ABSTRACT
Delicate variances in the translational machinery affect how efficiently different organisms approach protein synthesis. Determining the scale of this effect, however, requires knowledge on the differences of mistranslation levels. Here, we used a dual-luciferase reporter assay cloned into a broad host range plasmid to reveal the translational fidelity profiles of Pseudomonas putida, Pseudomonas aeruginosa and Escherichia coli. We observed that these profiles are surprisingly different, whereas species more prone to translational frameshifting are not necessarily more prone to stop codon readthrough. As tRNA modifications are among the factors that have been implicated to affect translation accuracy, we also show that translational fidelity is context-specifically influenced by pseudouridines in the anticodon stem-loop of tRNA, but the effect is not uniform between species.
Introduction
Translation is a well-conserved process responsible for accurately synthesizing proteins based on genetic information and necessary for all cellular processes. As demand for proteins in a cell is high, trade-off between speed and accuracy leads to some aberrations in nascent peptides, either because of errors during decoding or failure to maintain the correct reading frame. Missense error frequency during translation is usually lower than 1/1000 [Citation1–3], whereas frameshift errors are by two orders of magnitude less frequent [Citation4]. However, misincorporation of amino acids and programmed frameshift can occur in special cases at frequency as high as 10–20% [Citation2,Citation5]. Translation accuracy has been measured at quantitative level mostly in few model species, such as E. coli and Saccharomyces cerevisae, and it is evident that it varies significantly between different organisms [Citation2]. Differences in translational fidelity can shape the proteome and contribute to the adaptation with stressful conditions, however, according to our knowledge, no data for systematic comparison of error frequency of different bacterial species is available.
tRNA is involved in several cellular processes in addition to its central role in translation of genetic message. During translation tRNA molecules transfer activated amino acids from aminoacyl-tRNA synthetases to the ribosome for the mRNA directed protein synthesis. Non-canonical roles of tRNAs beyond translation involve biosynthesis of amino acids, modification of peptides and lipids, nucleotide alarmone synthesis, regulation of gene expression, etc., and the list of tRNA mediated cellular functions is still growing [Citation6]. tRNA is the most heavily modified RNA molecule in bacteria; about 10% of its nucleotides contain chemical modifications [Citation7]. The modifications are often found at certain regions, e.g. anticodon stem and loop structure (ASL), and have been implicated to determine codon reading specificity and translation fidelity [Citation8–11].
A large variety of modified nucleotides are found at position 34 (first position of anticodon) of elongator tRNAs. Nucleotide at the position 37, just 3´ of anticodon, contains complex modifications, which are involved in stabilizing codon – anticodon pairing and in improvement of reading frame maintenance [Citation10,Citation12]. Another set of modifications bracketing the anticodon loop are pseudouridines (positions 32, 38–40). These understudied Ψs are found in a subset of tRNA species and are introduced into tRNAs by pseudouridine synthases. Deletion of enzyme RluA gene leads to loss of isomerization of U746 of 23S rRNA and U32 in four E. coli tRNA species [Citation13]. Uridine residues at tRNA positions 38–40 are not isomerized in about half of tRNA species in the absence of TruA [Citation14,Citation15]. Similar loss of uridine isomerization around tRNA ASL is observed in P. putida upon deletion of RluA and TruA genes [Citation16]. Both TruA and RluA have been suggested to have a tRNA chaperone activity [Citation17]. In spite of the possible double function both genes are nonessential in E. coli [Citation18]. Loss of TruA function leads to inefficient reading of consecutive codons by its substrate tRNAs [Citation19] and decreased mistranslation at His codons under histidine starvation [Citation20]. Increased frameshifting for Ψ38-deficient tRNALeu has been observed [Citation21]. These results indicate that TruA has a function in maintaining translational fidelity. Systematic analysis of TruA in respect of codon reading accuracy and reading frame maintenance is still missing.
Structural and comparative studies have revealed that the first and the last nucleobase of the anticodon loop (bases 32 and 38) tend to form a non-Watson-Crick pair with a single hydrogen bond [Citation22]. This interaction can be facilitated by a pseudouridine, as it forms more stable base pairs and stacking interactions compared to the unmodified uridine [Citation23–25]. Interaction between bases 32 and 38 is transiently lost during +1 frameshifting [Citation26,Citation27]. Thus, pseudouridine at position 38/39 (isomerized by TruA) or 32 (isomerized by RluA) could help to keep codon reading frame by local stabilization of anticodon loop structure. If this is true, deletion of TruA or RluA is expected to increase frameshifting frequency. We aimed to test this possibility by combining TruA or RluA deletion strains of three different bacterial species with a series of frameshift and stop codon reporters.
So far, E. coli has served as the primary model in investigating molecular mechanisms affecting translation accuracy in bacteria. The genus Pseudomonas represents one of the largest group of bacteria that are known for their versatility and adaptability in hostile and fluctuating habitats [Citation28]. Pseudomonas putida, e.g., is found mostly in temperate soil and water habitats. The metabolic versatility and high tolerance to toxic and harsh conditions makes P. putida attractive for biotechnological applications such as biodegradation of environmental pollutants and synthesis of added-value chemicals [Citation29–31]. Pseudomonas aeruginosa, on the other hand, although found widely in the environment, is mostly known as an opportunistic hospital-acquired human pathogen, responsible for both chronic and acute infections [Citation32,Citation33].
Recently we have demonstrated that the tRNA modification enzymes TruA and RluA in soil bacterium P. putida possess similar targets as have been shown in E. coli and the absence of said enzymes increases mutation frequency in P. putida via lack of tRNA pseudouridylation [Citation16,Citation34]. Loss of translation fidelity has been suggested to affect error frequency of DNA replication [Citation35–37]. In this respect we wanted to analyse whether the deletion of pseudouridine synthases TruA and RluA influences the translation fidelity and frameshifting in particular. Comparative analysis of translation error frequency was performed in three bacterial species Pseudomonas putida, Pseudomonas aeruginosa and Escherichia coli using the same reporter constructs encoded by a shuttle vector.
Results
Translational accuracy within the same genetic context differs between Pseudomonas putida, Pseudomonas aeruginosa and Escherichia coli
To estimate error frequency of translation in three bacterial species, we exploited a dual-luciferase assay system. This assay is based on a fusion protein consisting of two luciferases (Rluc and Fluc). Test sequence containing slippery signal is inserted between the two cistrons. In this way Fluc is synthesized only after a frameshift event has occurred during mRNA translation. Thanks to the sensitivity and internal control of the system it is an efficient tool for analysis of translation accuracy at various sequence contexts. This reporter (), with its different derivatives carrying mutations in the linker-region to detect translational frameshifting or premature stop codons in the fluc gene to detect stop codon readthrough [Citation38], was inserted into a broad host range plasmid pSEVA/lacItac [Citation16] to allow its simultaneous usage in three bacterial species: P. putida, P. aeruginosa and E. coli.
Figure 1. Different constructs of the dual-luciferase assay system used to measure the translational fidelity. Rluc (blue) and Fluc (green) form a fusion protein connected by a short linker region. Nucleotide and amino acid sequence of the linker region is shown with adjacent regions of Rluc and Fluc highlighted. Different insertions and deletions introduced into the system to measure frameshifting are shown with red lines above the sequence, premature stop codons with their positions are shown below.
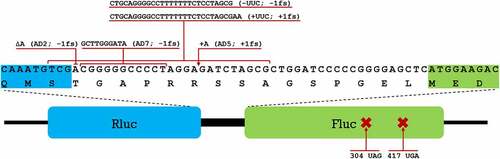
Using a set of plasmids encoding different derivatives of the Rluc-Fluc assay system, we measured the translational fidelity in seven different genetic contexts () to compare the basic translation fidelity profiles of the three bacterial species. In total there were 5 frameshifts, of which AD2 (−1 FS), AD5 (+1 FS) and AD7 (−1FS) have been described previously [Citation38], whereas slippery site containing -UUC (−1FS) and +UUC (+1FS) were constructed in this work. Stop codon readthrough was observed using plasmids where fluc gene contained premature stop codons at positions 304 (304 UAG) and 417 (417 UGA) [Citation38]. Detailed explanation with corresponding codons and tRNAs targeted by TruA and RluA within each of these contexts is available in supplementary materials (Tables S1-S8).
Results of the dual-luciferase assays in wild-type bacterial species, as the basic translation fidelity profiles of the three bacterial species, are shown in . Firstly, it is evident that error profiles of the three species are notably different. Translation error frequency in P. putida cells varies significantly, depending on the codon context and the type of error-prone sequence. Out of three −1 FS reporters UUC exhibited significantly higher frameshifting as compared to AD2 and AD7. +1 FS in the AD5 context was the most frequent of all frameshift events tested, occurring in over 15% of the cases when Rluc was translated, about two times or more frequently than other frameshift errors. −1 FS reporter AD2 has similar sequence as AD5 except an A is deleted while in AD5 an A is inserted (). FS frequency was more than an order of magnitude higher in AD5 as compared to AD2. In contrast, at the slippery sequence UUC, −1 and +1 frameshift occurred with nearly equal frequency. UGA stop codon was suppressed more often than UAG, however, compared to translational frameshifting, stop codon readthrough levels were orders of magnitude lower, at 0.08% (UGA) and 0.01% (UAG) compared to 0.6% (AD2, −1FS) to 17% (AD5, +1FS).
Figure 2. Translation errors among different genetic contexts (shown on the x axis) in wild-type strains of three bacterial species: Pseudomonas putida strain PaW85 (depicted in green), Pseudomonas aeruginosa strain PAO1-L (Orange) and Escherichia coli strain MG1655 (yellow). For the frameshift events both the name used to refer to specific context and the direction (either −1 or +1 frameshift) is shown; for the stop codon readthrough events the position of the codon within the assay system that has been mutated to produce a premature stop codon and the resulting stop codon are shown. In all cases n ≥ 11. Boxplots show the Fluc/Rluc values normalized against the Fluc/Rluc value of an unmutated (wild-type) test system in the corresponding species. Line in the box denotes the median value (also shown in table under the graph), the upper and lower borders of the box represent first and third quartile, the whiskers show the non-outlier range and circles are outliers.
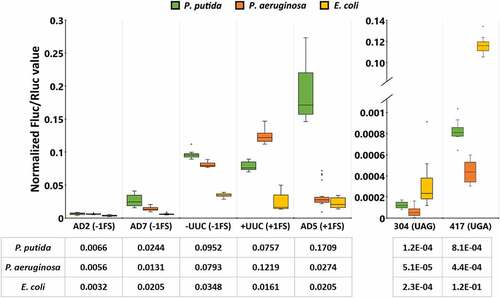
In P. aeruginosa, translational frameshifting occurred with low frequency except at the slippery sequence (UUC) when +1 frameshifting took place at a higher frequency – around 12% of all translation events, compared to 8% in the case of −1 FS. Translational readthrough at stop codons was once again very rare, with readthrough event taking place at a frequency of 1/2500 (UGA) or 1/20,000 (UAG) ().
In E. coli, translational frameshifting stayed below 5% in all reporters. UGA stop codon suppression level was around 12%, 500 times higher than UAG stop codon suppression (). Compared to P. putida and P. aeruginosa, UGA suppression was 148- or 272-fold higher, respectively.
Although the translational machinery is well conserved in all prokaryotes, significant variation still exists between species regarding errors made during protein synthesis. P. putida appeared to be slightly more error-prone than P. aeruginosa and E. coli, at least in the investigated contexts, however no clear and universal patterns emerged. Both Pseudomonas species exhibited frequent frameshifting at UUC slippery sites in both directions. Interestingly, while E. coli tended to be more frameshift-resistant across all studied contexts, it exhibited higher stop codon readthrough rate on both UAG and especially on UGA stop codons, than in P. putida and P. aeruginosa.
Effect of tRNA anticodon stem-loop pseudouridines on mistranslation
In order to detect whether the lack of pseudouridines in the ASL has an effect on translational fidelity, pseudouridine synthases-deficient ∆truA and ∆rluA strains of P. putida, P. aeruginosa and E. coli were analysed with the Rluc-Fluc assay system.
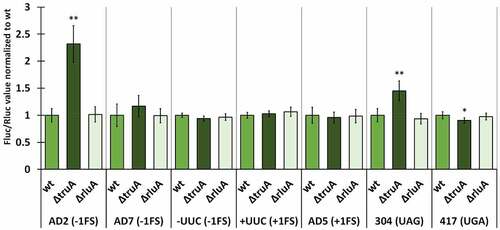
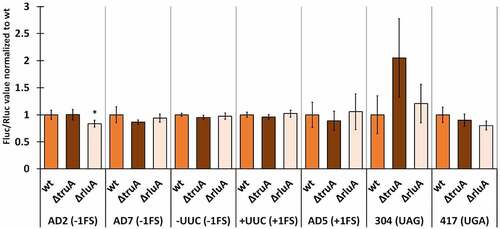
We normalized the Fluc/Rluc ratios of the deletion strains to the mean of Fluc/Rluc ratio of the respective wild-type strain (same reporter derivate). In P. putida, the lack of TruA increased −1 frameshifting in the AD2 context ~2.2 times and UAG stop codon readthrough ~1.4 times, both effects being statistically significant (, Table S9). In other cases, translation fidelity was similar to that observed in wild-type bacteria, even though slightly decreased in the case of UGA stop codon (0.9-fold). P. putida strain deficient in RluA displays comparable translational accuracy to the wild-type strain with reporters containing codons that are decoded by RluA substrate tRNAs (AD7 and UUC).
Figure 3. Effect of pseudouridines on translation error frequency using ΔtruA and ΔrluA strains relative to wild-type in P. putida PaW85. Deletion strains have been normalized against the wild-type strain within each genetic context. Error bars represent CI 95%, ‘*’ indicates p-value <0.05, ‘**’ indicates p-value <0.001 compared to the respective wild-type, n ≥ 11.
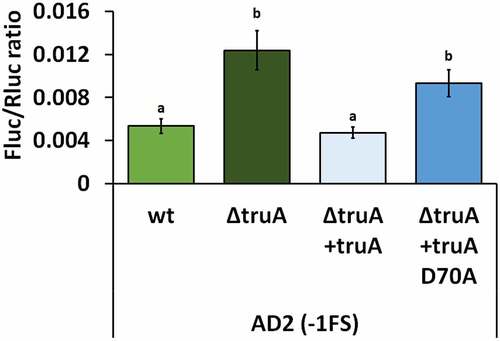
As catalytically inactive Ψ synthases can retain their substrate binding ability and in the case of TruB it has been shown that in E. coli the disruption of substrate binding ability affects the growth rate more than the disruption of pseudouridylation activity [Citation39], we set out to determine, whether the effect on translational fidelity was due to missing pseudouridines from the tRNA or some other, possibly unknown, function of TruA. Catalytic activity of TruA has been shown to stem from aspartatic acid residue, in P. putida this is D70 [Citation16,Citation40]. When the P. putida ∆truA strain was chromosomally complemented with the functional truA gene (∆truA+truA), the −1 FS frequency at AD2 reporter was reduced, while complementation with a catalytically inactive TruA (∆truA+truA D70A) did not rescue frameshifting (). Thus, high −1 FS at AD2 in P. putida was indeed due to the absence of Ψ-residues from the anticodon stem-loop of tRNAs.
Figure 4. Frequency of translation errors in P. putida wild-type strain (wt), truA deletion strain (∆truA), truA deletion strain complemented with functional truA (∆truA+truA), and truA deletion strain complemented with catalytically inactive truA (∆truA+truA D70A). Letters a and b indicate homogeneity groups, different letters denote a statistically significant difference (p-value <0.001), n ≥ 12.
In P. aeruginosa cells, lack of pseudouridines in the ASL had no significant effect on translational fidelity with one exception. UAG stop codon readthrough was increased in the absence of TruA, resulting in a 2-fold effect, similarly to P. putida. However, the effect was not statistically significant (Table S10). Lack of RluA slightly decreased (0.83-fold) −1 frameshifting on the AD2 sequence, but otherwise ΔrluA strain performed similarly to the wild-type P. aeruginosa ().
Figure 5. Effect of pseudouridines on translation error frequency using ΔtruA and ΔrluA strains relative to wild-type in P. aeruginosa PAO1-L. Deletion strains have been normalized against the wild-type strain within each genetic context. Error bars represent CI 95%, ‘*’ indicates p-value <0.05 compared to the respective wild-type, n ≥ 11.
In E. coli, translational fidelity was affected by the lack of either pseudouridine synthase in nearly all tested reporter constructs (, statistical significance presented in Table S11). TruA-deficient strain exhibited a ~ 1.5-fold increase in −1 frameshifting in all genetic contexts (AD2, AD7 and -UUC) while +1 frameshifting was affected in the AD5 context (1.3-fold) but not in +UUC context (). Readthrough was also increased at both stop codons, ~2.2 times at UAG and ~1.3 times at UGA. Interestingly, RluA-deficient strain exhibited decreased frameshifting and stop codon readthrough compared to wild-type in AD2 context (0.8-fold) and both stop codons (0.7-fold – UAG; 0.8-fold – UGA). In addition to this, −1 frameshifting at AD7 sequence was increased ~1.2 times in ∆rluA strain in comparison to wild-type ().
Figure 6. Effect of pseudouridines on translation error frequency using ΔtruA and ΔrluA strains relative to wild-type in E. coli MG1655. Deletion strains have been normalized against the wild-type strain within each genetic context. Error bars represent CI 95%, ‘*’ indicates p-value <0.05, ‘**’ indicates p-value <0.001 compared to the respective wild-type, n ≥ 11.
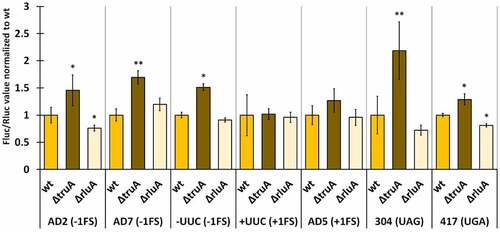
Taken together, only the increased UAG stop codon readthrough in the absence of TruA emerged as a universal characteristic when comparing the fidelity profiles between the pseudouridine-deficient strains of all three species. It was also evident that among the three species, the lack of TruA has a bigger effect on translational fidelity than the lack of RluA. Moreover, their effect was opposite, as translational errors were increased in ∆truA strains and decreased in ∆rluA strains. Lack of either pseudouridine synthase affected the translational fidelity in E. coli more than in P. putida and P. aeruginosa. Interestingly, on the AD2 sequence, the Pseudomonas sp. bacteria, which are more related and share more similarities in translational machinery and codon usage to each other than either does to E. coli, performed in dissimilar manner to each other, as P. aeruginosa was unaffected by the lack of TruA, while the translational fidelity was reduced in the TruA-deficient strains in P. putida and E. coli.
Discussion
Variation of mistranslation profiles between the investigated species
Programmed ribosomal frameshifting allows production of several proteins from overlapping coding sequences. It is a necessary event for propagation of some viruses [Citation41,Citation42], although programmed frameshift on genomic genes has also been described in prokaryotes [Citation43,Citation44] and eukaryotes [Citation45]. Translocation to the new reading frame is usually directed at specific sites by secondary structures in the mRNA. Spontaneous frameshifting taking place at random locations, however, leads to synthesis of aberrant or truncated polypeptide sequences. In E. coli spontaneous ribosomal frameshifting as a translational error has been shown to occur at an efficiency of up to 16% at argI mRNA [Citation46]. While studies regarding translational fidelity are common, they are generally focused on measuring translational error signals in a single species, thus making it difficult to assess whether the frequency of the effect is species-specific or common for other bacteria as well. Eukaryotic systems have been previously compared [Citation47], but prokaryotic studies have centred on a single species, usually E. coli. Whether and to what extent translational fidelity can vary between bacterial species has not been thoroughly studied.
In the current study we show that frequency of mistranslation events, frameshifting and stop codon readthrough, are markedly different in three bacterial species. The three investigated species all belong to the class of Gammaproteobacteria but occupy different habitats in nature. P. putida is a soil bacterium [Citation29], while P. aeruginosa (belonging to the same genus) is mostly known as an opportunistic human pathogen [Citation48]. Facultative anaerobe E. coli is well-known prokaryotic model organism, commonly found in lower intestine of warm-blooded animals [Citation49].
From our experiments it is seen that frameshifting is more frequent in Pseudomonas species. In particular, +1 FS is an order of magnitude higher in P. putida in comparison to P. aeruginosa and E. coli (). Both −1 and +1 FS frequency at UUC slippery site is 2–7 times higher in Pseudomonas species than in E. coli. Stop codon readthrough, on the other hand, is more frequent in E. coli. One reason behind this variation in error frequency between bacterial species can be modification pattern of tRNA around ASL. The results of these studies will be discussed below.
UGA translational readthrough is a known phenomenon in E. coli, occurring at a frequency of up to 20% [Citation50], which is consistent with our experiments where we measured the readthrough efficiency at 12% (). During UGA stop codon readthrough either tryptophan or selenocysteine is incorporated into the growing polypeptide chain. As a SECIS element required for the selenocysteine recoding event is not present in our Rluc-Fluc mRNA, we can presume that UGA stop codon is likely to be suppressed by tRNATrp. In E. coli, the tRNATrp is hypermodified at the position 37 by enzymes MiaA (A to i6A) and MiaB (i6A to ms2i6A) [Citation51]. In P. putida enzyme MiaE has been identified, which further modifies ms2i6A to ms2io6A [Citation52]. Based on genome annotation, MiaE is also present in P. aeruginosa (strain PAO-1) [Citation53], but absent from E. coli (strain K-12) [Citation54]. tRNA position 37 modifications have been shown to affect readthrough as lack of MiaB decreases UGA readthrough in E. coli [Citation55]. Therefore, it is possible that the significant difference regarding UGA readthrough in our experiments between E. coli (12%) and Pseudomonas sp. (0.08% in P. putida, 0.04% in P. aeruginosa) could be due to differences in modifications at the position 37 of tRNATrp in Pseudomonas sp. and E. coli (). Based on our results, UAG stop codon is suppressed at a lower rate than UGA in all of the investigated species, which is consistent with what has been previously reported in E. coli [Citation50].
Effect of Ψ32/38–40 on mistranslation
Pseudouridines in the ASL of tRNAs have been previously shown to affect reading frame maintenance during translation. Interestingly, while +1 frameshifting was increased in Salmonella enterica serovar Typhimurium cells lacking Ψ38–40 [Citation10,Citation56], in S. cerevisiae stop codon readthrough and +1 frameshifting were decreased in cells missing Ψ38–39 [Citation57]. In our experiments, lack of Ψ residues in the ASL affects translational frameshifting in three bacterial species in a different way (). Specifically, −1 FS frequency in the AD2 context in ∆truA strains is increased 2.2-fold in P. putida and 1.5-fold in E. coli. ∆truA strain of E. coli has elevated −1 FS frequency in all three reporters. P. putida ∆truA strain has increased −1 FS only in AD2 context and in P. aeruginosa the absence of TruA does not seem to affect −1 FS frequency at all. It is important to note that wild-type E. coli has a lower level −1 FS as compared to Pseudomonas species. Absence of Ψ residues does not cause altered +1 frameshifting in any of the investigated bacterial species, which is different from what has been reported in S. enterica strain missing Ψ38–40 [Citation10] demonstrating importance of the specific sequence context. There is no simple explanation why −1 FS frequency depends on the presence of Ψ residues at positions 38–40 of ASL. Structural studies on the E. coli 70S ribosomes in complex with tRNASufA6 (a frameshift suppressor derivative of tRNAPro) have revealed ASL conformational change during +1 frameshifting. Interaction between tRNA bases 32 and 38 is lost in the ribosomal P and E sites [Citation26]. Note that uridines 32 and 38 are substrates for RluA and TruA, respectively. Moreover, tRNASufA6 in the P site shows extensive conformational rearrangements of the 30S head and body domains [Citation26]. Our results on the FS frequency in TruA defective strains suggest that ASL conformational change can occur also during −1 FS event. If this is true, pseudouridine-dependent conformational rigidity helps to keep the reading frame during translation by preventing conformational change in ASL. We cannot exclude a possibility that the ribosomes of different bacterial species use alternative ways to keep mRNA reading frame during translation.
Regarding the changes in translational fidelity in Ψ-synthase-deficient strains, we noticed that UAG stop codon readthrough was enhanced in ∆truA strains in all three bacterial species (). Previously it has been shown that glutamine is incorporated at the UAG codon during mistranslation [Citation58]. There are two glutamine tRNAs in E. coli (with anticodons CUG and UUG) and a single tRNA in P. putida and P. aeruginosa (anticodon UUG) and they are all targets for the pseudouridine synthase TruA [Citation59]. More specifically, the U at position 38 in tRNAGln is isomerized. Our results indicate that a Ψ38 helps to make the decoding stricter, i.e. decrease the frequency of stop codon readthrough. Similar effect of Ψ deficiency at position 38 was previously shown to decrease mistranslation at His codons [Citation20]. tRNAGln with the anticodon CUG, that normally decodes CAG codon can base pair with the UAG stop codon if a G–U wobble base pair is formed between the first nucleotide in the stop codon and the third nucleotide in the anticodon. This is also required for the tRNA with the anticodon UUG, which would additionally require non-canonical base pairing between the last G of the stop codon and the first U of anticodon. It is likely that a Ψ38 stabilizes the structure of the anticodon stem-loop due to stronger stacking interactions and thereby reducing conformational flexibility. Stable ASL structure would prevent conformational flexibility required for the frameshift event. With U38, the stem-loop allows for more flexibility leading to increased frequency of mistranslation events.
Concluding remarks
Based on the results obtained we make the following conclusions:
Frameshift frequency at known slippery sequence sites varies in Pseudomonas species and in E. coli up to 10 times. General order of FS frequency is P.putida > P. aeruginosa > E. coli.
FS frequency in both directions at UUU UUC site is 3 times more frequent in Pseudomonas species as compared to E. coli.
Stop codon readthrough at UAG codon is similar in the three bacterial species. In contrast, readthrough at UGA is three orders of magnitude more frequent in E. coli as compared to Pseudomonas species.
TruA supports reading frame maintenance and reduces stop codon readthrough in E. coli. In P. putida, TruA has a context-specific effect on −1 frameshifting and decreases misreading at UAG. In P. aeruginosa TruA does not appear to affect translation fidelity.
Materials and methods
Bacterial strains, plasmids, and media
The bacterial strains and plasmids used in this study are listed in . All P. putida strains are derivatives of PaW85 which is isogenic to KT2440 [Citation60,Citation61], P. aeruginosa strains are derivatives of PAO1-L [Citation62], and E. coli strains are derivatives of MG1655 [Citation63]. P. putida strains were grown at temperature 30°C, E. coli and P. aeruginosa strains were grown at 37°C, except for measurement of translational fidelity using the Rluc-Fluc system, when all the cells were grown at 30°C.
Table 1. Bacterial strains and plasmids used in this study.
For complete medium either LB, or glc + CAA was used. For glc + CAA M9 buffer was supplemented with casamino acids (CAA) with tryptone and glucose both at final concentration 0.2%. Solid medium contained 1.5% Difco agar. Antibiotics were added at final concentrations: Kanamycin (Km) 50 µg/mL, gentamycin (Gm) 10 µg/mL.
Construction of strains and plasmids
For construction of MG1655 TruA and RluA deletion strains, BWΔtruA::km and BWΔrluA::km strains were obtained from the Keio collection [Citation18]. MG1655 deletion strains were generated by transferring the ΔtruA::km or ΔrluA::km fragment into MG1655 by P1 phage transduction. In the next step, kanamycin resistant colonies were selected, and the kanamycin resistance gene was removed by expressing flippase from a temperature sensitive plasmid pCP20 at 30°C, which was afterwards removed from cells by cultivation at 37°C [Citation64]. Successful deletion was confirmed by PCR.
Broad-host range plasmid pSEVA/lacItac [Citation16] was chosen as carrier for the dual-luciferase assay system. Frameshift reporters AD2, AD5 and AD7, and stop codon readthrough reporters 304UAG and 417UGA were based on previously published reporters [Citation38], transferred under the control of the Tac-promoter of pSEVA/lacItac by circular polymerase extension cloning [Citation65]. To generate UUC ±FS reporters, we first constructed pSEVA RF NcoI/PstI plasmid that allows cloning of wide range of sequences between the rluc and fluc genes. Site directed mutagenesis and Eco81I and MluI fragment replacement on the initial plasmid (pSEVA RFwt) were used to generate NcoI and PstI restriction sites to the ends of the linker region of the Rluc and Fluc. Oligonucleotides containing the UUC sequence were designed with NcoI and PstI sites in the flanking regions. Then, the oligonucleotides were annealed, cleaved with NcoI and PstI and inserted into the pSEVA RF NcoI/PstI plasmid.
Dual luciferase translation assay
Cells carrying the reporter were grown overnight at 30°C in 1.5 mL glc + CAA medium supplemented with either kanamycin (50 μg/mL for P.putida and E.coli) or gentamycin (10 μg/mL for P.aeruginosa). Cultures were diluted to OD580 ~ 0.1 into fresh glc + CAA medium containing the appropriate antibiotic and IPTG (0.5 mM). Cells carrying frameshift reporters were grown in 2 ml and cells carrying stop codon readthrough reporters were grown in 4 mL aliquots. After 3 hours of growth at 30°C, cells were collected, pelleted and flash frozen in liquid nitrogen. Frozen cells were resuspended in 400 μL of Passive Lysis buffer of Dual-Luciferase Reporter Assay System (Promega) and placed on ice for 10 min.
For measurement, 50 μL of cell extract was assayed for Fluc activity; the reaction mixture was diluted 50-fold using Passive Lysis buffer and 50 μL of the diluted reaction mixture was assayed for Rluc activity. Luminescence was measured using a TECAN Infinite ProM200 plate reader. Fluc activity was measured over a 10 second interval 2 minutes after starting the reaction, Rluc activity was measured over a 10 second interval 4 minutes after Fluc activity was measured. Raw values of luciferase assays are shown in the Table S12. Taking the dilution factor into account, the ratio of Fluc activity to Rluc activity was calculated and the Fluc/Rluc ratio of each replicate was normalized against the mean value of the reporter system, where both luciferase genes were functional and in the same frame (pSEVA RFwt). The normalization standard (Fluc/Rluc ratio using pSEVA RFwt construct) was determined separately in the wild-type strain for each species.
Statistical analysis
Shapiro-Wilk test was used to determine the normality of the dataset. As the data did not follow normal distribution, non-parametric method was used to compare different datasets. Kruskal-Wallis test was performed, followed by Dunn’s post-hoc test. Calculations were performed using Statistica software (TIBCO Software).
Author contributions
Conceived and designed the experiments: K.J., J.R., M.K.
Performed the experiments: K.J., M.T., H.I., M.L.
Analysed the data: K.J., M.T., M.L., J.R., M.K.
Wrote the initial draft K.J.
Manuscript finalization: K.J., J.R., M.K.
Supplemental Material
Download MS Word (34.9 KB)Acknowledgments
This study was supported by the Institutional Research Funding PRG707 from the Estonian Research Council (Eesti Teadusagentuur) to MK, by the Institutional Research Funding PRG669/1179 from the Estonian Research Council to JR, and by the Estonian Research Council grant PSG295 to ML
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15476286.2022.2121447
References
- Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32(1):223–238.
- de Pouplana LR, Santos MA, Zhu J-H, et al. Protein mistranslation: friend or foe?. Trends Biochem Sci. 2014;39(8):355–362.
- Mordret E, Dahan O, Asraf O, et al. Systematic detection of amino acid substitutions in proteomes reveals mechanistic basis of ribosome errors and selection for translation fidelity. Mol Cell. 2019;75(3):427–441.
- Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26(1):29–50.
- Rodnina MV, Korniy N, Klimova M, et al. Translational recoding: canonical translation mechanisms reinterpreted. Nucleic Acids Res. 2019;48(3):1056–1067.
- Katz A, Elgamal S, Rajkovic A, et al. Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Mol Microbiol. 2016;101(4):545–558.
- Björk GR, Hagervall TG. Transfer RNA modification. EcoSal Plus. 2005;1(2). DOI:10.1128/ecosalplus.4.6.2
- Grosjean H, Westhof E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016;44(17):8020–8040.
- Smith TJ, Tardu M, Khatri HR, et al. mRNA and tRNA modification states influence ribosome speed and frame maintenance during poly (lysine) peptide synthesis. J Biol Chem. 2022;298(6):102039.
- Urbonavičius J, Qian Q, Durand JM, et al. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20(17):4863–4873.
- Yarian C, Townsend H, Czestkowski W, et al. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem. 2002;277(19):16391–16395.
- Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon‐domain modifications. EMBO Rep. 2008;9(7):629–635.
- Wrzesinski J, Nurse K, Bakin A, et al. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for psi 746 in 23S RNA is also specific for psi 32 in tRNA (phe). Rna. 1995;1(4):437–448.
- Boccaletto P, Stefaniak F, Ray A, et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50(D1):D231–D235.
- Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell. 2007;26(2):189–203.
- Tagel M, Ilves H, Leppik M, et al. Pseudouridines of tRNA anticodon stem-loop have unexpected role in mutagenesis in Pseudomonas sp. Microorganisms. 2020;9(1):25.
- Porat J, Kothe U, Bayfield MA. Revisiting tRNA chaperones: new players in an ancient game. RNA. 2021;27(5):543–559.
- Baba T, Ara T, Hasegawa M, et al. Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2(1):2006–2008.
- Johnston HM, Barnes WM, Chumley FG, et al. Model for regulation of the histidine operon of Salmonella. Proc Nat Acad Sci. 1980;77(1):508–512.
- Parker J. Specific mistranslation in hisT mutants of Escherichia coli. Mol Gen Genet MGG. 1982;187(3):405–409.
- Urbonavicius J, Stahl G, Durand JM, et al. Transfer RNA modifications that alter+ 1 frameshifting in general fail to affect− 1 frameshifting. Rna. 2003;9(6):760–768.
- Auffinger P, Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J Mol Biol. 1999;292(3):467–483.
- Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23(24):5020–5026.
- Jiang J, Aduri R, Chow CS, et al. Structure modulation of helix 69 from Escherichia coli 23S ribosomal RNA by pseudouridylations. Nucleic Acids Res. 2014;42(6):3971–3981.
- Kierzek E, Malgowska M, Lisowiec J, et al. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014;42(5):3492–3501.
- Hong S, Sunita S, Maehigashi T, et al. Mechanism of tRNA-mediated+ 1 ribosomal frameshifting. Proc Nat Acad Sci. 2018;115(44):11226–11231.
- Maehigashi T, Dunkle JA, Miles SJ, et al. Structural insights into+ 1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc Nat Acad Sci. 2014;111(35):12740–12745.
- Silby MW, Winstanley C, Godfrey SAC, et al. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev. 2011;35(4):652–680.
- Kivisaar M. Narrative of a versatile and adept species Pseudomonas putida. J Med Microbiol. 2020;69(3):324–338.
- Nikel PI, Martínez-García E, De Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nature Rev Microbiol. 2014;12(5):368–379.
- Poblete-Castro I, Becker J, Dohnt K, et al. Industrial biotechnology of Pseudomonas putida and related species. Appl Microbiol Biotechnol. 2012;93(6):2279–2290.
- Bodey GP, Bolivar R, Fainstein V, et al. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5(2):279–313.
- Rossi E, La Rosa R, Bartell JA, et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nature Rev Microbiol. 2021;19(5):331–342.
- Tagel M, Tavita K, Hõrak R, et al. A novel papillation assay for the identification of genes affecting mutation rate in Pseudomonas putida and other pseudomonads. Mutat Res. 2016;790:41–55.
- Al Mamun AAM, Gautam S, Humayun MZ. Hypermutagenesis in mutA cells is mediated by mistranslational corruption of polymerase, and is accompanied by replication fork collapse. Mol Microbiol. 2006;62(6):1752–1763.
- Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50(1):625–644.
- Slupska MM, Baikalov C, Lloyd R, et al. Mutator tRNAs are encoded by the Escherichia coli mutator genes mutA and mutC: a novel pathway for mutagenesis. Proc Nat Acad Sci. 1996;93(9):4380–4385.
- Devaraj A, Shoji S, Holbrook ED, et al. A role for the 30S subunit E site in maintenance of the translational reading frame. Rna. 2009;15(2):255–265.
- Gutgsell N, Englund N, Niu L, et al. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. Rna. 2000;6(12):1870–1881.
- Huang L, Pookanjanatavip M, Gu X, et al. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37(1):344–351.
- Kendra JA, Advani VM, Chen B, et al. Functional and structural characterization of the chikungunya virus translational recoding signals. J Biol Chem. 2018;293(45):17536–17545.
- Melian EB, Hall-Mendelin S, Du F, et al. Programmed ribosomal frameshift alters expression of west Nile virus genes and facilitates virus replication in birds and mosquitoes. PLoS Pathog. 2014;10(11):e1004447.
- Chandler M, M. Fayet OJM. Translational frameshifting in the control of transposition in bacteria. Molecular Microbiology. 1993;7(4):497–503.
- Flower AM, McHenry CS. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Nat Acad Sci. 1990;87(10):3713–3717.
- Manktelow E, Shigemoto K, Brierley I. Characterization of the frameshift signal of Edr, a mammalian example of programmed− 1 ribosomal frameshifting. Nucleic Acids Res. 2005;33(5):1553–1563.
- Fu C, Parker J. A ribosomal frameshifting error during translation of the argl mRNA of Escherichla coli. Mol Gen Genet MGG. 1994;243(4):434–441.
- Plant EP, Dinman JDJR. Comparative study of the effects of heptameric slippery site composition on− 1 frameshifting among different eukaryotic systems. RNA. 2006;12(4):666–673.
- Bodey GP, Bolivar R, Fainstein V, et al. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5(2):279–313.
- Blount ZD. The natural history of model organisms: the unexhausted potential of E. coli. Elife. 2015;4:e05826.
- Cridge AG, Major LL, Mahagaonkar AA, et al. Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2006;34(7):1959–1973.
- Zhao J, Leung H-CE, Winkler ME. The miaA mutator phenotype of Escherichia coli K-12 requires recombination functions. J Bacteriol. 2001;183(5):1796–1800.
- Carpentier P, Lepretre C, Basset C, et al. Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction. Nucleic Acids Res. 2020;48(17):9918–9930.
- Winsor GL, Griffiths EJ, Lo R, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D653.
- Keseler IM, Gama-Castro S, Mackie A, et al. The EcoCyc database in 2021. Front Microbiol. 2021;12:2098.
- Vacher J, Grosjean H, Houssier C, et al. The effect of point mutations affecting Escherichia coli tryptophan tRNA on anticodon-anticodon interactions and on UGA suppression. J Mol Biol. 1984;177(2):329–342.
- Li J, Esberg B, Curran JF, et al. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J Mol Biol. 1997;271(2):209–221.
- Lecointe F, Namy O, Hatin I, et al. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Biol Chem. 2002;277(34):30445–30453.
- Nilsson M, Rydén-Aulin M. Glutamine is incorporated at the nonsense codons UAG and UAA in a suppressor-free Escherichia coli strain. Biochim Biophys Acta. 2003;1627(1):1–6.
- Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44(D1):D184–D189.
- Bayley SA, Duggleby CJ, Worsey MJ, et al. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet. 1977;154(2):203–204.
- Regenhardt D, Heuer H, Heim S, et al. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ Microbiol. 2002;4(12):912–915.
- Sidorenko J, Jatsenko T, Kivisaar M. Ongoing evolution of Pseudomonas aeruginosa PAO1 sublines complicates studies of DNA damage repair and tolerance. Mutat Res. 2017;797-799:26–37.
- Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia Coli and related bacteria. Cold Spring Harbor: CSHL Press; 1992.
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Nat Acad Sci. 2000;97(12):6640–6645.
- Quan J, Tian J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat Protoc. 2011;6(2):242–251.
