirA pair appears as paradigm for the function of modules consisting of a sRNA and one or several small protein regulators that affect major metabolic enzymes.
Material and methods
Strains and growth conditions
Synechocystis wild type and derivative strains were grown photoautotrophically at 30°C on Cu2+-free BG11 medium [Citation45], supplemented with 1 g l−1 NaHCO3 (BG11C) and bubbled with 1% (v/v) CO2 in air, under continuous illumination (50–70 µmol of photons m−2 s−1; 4000 K LED lights). Prior to the experiments investigating the impact of altered NsiR4 abundance the cultures were supplemented with 1 µM CuSO4. Ammonium treatment was carried out by addition of 10 mM NH4Cl and 20 mM N-tris(hydroxymethyl)-methyl-2-aminoethane-sulphonic acid (TES) buffer, pH 7.5. The recombinant Synechocystis strains used in this study were generated and described previously [Citation40].
Verification of sRNA:mRNA interaction
For the verification of sRNA:mRNA interactions in vivo a heterologous reporter system and the pXG10-SF plasmid were used as described [Citation46,Citation47]. Here, co-expression of a respective sRNA and an mRNA harbouring the 5ʹUTR of the potential target and an open reading frame encoding a reporter gene allows monitoring of the sRNA:mRNA interaction and its regulatory impact on gene expression. For the reporter assay E. coli TOP 10 were transformed with different plasmid combinations that harboured either a superfolder gfp gene transcriptionally fused to the 5ʹUTR of putative target mRNAs or respective sRNAs and negative controls (nonsense RNA). The primers used for cloning and the resulting plasmids are given in Supplementary Tables S1 and S2. Briefly, the entire 5ʹUTR of pirA containing the predicted NsiR4 interaction sequence and a part of the coding region were amplified from Synechocystis gDNA using the primer combinations ssr0692_5_NsiI/ssr0692_3_NheI. Further processing was performed as described [Citation40]. For every plasmid combination, six clones were picked to inoculate a liquid culture grown in LB medium with the corresponding antibiotics at 37°C overnight. A volume of 30 μl of each pre-culture were diluted in 170 μl fresh LB medium and fixed with 22 μl 10% Histofix (Roti-Histofix 10%, Carl Roth GmbH). The GFP fluorescence was measured with an Accuri C6 flow cytometer (BD Biosciences) as described [Citation48].
Western blot analysis
For the analysis of protein accumulation, crude extracts from 2 ml culture were prepared as previously described [Citation49]. Subsequently, proteins were separated via SDS-PAGE [Citation50] and transmitted to nitrocellulose membranes (Bio-Rad). Antibodies against PirA and the loading control TrxA [Citation51] were diluted 1:5,000 (anti-PirA) or 1:10,000 (anti-TrxA). Blocking, incubation and washing of membranes was performed as described [Citation32].
Determination of arginine
To monitor arginine levels, cells were cultivated in BG11 with conventional nitrate content until an OD750 of 0.8 was achieved. Shortly before and after the addition of 10 mM ammonium chloride cells were harvested by centrifugation and snap-frozen in liquid N2 followed by ethanolic extraction. Arginine was quantified via a high-performance liquid chromatography as described [Citation52].
Results
In a previous study, we performed transcript profiling of Synechocystis strains overexpressing the regulatory RNA NsiR4 [Citation40]. This study led to the confirmation of two different target genes, gifA and ssr1528, both of which showed reduced transcript levels due to the diminished loading with ribosomes caused by high coverage of NsiR4 with its respective binding sites on each mRNA. However, the entire NsiR4 regulon in Synechocystis is not known and likely comprises many other genes in addition to gifA and ssr1528, consistent with recent findings for the NsiR4 homolog in other cyanobacteria. In Nostoc sp. PCC 7120 NsiR4 also controls the expression of glpX encoding bifunctional sedoheptulose-1,7-bisphosphatase/fructose-1,6-bisphosphatase and pgk encoding phosphoglycerate kinase [Citation53]. One gene that also showed decreased transcript levels in response to pulse overexpression of NsiR4 in Synechocystis was ssr0692 [Citation40]. This gene was recently elucidated to encode the small protein PirA, a key regulator of arginine synthesis [Citation32]. While secondary effects could not be excluded, we speculated that the pirA gene might represent an additional NsiR4 target, especially considering the differential arginine accumulation kinetics of the NsiR4 overexpression strain [Citation40].
NsiR4 interacts with the 5ʹUTR of pirA and diminishes translation
To investigate whether NsiR4 bears the potential to bind the pirA mRNA, biocomputational RNA–RNA interaction prediction was performed utilizing the intaRNA tool [Citation54,Citation55]. This analysis yielded a complementing region of NsiR4 and the pirA 5ʹUTR which stretches from positions −4 to −18 upstream of the pirA start codon (see ). This arrangement most likely blocks access to the ribosomal binding site by NsiR4 and might interfere with pirA translation.
Figure 1. In vivo reporter assays for the verification of direct interaction between the 5ʹUTR of pirA and NsiR4. A: Computational interaction prediction between the 5ʹUTR of pirA and NsiR4 using the IntaRNA tool [Citation54,Citation55]. The numbers refer to the TSS +1. The start codon of pirA is highlighted in green while inserted mutations are highlighted in red. B: GFP fluorescence in E. coli TOP 10 strains with different plasmid combinations expressing NsiR4 or a nonsense RNA (plasmid pJV300) in presence of the 5’UTR of pirA which was fused to a sfgfp gene, as well as a negative control accounting for the autofluorescence of the cells (pXG0). It should be noted that the mutation, which was introduced into the pirA 5ʹUTR affected translation and hence, the GFP fluorescence detected for these constructs (column 5–6) was generally lower as observed for the WT version. Data are the mean ± SD of two independent experiments each using six independent cultures/clones. Asterisks label statistically different mean values, confirmed by single factor analysis of variance (ANOVA).
![Figure 1. In vivo reporter assays for the verification of direct interaction between the 5ʹUTR of pirA and NsiR4. A: Computational interaction prediction between the 5ʹUTR of pirA and NsiR4 using the IntaRNA tool [Citation54,Citation55]. The numbers refer to the TSS +1. The start codon of pirA is highlighted in green while inserted mutations are highlighted in red. B: GFP fluorescence in E. coli TOP 10 strains with different plasmid combinations expressing NsiR4 or a nonsense RNA (plasmid pJV300) in presence of the 5’UTR of pirA which was fused to a sfgfp gene, as well as a negative control accounting for the autofluorescence of the cells (pXG0). It should be noted that the mutation, which was introduced into the pirA 5ʹUTR affected translation and hence, the GFP fluorescence detected for these constructs (column 5–6) was generally lower as observed for the WT version. Data are the mean ± SD of two independent experiments each using six independent cultures/clones. Asterisks label statistically different mean values, confirmed by single factor analysis of variance (ANOVA).](/cms/asset/2d473ab5-0006-496f-9926-4b71a4dbc52e/krnb_a_2082147_f0001_oc.jpg)
To test whether NsiR4 interacts with the 5ʹUTR of pirA, heterologous reporter gene assays developed for RNA–RNA interaction verification in E. coli [Citation47] were performed. For that, the 5ʹUTR of pirA was fused to a superfolder gfp gene and co-expressed with NsiR4. Indeed, co-expression of NsiR4 caused a decrease in GFP fluorescence compared to a negative control omitting NsiR4 expression (). To test whether the observed reduction in GFP fluorescence was solely based on interaction via the predicted RNA–RNA binding region, the respective sequences were mutated in both NsiR4 and the pirA 5ʹUTR (see ) and different combinations were tested. Mutation of either NsiR4 or the pirA 5ʹUTR in combination with the WT version of the respective interaction partner prevented the reduction in fluorescence. However, the reduction in fluorescence was restored as soon as the two mutually compatible versions of mutated NsiR4 and pirA 5ʹUTR were combined (). These results unambiguously illustrated that NsiR4 binding to the 5ʹUTR of pirA is specific, occurs at the predicted site and interferes with protein synthesis. Given that post-transcriptional regulation of FBP/SBPase and PGK, both key players of the CBB cycle, by NsiR4 was recently demonstrated in the filamentous, diazotrophic strain Nostoc sp. PCC 7120 [Citation53] we also tested these interactions in Synechocystis. Surprisingly, utilizing the same reporter assays, none of the potential interaction partners of NsiR4 could experimentally be confirmed for any of the variants from Synechocystis. Thus, we tested mRNA:sRNA pairs from Nostoc sp. PCC 7120 as well. Indeed, we were able to confirm the interaction of this NsiR4 variant with pgk, contrasting the situation in Synechocystis (Supplementary Figure S1).
NsiR4 interferes with PirA accumulation in Synechocystis
At this stage, it remained obscure whether the confirmed interference of NsiR4 with pirA translation had any relevance in Synechocystis as the reporter assays were performed in E. coli and a heterologous system might lack factors required to achieve full control. To test if the determined interaction between NsiR4 and the pirA mRNA had any effect on the accumulation of the PirA protein in Synechocystis, a previously generated PirA-specific antiserum [Citation32] was used. In particular, we investigated three available strains [Citation40] in comparison to the WT, an NsiR4 deletion mutant (ΔnsiR4) and two strains ectopically expressing NsiR4 from a plasmid via the Cu2+-activated petE promoter, either in WT (NsiR4oex) or ΔnsiR4 background (ΔnsiR4::oex). PirA was described to strongly accumulate in response to ammonium shocks [Citation32]. Interestingly, compared to WT a delayed PirA accumulation in response to ammonium shock was observed for strain NsiR4oex (). By contrast, ΔnsiR4 showed a slightly higher accumulation of PirA 30 minutes after ammonium addition, which was clearly abrogated in strain ΔnsiR4::oex (). In the light of the reporter assays verifying interaction between NsiR4 and the 5ʹUTR of pirA, these results clearly demonstrate that post-transcriptional control by NsiR4 has biological relevance for PirA accumulation kinetics in vivo.
Figure 2. PirA accumulation kinetics in different NsiR4 mutant strains. A: Representative Western blots using antibodies specific against PirA [Citation32]. Equal loading is confirmed by the simultaneous detection of thioredoxin (TrxA) using specific TrxA antibodies. At time point 0, PirA accumulation was stimulated by the addition of 10 mM ammonium (provided as ammonium chloride) to cells that were pre-cultivated in BG11 supplemented with nitrate as sole N source. Prior to sampling all cultures were supplemented with 1 µM CuSO4 and further cultivated for 12 hours to ensure sufficient overexpression of NsiR4 in strains NsiR4oex and ΔnsiR4::oex as described previously [Citation40]. B: Densitometric analysis of the obtained signal intensity for PirA using ImageJ software. All values were normalized to the corresponding signal for TrxA of the same blot and time point. The data are shown as relative intensities compared to the signal for the WT at 360 min (set as 1).
![Figure 2. PirA accumulation kinetics in different NsiR4 mutant strains. A: Representative Western blots using antibodies specific against PirA [Citation32]. Equal loading is confirmed by the simultaneous detection of thioredoxin (TrxA) using specific TrxA antibodies. At time point 0, PirA accumulation was stimulated by the addition of 10 mM ammonium (provided as ammonium chloride) to cells that were pre-cultivated in BG11 supplemented with nitrate as sole N source. Prior to sampling all cultures were supplemented with 1 µM CuSO4 and further cultivated for 12 hours to ensure sufficient overexpression of NsiR4 in strains NsiR4oex and ΔnsiR4::oex as described previously [Citation40]. B: Densitometric analysis of the obtained signal intensity for PirA using ImageJ software. All values were normalized to the corresponding signal for TrxA of the same blot and time point. The data are shown as relative intensities compared to the signal for the WT at 360 min (set as 1).](/cms/asset/f5cd12e9-da38-4780-a0b9-f2795ac0a24a/krnb_a_2082147_f0002_oc.jpg)
NsiR4-dependent regulation of PirA accumulation affects arginine metabolism
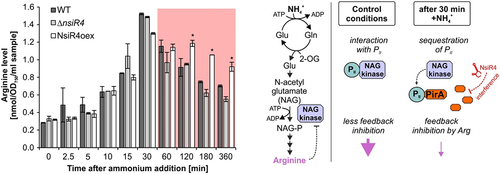
Our western blots revealed that the regulation of pirA by NsiR4 was effective at protein level. Therefore, we set out to determine if this interaction had any relevance for the metabolism of Synechocystis. PirA was recently shown to inhibit arginine synthesis by interaction with the regulatory PII protein [Citation32] thereby interfering with NAGK activity. Unfortunately, we were unable to perform reproducible in vivo measurement of NAGK activity, thus we monitored arginine accumulation after ammonium shock in the WT and strains ΔnsiR4 and NsiR4oex (). As expected, the internal arginine pool steadily increased in all strains within 30 min. Afterwards, the arginine level began to drop, likely due to the simultaneously accumulated PirA protein that interferes with NAGK activity. This is supported by the fact that a differential accumulation of arginine 60 minutes after ammonium addition was clearly observable for strain NsiR4oex when compared to WT and ΔnsiR4, matching the differential PirA protein accumulation determined by western blot (). As both WT and ΔnsiR4 featured a significantly increased PirA content compared to NsiR4oex after 60 minutes, these data nicely illustrate the regulatory impact of NsiR4 on arginine metabolism via post-transcriptional regulation of pirA.
Figure 3. Implications of enhanced NsiR4 accumulation on arginine metabolism. Left panel: Shown are arginine accumulation kinetics in WT and strains ΔnsiR4 and NsiR4oex in response to addition of 10 mM ammonium. Data are the mean ± SD of three independent replicates. Asterisks label significant deviation from the WT controls, confirmed by single factor analysis of variance (ANOVA). Right panel: Schematic of the effects of the NsiR4-PirA module on arginine synthesis by modulating NAGK activity before and after ammonium shock.
Discussion
The synthesis of arginine is a highly regulated process as this N-rich amino acid represents a central hub for N storage and redistribution in cyanobacteria [Citation56,Citation57]. Beyond its role as a proteinogenic amino acid, arginine serves as building block of N storage polymers, such as cyanophycin [Citation57], polyamines as well as combined N buffer [Citation58]. In cyanobacteria, the process of arginine synthesis is mainly regulated via its rate-limiting step, the reaction catalysed by NAGK which commits N-acetylglutamate towards arginine synthesis via irreversible phosphorylation. This enzyme is feedback-inhibited by arginine and is activated by the master regulator of C/N balancing, the PII protein, in a sophisticated regulatory interplay. This regulation integrates 2-OG levels and ATP/ADP ratios, that is, small-molecule effectors of N and energy metabolism, which are utilized to probe metabolic states of the cell [Citation29,Citation30,Citation59]. Given the vast N and energy requirements of arginine synthesis, it appears that the core principle of its tight regulation in cyanobacteria is to only permit synthesis when sufficient N and energy resources are available (for a review see [Citation58]). Consistently, the recently discovered small protein PirA accumulates in response to high ammonium concentrations and acts as a competitor of NAGK for PII binding in presence of sufficient concentrations of ADP as a measure of low cellular energy status [Citation32].
Interestingly, the ever growing number of characterized RNA regulators has corroborated assumptions that this class of regulators might surpass protein-based regulation in number and diversity [Citation60,Citation61]. Unfortunately, little is known about sRNAs that exert post-transcriptional control on genes involved in arginine metabolism. One of the few examples is the recently characterized 3ʹUTR-derived sRNA argX in Lactobacillus lactis, which modulates expression of the arc operon encoding arginine catabolism genes [Citation62]. The expression of this sRNA is directly regulated by arginine levels and it inhibits the synthesis of arginine diiminase by post-transcriptional regulation, thus directly contributing to control of arginine accumulation. Another example is the dual-function sRNA SR1 of Bacillus subtilis, with a multitude of homologs in closely related species, which serves both as regulatory RNA and messenger RNA. In its untranslated state, SR1 inhibits translation initiation of the ahrC mRNA, encoding an transcriptional activator of the arginine catabolic operons rocABC and rocDEF [Citation63,Citation64].
In this study, we show that the cyanobacterial sRNA NsiR4 provides another example as it controls the synthesis of the small protein PirA which in turn interferes with the activity of NAGK. This finding establishes NsiR4 as central regulatory element that coherently executes control on N entry via GS-GOGAT [Citation40] and N stockpiling via NAGK. Interestingly, NsiR4 does not target the corresponding genes directly but controls the expression of two regulatory proteins (IF7 and PirA) that in turn target the activity of these enzymes.
NsiR4 supports metabolic flexibility during fluctuating N supply via post-transcriptional control of small regulatory proteins targeting amino acid synthesis
Nitrogen is a vital nutrient and its bioavailable forms are subject to constant fluctuations in many natural environments. When cyanobacteria are exposed to N limitation, 2-OG levels rise and cause a dissociation of PII from NAGK, which enhances the effect of arginine feedback inhibition and in turn decreases enzyme activity [Citation29,Citation30,Citation58]. In Synechocystis, NsiR4 expression is strongly induced while both gifA and pirA genes are repressed by NtcA to promote transition from a resource-conserving to an N scavenging mode (see ). NsiR4 interferes with translation of its target mRNAs by base-pairing around the RBS, which interferes with ribosome binding and may also foster degradation of the corresponding transcript. This mode of regulatory interplay, the combination of a transcriptional regulator (NtcA) and an sRNA (NsiR4) which share a target protein (IF7 or PirA) are common to bacterial systems and in that case result in an multi-output AND-gated type 3 coherent feed-forward loop with a reversed sign-sensitive behaviour (see ) [Citation65]. This type of regulatory node allows a delayed response of target gene expression to a transcription factor off step, which in this case is the inactivation of NtcA during N excess (see ). As cells experience N oversupply, 2-OG levels decline and NtcA becomes inactivated by PII-mediated PipX interaction. This results in a de-repression of the gifA and pirA genes, while the NtcA-mediated activation of NsiR4 is abolished. As a result, residual NsiR4 partially prevents the translation of target mRNA in this transition state, causing delays in the expression of PirA and IF7 during shifts to N-excess conditions. Given that brackish water cyanobacteria such as Synechocystis thrive in habitats with strongly fluctuating N supply [Citation66], this mechanism could provide additional metabolic flexibility. Consistently, genomes of the marine genus Prochlorococcus which lack NsiR4 [Citation40] appear to also miss homologs of PirA and IF7 [Citation32,Citation67,Citation68]. As these genome-streamlined organisms flourish in nutrient-deficient ultraoligotrophic realms of the ocean, which lack strong fluctuations of nutrient availability, N-cost intensive regulatory systems that fine-tune N assimilation are not required [Citation69–71].
Figure 4. The current status of the regulatory network of NsiR4 in Synechocystis. A: Schematic overview of the NtcA-NsiR4-IF7-PirA network and its impact on N metabolism. The network has been built based on data from this study and previous work [Citation25,Citation32,Citation40]. B: Proposed model of the NtcA-NsiR4 feed-forward loop and its effect on the accumulation of corresponding gene products in response to fluctuating N supply (data based on [Citation25,Citation32,Citation40,Citation65,Citation67,Citation72]). The solid line represents target protein expression in a system omitting NsiR4, the dotted line describes a system including NsiR4. C: Regulatory relations between the different players of the NtcA-NsiR4-IF7-PirA network resulting in a multi-output AND-gated type 3 coherent feed-forward loop with a reversed sign-sensitive behaviour [Citation65].
![Figure 4. The current status of the regulatory network of NsiR4 in Synechocystis. A: Schematic overview of the NtcA-NsiR4-IF7-PirA network and its impact on N metabolism. The network has been built based on data from this study and previous work [Citation25,Citation32,Citation40]. B: Proposed model of the NtcA-NsiR4 feed-forward loop and its effect on the accumulation of corresponding gene products in response to fluctuating N supply (data based on [Citation25,Citation32,Citation40,Citation65,Citation67,Citation72]). The solid line represents target protein expression in a system omitting NsiR4, the dotted line describes a system including NsiR4. C: Regulatory relations between the different players of the NtcA-NsiR4-IF7-PirA network resulting in a multi-output AND-gated type 3 coherent feed-forward loop with a reversed sign-sensitive behaviour [Citation65].](/cms/asset/d20b98e9-917b-4ed9-abe1-0df6445e8d20/krnb_a_2082147_f0004_oc.jpg)
The NsiR4 regulon beyond cyanobacterial N metabolism
N metabolism is tightly linked to primary C metabolism mainly via 2OG, which is an intermediate of both GS-GOGAT and the tricarboxylic acid (TCA) cycle. Interestingly, based on in silico analyses NsiR4 was predicted to interact with several further mRNAs that encode central enzymes of primary C metabolism, for example, involved in the CBB cycle as well as glycogen synthesis (Supplementary Table S3). These included fructose-1,6-/sedoheptulose-1,7-bisphosphatase (FBP/SBPase), phosphoglycerate kinase (PGK), ADP-glucose pyrophosphorylase (AGPase) as well as central components of the photosynthetic electron transport chain that provides reduction equivalents for C metabolism. All these enzymes appear to represent central hubs determining specific routes of C metabolism. By targeting these reactions directly, it could be assumed that NsiR4 might co-determine C flux in response to varying C:N ratios in cyanobacteria. Indeed, in the study by Brenes-Álvarez and colleagues the biological impact of post-transcriptional regulation of the two CBB cycle enzymes FBP/SBPase and PGK by NsiR4 was demonstrated in the filamentous, diazotrophic strain Nostoc sp. PCC 7120 representing an additional link between N control and CO2 assimilation [Citation53]. Astonishingly, using all variants from Synechocystis, none of the potential interaction partners of NsiR4 could experimentally be confirmed using the same reporter assay. Therefore, we also tested the mRNA:sRNA pairs from Nostoc sp. PCC 7120. Indeed, interaction of this NsiR4 variant with pgk could be confirmed, which is in contrast to the situation in Synechocystis (Supplementary Figure S1). These results indicate that NsiR4 may need additional factors to interact with its targets in Synechocystis or has different target sets in the two strains.
Author contribution
Conceptualization, S.K., M.I.M.-P.; Original draft preparation, P.B.; Reporter assays, L.H.; Western blot analysis, M.I.M.-P.; Arginine accumulation kinetics; S.K.; Review & Editing, P.B., M.I.M.-P., L.H., W.R.H., F.J.F. and S.K.
Data availability
The data that support the findings of this study are available from the corresponding authors S.K. and M.I.M.P. upon reasonable request.
Supplemental Material
Download MS Word (150.6 KB)Acknowledgments
The project was funded by a grant of the German Science Foundation (DFG, grant KL3114/2-1 to S.K.), the graduate school GRK2344 MeInBio (grant 322977937 to LH and WRH) and PID2019-104513GB-100/AEI/10.13039/501100011033 from Agencia Estatal de Investigación (AEI) to F.J.F. and M.I.M.P. We thank Martin Hagemann (University of Rostock) for providing the analytics equipment for the amino acid quantification. We also acknowledge the use of the facilities of the Centre for Biocatalysis (MiKat) at the Helmholtz Centre for Environmental Research (UFZ). The UFZ is supported by the Helmholtz Association.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2022.2082147.
Additional information
Funding
References
- Zehr JP, Ward BB. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl Environ Microbiol. 2002;68(3):1015–1024.
- Forchhammer K. Glutamine signalling in bacteria. Front Biosci J Virtual Libr. 2007;12(1):358–370.
- Stadtman ER. The story of glutamine synthetase regulation. J Biol Chem. 2001;276(48):44357–44364.
- Brown MS, Segal A, Stadtman ER. Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the PII-regulatory protein. Proc Natl Acad Sci. 1971;68:2949–2953.
- Mangum JH, Magni G, Stadtman ER. Regulation of glutamine synthetase adenylylation and deadenylylation by the enzymatic uridylylation and deuridylylation of the PII regulatory protein. Arch Biochem Biophys. 1973;158(2):514–525.
- Anderson WB, Hennig SB, Ginsburg A, et al. Association of ATP: glutamine synthetase adenylyltransferase activity with the P1 component of the glutamine synthetase deadenylylation system. Proc Natl Acad Sci. 1970;67(3):1417–1424.
- Anderson WB, Stadtman ER. Glutamine synthetase deadenylylation: a phosphorolytic reaction yielding ADP as nucleotide product. Biochem Biophys Res Commun. 1970;41(3):704–709.
- Woolfolk CA, Stadtman ER. Cumulative feedback inhibition in the multiple end product regulation of glutamine synthetase activity in Escherichia coli. Biochem Biophys Res Commun. 1964;17(4):313–319.
- Woolfolk CA, Stadtman ER. Regulation of glutamine synthetase: III. Cumulative feedback inhibition of glutamine synthetase from Escherichia coli. Arch Biochem Biophys. 1967;118(3):736–755.
- Weiss V, Kramer G, Dünnebier T, et al. Mechanism of regulation of the bifunctional histidine kinase NtrB in Escherichia coli. J Mol Microbiol Biotechnol. 2002;4(3):229–233.
- Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol Rev. 1995;59(4):604–622.
- Flombaum P, Gallegos JL, Gordillo RA, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci. 2013;110(24):9824–9829.
- Montoya JP, Holl CM, Zehr JP, et al. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature. 2004;430(7003):1027–1032.
- Canfield DE, Des Marais DJ. Biogeochemical cycles of carbon, sulfur, and free oxygen in a microbial mat. Geochim Cosmochim Acta. 1993;57(16):3971–3984.
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive earth’s biogeochemical cycles. Science. 2008;320(5879):1034–1039.
- Falkowski PG, Barber RT, Smetacek V. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281(5374):200–206.
- Capone DG, Zehr JP, Paerl HW, et al. Trichodesmium, a globally significant marine cyanobacterium. Science. 1997;276(5316):1221–1229.
- Fisher R, Tuli R, Haselkorn R. A cloned cyanobacterial gene for glutamine synthetase functions in Escherichia coli, but the enzyme is not adenylylated. Proc Natl Acad Sci U S A. 1981;78(6):3393–3397.
- Mérida A, Leurentop L, Candau P, et al. Purification and properties of glutamine synthetases from the cyanobacteria Synechocystis sp. strain PCC 6803 and Calothrix sp. strain PCC 7601. J Bacteriol. 1990;172(8):4732–4735.
- Orr J, Haselkorn R. Regulation of glutamine synthetase activity and synthesis in free-living and symbiotic Anabaena spp. J Bacteriol. 1982;152(2):626–635.
- Stacey G, Tabita FR, Baalen CV. Nitrogen and ammonia assimilation in the cyanobacteria: purification of glutamine synthetase from Anabaena sp. strain CA. J Bacteriol. 1977;132(2):596–603.
- Reyes JC, Florencio FJ. A novel mechanism of glutamine synthetase inactivation by ammonium in the cyanobacterium Synechocystis sp . PCC 6803. Involvement of an inactivating protein. FEBS Lett. 1995;367(1):45–48.
- García-Domínguez M, Reyes JC, Florencio FJ. Glutamine synthetase inactivation by protein–protein interaction. Proc Natl Acad Sci. 1999;96(13):7161–7166.
- Giner-Lamia J, Robles-Rengel R, Hernández-Prieto MA, et al. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017;45. 11800–11820.
- García-Domínguez M, Reyes JC, Florencio FJ. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol Microbiol. 2000;35:1192–1201.
- Espinosa J, Rodríguez-Mateos F, Salinas P, et al. PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc Natl Acad Sci U S A. 2014;111(23):E2423–2430.
- Forcada-Nadal A, Llácer JL, Contreras A, et al. The PII-NAGK-PipX-NtcA regulatory axis of Cyanobacteria: a tale of changing partners, allosteric effectors and non-covalent interactions. Front Mol Biosci. 2018;5:91.
- Leigh JA, Dodsworth JA. Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol. 2007;61(1):349–377.
- Burillo S, Luque I, Fuentes I, et al. Interactions between the nitrogen signal transduction protein PII and N -acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J Bacteriol. 2004;186(11):3346–3354.
- Heinrich A, Maheswaran M, Ruppert U, et al. The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-l-glutamate kinase. Mol Microbiol. 2004;52(5):1303–1314.
- Brandenburg F, Klähn S. Small but smart: on the diverse role of small proteins in the regulation of cyanobacterial metabolism. Life. 2020;10(12):322.
- Bolay P, Rozbeh R, Muro-Pastor MI, et al. The novel PII-interacting protein PirA controls flux into the cyanobacterial ornithine-ammonia cycle. mBio. 2021;12(2):e00229–21.
- Ionescu D, Voss B, Oren A, et al. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J Mol Biol. 2010;398(2):177–188.
- Muro-Pastor AM. The heterocyst-specific NsiR1 small RNA is an early marker of cell differentiation in cyanobacterial filaments. mBio. 2014;5(3):e01079–14.
- Klähn S, Bolay P, Wright PR, et al. A glutamine riboswitch is a key element for the regulation of glutamine synthetase in cyanobacteria. Nucleic Acids Res. 2018;46(19):10082–10094.
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628.
- Sun D, Chen J, Wang Y, et al. Metabolic engineering of Corynebacterium glutamicum by synthetic small regulatory RNAs. J Ind Microbiol Biotechnol. 2019;46(2):203–208.
- Na D, Yoo SM, Chung H, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol. 2013;31(2):170–174.
- Kopf M, Hess WR. Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol Rev. 2015;39(3):301–315.
- Klähn S, Schaal C, Georg J, et al. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci U S A. 2015;112(45):E6243–6252.
- Landt SG, Lesley JA, Britos L, et al. CrfA, a small noncoding RNA regulator of adaptation to carbon starvation in Caulobacter crescentus. J Bacteriol. 2010;192(18):4763–4775.
- Romeo A, Sonnleitner E, Sorger-Domenigg T, et al. Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology. 2012;158(6):1543–1552.
- Georg J, Dienst D, Schürgers N, et al. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in Cyanobacteria. Plant Cell. 2014;26(9):3661–3679.
- Georg J, Kostova G, Vuorijoki L, et al. Acclimation of oxygenic photosynthesis to iron starvation is controlled by the sRNA IsaR1. Curr Biol CB. 2017;27(10):1425–1436.e7.
- Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988 January 1 ed., 1988;167:3–27.
- Corcoran CP, Podkaminski D, Papenfort K, et al. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol. 2012;84(3):428–445.
- Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35(3):1018–1037.
- Wright PR, Richter AS, Papenfort K, et al. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci U S A. 2013;110(37):E3487–3496.
- Reyes JC, Crespo JL, Garcia-Dominguez M, et al. Electron transport controls glutamine synthetase activity in the facultative heterotrophic cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 1995;109. 899–905.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature. 1970;227(5259):680–685.
- Navarro F, Martín-Figueroa E, Florencio FJ. Electron transport controls transcription of the thioredoxin gene trxA in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 2000;43:23–32.
- Hagemann M, Vinnemeier J, Oberpichler I, et al. The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Biol Stuttg Ger. 2005;7(1):15–22.
- Brenes-Álvarez M, Olmedo-Verd E, Vioque A, et al. A nitrogen stress-inducible small RNA regulates CO2 fixation in Nostoc. Plant Physiol. 2021;187(2):787–798.
- Busch A, Richter AS, Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinforma Oxf Engl. 2008;24(24):2849–2856.
- Wright PR, Georg J, Mann M, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42(W1):W119–W123.
- Llácer JL, Fita I, Rubio V. Arginine and nitrogen storage. Curr Opin Struct Biol. 2008;18(6):673–681.
- Flores E, Arévalo S, Burnat M. Cyanophycin and arginine metabolism in cyanobacteria. Algal Res. 2019;42:101577.
- Forchhammer K, Selim KA. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol Rev. 2020;44(1):33–53.
- Llácer JL, Contreras A, Forchhammer K, et al. The crystal structure of the complex of PII and acetylglutamate kinase reveals how P controls the storage of nitrogen as arginine. Proc Natl Acad Sci U S A. 2007;104(45):17644–17649.
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43(6):880–891.
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3(12):a003798.
- van der Meulen SB, Hesseling-Meinders A, de Jong A, et al. The protein regulator ArgR and the sRNA derived from the 3’-UTR region of its gene, ArgX, both regulate the arginine deiminase pathway in Lactococcus lactis. PloS One. 2019;14(6):e0218508.
- Gimpel M, Preis H, Barth E, et al. SR1—a small RNA with two remarkably conserved functions. Nucleic Acids Res. 2012;40(22):11659–11672.
- Heidrich N, Chinali A, Gerth U, et al. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol Microbiol. 2006;62(2):520–536.
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci. 2003;100(21):11980–11985.
- Reed RH, Warr SRC, Richardson DL, et al. Multiphasic osmotic adjustment in a euryhaline cyanobacterium. FEMS Microbiol Lett. 1985;28(3):225–229.
- Bolay P, Muro-Pastor MI, Florencio FJ, et al. The distinctive regulation of cyanobacterial glutamine synthetase. Life Basel Switz. 2018;8(4): 52
- Saelices L, Galmozzi CV, Florencio FJ, et al. Mutational analysis of the inactivating factors, IF7 and IF17 from Synechocystis sp. PCC 6803: critical role of arginine amino acid residues for glutamine synthetase inactivation. Mol Microbiol. 2011;82(4):964–975.
- Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6(2):R14.
- Giovannoni SJ, Thrash JC, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8(8):1553–1565.
- Steglich C, Futschik ME, Lindell D, et al. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLOS Genet. 2008;4(8):e1000173.
- Muro-Pastor MI, Reyes JC, Florencio FJ. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J Biol Chem. 2001;276(41):38320–38328.
