ABSTRACT
Angiotensin-converting enzyme 2 (ACE2) and several proteins have been identified as entry factors for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, whether long noncoding RNAs are involved in SARS-CoV-2 entry remains unknown. In this study, we investigated the role of small nucleolar RNA host gene 15 (SNHG15) in SARS-CoV-2 entry using a SARS-CoV-2 spike pseudotyped lentivirus with a luciferase reporter. Overexpression of SNHG15 promoted but SNHG15 knockdown limited SARS-CoV-2 entry in a dose- and time-dependent manner. SNHG15 interacted with Rab-like protein 2A (RABL2A). Overexpression and knockdown of RABL2A produced similar effects on SARS-CoV-2 entry as those of SNHG15. Furthermore, RABL2A knockdown abolished the SNHG15-mediated increase in SARS-CoV-2 entry. In conclusion, SNHG15 is a critical regulatory factor that aids SARS-CoV-2 entry through RABL2A.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), which was declared a global pandemic in March 2020 by the WHO [Citation1,Citation2]. SARS-CoV-2 was first identified in 2019 as a novel coronavirus from patients with pneumonia in China [Citation3]. As of 22 August, more than 596 million confirmed cases and 6.45 million COVID-19-caused deaths worldwide have been reported. It is still continuing to be a major public health threat [Citation4].
SARS-CoV-2 is an enveloped, positive-sense single-stranded RNA virus belonging to the family Coronaviridae [Citation5]. SARS-CoV-2 has a nonsegmented genome that encodes 16 nonstructural proteins (nsps) and four major structural proteins: the spike (S) protein, nucleocapsid (N) protein, membrane (M) protein and envelope (E) protein [Citation6]. The SARS-CoV-2 S protein has two functional domains: an S1 receptor-binding domain (RBD), which initially binds to the angiotensin converting enzyme 2 (ACE2) receptor on the cell surface, and an S2 domain, which causes fusion of the viral and host cell membranes [Citation7,Citation8]. Cleavage of the S protein is mediated by the proteases furin and transmembrane serine protease 2 (TMPRSS2), resulting in exposure of the S1 and S2 domains and enabling virus entry into the cell [Citation9–12]. ACE2 is the primary receptor through which SARS-CoV-2 binds and enters cells [Citation13]. Several cofactors that enhance the binding of SARS-CoV-2 to ACE2 receptors have been identified [Citation14]. Neuropilin-1 acts as an entry cofactor by binding to the cleaved form of the SARS-CoV-2 S protein [Citation15,Citation16]. A few studies have shown that the RBD domain of the SARS-CoV-2 S protein interacts with heparan sulphate in close proximity to ACE2, causing an open conformation of the S protein to facilitate binding with the ACE2 receptor [Citation17,Citation18]. SARS-CoV-2 S protein can also effectively bind to the CD147 receptor and enter the cell. Blockage of the CD147 and S protein interaction using meplazumab, an anti-CD147 humanized antibody, led to a significant decrease in virus entry [Citation19]. AXL receptor tyrosine kinase has been reported to bind with the N-terminal domain of the SARS-CoV-2 spike protein and facilitate virus entry into pulmonary and bronchial epithelial cells [Citation20].
The SARS-CoV-2 virus can enter the cell through clathrin-mediated endocytosis, similar to influenza virus [Citation21]. The postreceptor binding entry mechanisms, such as caveolae, and clathrin- and caveolae-independent mechanisms involving lipid rafts are yet to be clarified. In general, coronaviruses, following entry, cause the release and uncoating of genomic viral RNA into the cytoplasm. Two large open reading frames, namely, ORF1a and ORF1b, are translated into pp1a and pp1ab, which are further processed into 16 nsps. These nsps together form the viral replication and transcription complex (RTC) [Citation6]. Viral RNA synthesis begins with the generation of full-length negative-sense genomic copies, which serve as templates for the synthesis of new positive-sense genomic RNAs. The newly synthesized genomes are further translated into more nsps, and RTCs are produced and packaged into new virions. The structural proteins in coronaviruses assist with virus assembly and budding by exocytosis. However, their role in SARS-CoV-2 replication has not been completely elucidated [Citation22].
Long noncoding RNAs (lncRNAs) have previously been identified to regulate the pathogenesis of various diseases, including viral infections [Citation23–27]. Although several studies have reported that lncRNAs are differentially expressed during SARS-CoV-2 infection through RNA sequencing [Citation28–32], their functional activities during SARS-CoV-2 infection remain unknown. Small nucleolar RNA host gene 15 (SNHG15) is one of the dysregulated lncRNAs during SARS-CoV-2 infection [Citation28,Citation33]. Our unpublished studies have shown that SNHG15 is a critical lncRNA regulating influenza virus entry. Since SARS-CoV-2 is an RNA virus that utilizes receptor-mediated endocytosis for entry into the cell, similar to the influenza virus, we chose SNHG15 to study SARS-CoV-2 entry. In this study, we found that SNHG15 promoted SARS-CoV-2 entry by interacting with RABL2A.
Results
SNHG15 promotes SARS-CoV-2 entry
The experimental design for this paper is shown in . There are 5 isoforms of SNHG15. We first determined the copy numbers of each isoform in HEK293T-ACE2 cells. As shown in , isoform 1 had the highest copy number, followed by isoform 2. The isoforms 3–5 expression levels were low. The copy number of small nucleolar RNA H/ACA Box 9 (SNORA9), a gene located at the intron of SNHG15 was comparable to that of SNHG15 isoform 1. The RABL2A expression level was lower than SNHG15 isoform 1.
Figure 1. SNHG15 overexpression enhances SARS-CoV-2 entry into cells. (A) an experimental design schematic diagram. (B) Copy numbers of SNHG15, SNORA9 and RABL2A in HEK293T-ACE2 cells as determined by droplet digital PCR. (C-F) HEK293T – ACE2 cells were transfected with 100 ng of vector control (VC) or SNHG15 isoform 1overexpression plasmids (C, D) for 24 h, followed by infection with SARS-CoV-2 pseudovirus at the indicated doses for 72 h (E) on the X-axis or at a dose of 6 × 105 relative luciferase units (RLUs) for the indicated time points (F). The transfection efficiency of VC and SNHG15 overexpression plasmids is indicated by GFP (C). Scale bar: 100 µm. SNHG15 levels were determined by real-time PCR and were normalized to β-actin (D). Luciferase activity levels representing SARS-CoV-2 entry (Y-axis) were measured in the infected cells, and the results are expressed as RLUs (E, F). Data are expressed as the means ± SEs. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. the VC of the respective doses or time points (n = 3). Student’s t test for D and two-way ANOVA followed by Sidak’s multiple comparisons test for E and F were used.
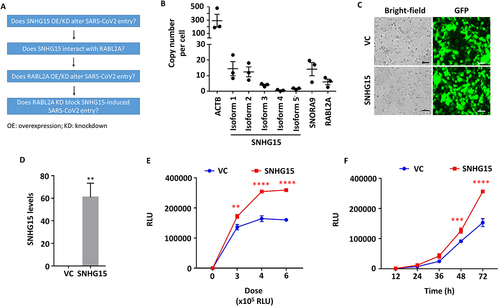
To determine the functional activity of SNHG15 on SARS-CoV-2 entry, we employed pseudotyped lentiviral particles with SARS-CoV-2 spike protein and luciferase reporter (SARS-CoV-2 pseudovirus) and a HEK293T-ACE2 engineered cell line that expresses human ACE2. As SNHG15 isoform 1 has the highest copy number among all of the isoforms in HEK293T-ACE2 cells, we overexpressed SNHG15 isoform 1 in HEK293T-ACE2 cells with an SNHG15 isoform 1 expression plasmid using Lipofectamine 3000. High transfection efficiencies of both vector control (VC) and SNHG15 plasmids were achieved, as shown by green fluorescent protein (GFP) expression (). Overexpression of SNHG15 was confirmed by real-time PCR (). The SNHG15-overexpressing cells were then infected with SARS-CoV-2 pseudovirus at various doses for 72 h or at a relative luciferase unit (RLU) of 6 × 105 for different times. The SARS-CoV-2 entry levels were measured by determining the luciferase activity in the infected cells. RLUs on the X-axis represent the amount of the virus added, while RLUs on the Y-axis represent SARS-CoV-2 entry. The results revealed that SNHG15 isoform 1 promoted SARS-CoV-2 entry in a dose- and time-dependent manner (). Whether other isoforms are also involved in SARS-CoV-2 entry remains to be determined.
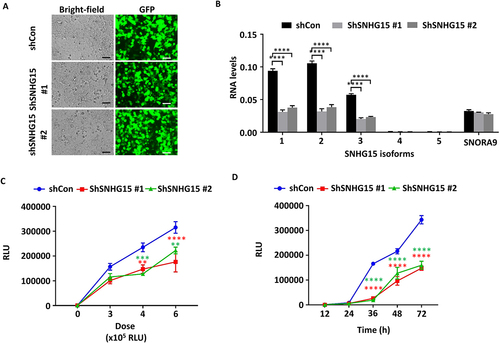
We further determined the effect of SNHG15 knockdown on SARS-CoV-2 entry. HEK293T-ACE2 cells were transfected with control and SNHG15-specific shRNAs using Lipofectamine 3000. GFP expression revealed that the transfection efficiency was high (). Real-time PCR results revealed that SNHG15 isoforms 1–3 were significantly reduced by both shRNAs (). The levels of SNORA9 were not altered by shRNAs (). The SNHG15-knockdown cells were then infected with SARS-CoV-2 pseudovirus at the indicated doses for 72 h or at an RLU of 6 × 105 for the indicated times. Luciferase activity levels indicated that shRNA-mediated knockdown of SNHG15 reduced SARS-CoV-2 entry in a dose- and time-dependent manner (), thus confirming the functional role of SNHG15 during SARS-CoV-2 entry.
Figure 2. SNHG15 knockdown limits SARS-CoV-2 entry into cells. HEK293T – ACE2 cells were transfected with 100 ng of shRNA vector control (shCon) or SNHG15 shRNA plasmids (shSNHG15) (A, B) for 24 h, followed by infection with SARS-CoV-2 pseudovirus at the indicated doses for 72 h on the X-axis (C) or at a dose of 6 × 105 relative luciferase units (RLUs) for the indicated time points (D). The transfection efficiency of the shCon and shSNHG15 plasmids is indicated by GFP (A). Scale bar: 100 µm. SNHG15 isoform and SNORA9 levels were determined by real-time PCR and were normalized to β-actin (B). Luciferase activity levels representing SARS-CoV-2 entry (Y-axis) were measured, and the results are expressed as RLUs (C, D). Data are expressed as the means ± SEs. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. the VC of the respective doses or time points (n = 3). Two-way ANOVA followed by Sidak’s multiple comparisons test was used.
SNHG15 interacts with RABL2A
We performed RNA pulldown coupled with proteomics to identify the proteins that interact with SNHG15. The proteomics analysis identified RABL2A, but not any other isoforms of RAB as a binding protein of SNHG15. To confirm the interaction of SNHG15 and RABL2A, we performed an RNA immunoprecipitation (RIP) assay using RABL2A antibodies or IgG control. The RIP assay results revealed that SNHG15 was specifically bound to RABL2A compared to the IgG control ().
Figure 3. RABL2A overexpression increases SARS-CoV-2 entry into cells. (A) SNHG15 levels in immunoprecipitates with anti-RABL2A or IgG. (B-E) HEK293T – ACE2 cells were transfected with 100 ng of vector control (VC) or RABL2A overexpression plasmids (B, C) for 24 h, followed by infection with SARS-CoV-2 pseudovirus at the indicated doses for 72 h on the X-axis (D) or at a dose of 6 × 105 relative luciferase units (RLUs) for the indicated time points (E). The transfection efficiency of the vector control and RABL2A plasmids is indicated by GFP (B). Scale bar: 100 µm. RABL2A protein levels were determined by western blotting (C). Luciferase activity levels representing SARS-CoV-2 entry (Y-axis) were measured, and the results are expressed as RLUs (D, E). The VC in Panels D and E is the same as that in Fig. 1E and F because both experiments were performed at the same time. Data are expressed as the means ± SEs. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. the VC of the respective doses or time points (n = 3). Student’s t test was used for A, and two-way ANOVA followed by Sidak’s multiple comparisons test was used for D and E.
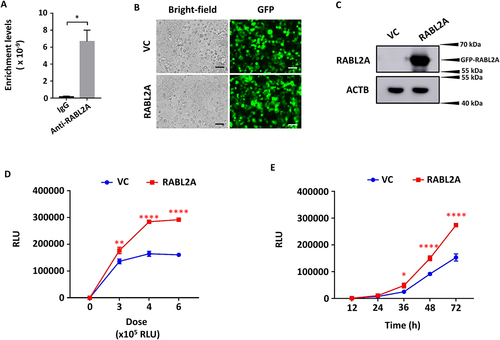
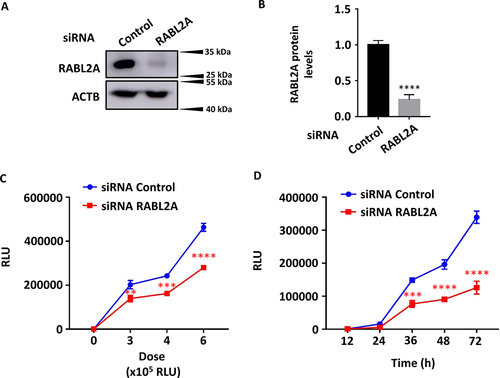
RABL2A increases SARS-CoV-2 entry
RAB GTPases are known to play important roles in the regulation of exocytotic and endocytotic pathways. Hence, we examined the role of RABL2A in SARS-CoV-2 entry. We overexpressed RABL2A in HEK293T-ACE2 cells with a RABL2A overexpression plasmid. A good level of transfection efficiency was achieved (). Western blot results revealed significant overexpression of a GFP-RABL2A fusion protein at ~ 56 kDa (). RABL2A-overexpressing cells were then infected with SARS-CoV-2 pseudovirus at the indicated doses and time points. Luciferase activity levels indicated that RABL2A enhanced SARS-CoV-2 entry (), similar to SNHG15 isoform 1 overexpression ().
We then transfected HEK293T-ACE2 cells with RABL2A siRNAs using Lipofectamine RNAi max reagent and determined the knockdown efficiency by western blotting. An endogenous RABL2A protein band was detected at 28 kDa (). siRNA RABL2A reduced RABL2A protein levels by 76.46 ± 0.07% compared to control cells (). RABL2A-knockdown cells were then infected with SARS-CoV-2 pseudovirus at various doses and time points, and luciferase activity was measured. RABL2A knockdown limited SARS-CoV-2 entry in a dose- and time-dependent manner ().
Figure 4. RABL2A knockdown inhibits SARS-CoV-2 entry into cells. HEK293T – ACE2 cells were transfected with 100 ng of control and RABL2A siRnas at 2 nM for 48 h (A, B), followed by infection with SARS-CoV-2 pseudovirus at the indicated doses for 72 h on the X-axis (C) or at a dose of 6 × 105 RLUs for the indicated time points (D). RABL2A protein levels were determined by western blotting, quantified and normalized to β-actin (A, B). Luciferase activity levels representing SARS-CoV-2 entry (Y-axis) were measured, and the results are expressed as relative luciferase units (RLUs) (C, D). Data are expressed as the means ± SEs. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. the siRNA control of the respective doses or time points (n = 3). Student’s t test for B and two-way ANOVA followed by Sidak’s multiple comparisons test for C and D were used.
SNHG15 acts through RABL2A
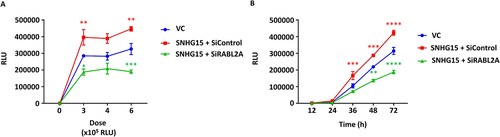
Since SNHG15 interacts with RABL2A and both proteins exhibit similar functional activities in SARS-CoV-2 entry, we tested whether SNHG15 promotes SARS-CoV-2 entry through RABL2A. We transfected HEK293T-ACE2 cells with control or RABL2A siRNAs and then transfected them with an SNHG15 isoform 1 overexpression plasmid. Cells were infected with SARS-CoV-2 pseudovirus at the indicated doses and time points, and luciferase activity levels were measured. As expected, SARS-CoV-2 entry was increased in SNHG15-overexpressing cells compared to vector control cells. RABL2A knockdown reduced SARS-CoV-2 entry in SNHG15-overexpressing cells, indicating that SNHG15 functional activity on SARS-CoV-2 entry is mediated through RABL2A ().
Figure 5. Knockdown of RABL2A abolishes SNHG15-mediated SARS-CoV-2 entry. HEK293T – ACE2 cells were transfected with control and RABL2A siRnas at 2 nM for 24 h and then were transfected with 100 ng of SNHG15 isoform 1 overexpression plasmids. The control cells were transfected with 100 ng of vector control (VC) overexpression plasmids for 24 h. Cells were then infected with SARS-CoV-2 pseudovirus at the indicated doses for 72 h on the X-axis (A) or at a dose of 6 × 105 relative luciferase units (RLUs) for the indicated time points (B), and luciferase activity levels representing SARS-CoV-2 entry (Y-axis) were measured. Data are expressed as the means ± SEs. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. the VC of the respective doses or time points (n = 3). Two-way ANOVA followed by Sidak’s multiple comparisons test was used.
Discussion
Our study is the first report of a functional role of SNHG15, a conserved lncRNA, in SARS-CoV-2 entry. Using pseudotyped lentiviral particles with SARS-CoV-2 spike protein, we determined the functional role of SNHG15 in SARS-CoV-2 entry. Overexpression and knockdown studies revealed that SNHG15 promotes SARS-CoV-2 entry into cells. RABL2A, an interacting partner of SNHG15, showed similar activity in SARS-CoV-2 entry. Knockdown of RABL2A significantly blocked the SNHG15-mediated entry of SARS-CoV-2, indicating that SNHG15 aids SARS-CoV-2 entry through RABL2A.
SNHG15 is a small nucleolar host gene encoding SNORA9 in its intron region according to the human genome assembly (GRCh38/hg38). SNHG15 has five isoforms that share similar sequences and secondary structures. SNHG15 is predicted to have no protein coding potential [Citation34]. Several studies have indicated that SNHG15 plays a critical role in the molecular regulation of various cancers through its interaction with host factors [Citation35,Citation36]. In this study, we identified that SNHG15 is a critical cellular lncRNA that aids the entry of SARS-CoV-2.
LncRNAs normally function via their interactions with protein, RNA, and DNA. SNHG15 was previously identified to bind to apoptosis-inducing factor (AIF) in LoVo cells [Citation34] and EZH2 in AsPC-1 and BxPC-3 cells [Citation37]. In this study, we identified RABL2A as a new binding partner of SNHG15. RABL2A is likely a downstream component of SNHG15 because siRNA knockdown of RABL2A reduced SNHG15 overexpression-induced SARS-CoV-2 entry. However, how SNHG15 affects RABL2A function remains to be determined. It is possible that SNHG15 enhances the recruitment and/or activity of RABL2A.
Our findings have demonstrated that RABL2A is a critical host factor that is required for SARS-CoV-2 entry. The exact molecular mechanism of RABL2A in SARS-CoV-2 viral entry has yet to be determined. Several studies have demonstrated the role of RAB GTPases in the virus life cycle [Citation38]. In general, RAB GTPases are well-known regulators of endocytosis pathways [Citation39]. Six RAB-like proteins, namely, RABL2A, RABL2B, RABL3, RABL4, RABL5, and RABL6, have been identified. RABL2B, RABL4, and RABL5 regulate intraflagellar transportation in cilia [Citation40–42]. RABL2A activity on viral entry was first reported in rotavirus infection, which utilizes endocytosis-mediated internalization and replicates in the cytoplasm, similar to SARS-CoV-2 [Citation43].
Our study identified the role of the conserved lncRNA SNHG15 in the regulation of SARS-CoV-2 entry. Through this study, we have highlighted the importance of identifying critical host factors, including noncoding RNAs, in the regulation of SARS-CoV-2 infection. Understanding the biology of SARS-CoV-2 replication and the critical host factors involved in aiding the replication machinery may lead to potential intervention strategies against SARS-CoV-2 infection.
Materials and methods
Cell culture
The Human Embryonic Kidney Cells Expressing Human Angiotensin-Converting Enzyme 2 (HEK293T-ACE2) cell line was obtained from BEI resources (Catalog No. NR-52511, MD, USA), cultured and maintained in DMEM containing 10% foetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA, USA) and 0.1% penicillin and streptomycin (PS) solution (Life Technologies Corporation, Carlsbad, CA, USA).
Viruses
We generated pseudotyped lentiviral particles with the SARS-CoV-2 spike (S) glycoprotein gene of the Wuhan-Hu-1 strain (GenBank: NC_045512), luciferase (Luc2) and green fluorescent protein (GFP) using the SARS-Related Coronavirus 2, Wuhan-Hu-1 Spike-Pseudotyped Lentiviral Kit (Catalog No. NR-52948, BEI resources, MD, USA) as previously described [Citation44].
Titer determination
HEK293T-ACE2 cells were seeded at a density of 1.25 × 104 cells/well in type I collagen-coated 96-well plates. Threefold serial dilutions of the SARS-CoV-2 pseudovirus were made in DMEM complete media containing polybrene at a final concentration of 8 μg/mL and were added to the cells. At 72 h post infection, cells were lysed using 50 µl of 1× passive lysis buffer, and luciferase activities were measured as described in the luciferase reporter assay. Viral titer results are presented as relative luciferase units (RLUs) as previously described [Citation44].
Real-time PCR
RNA sample collection was performed using 1 ml of TRI reagent (Catalog # TR118, Molecular Research Center, Cincinnati, OH, USA). RNA isolation was performed as previously described [Citation45]. Using MMLV enzyme (Thermo Fischer, Waltham, MA, USA), 1 µg of the RNA was reverse-transcribed into cDNA. The NCBI Primer designing tool was used to design primers for target genes. Primer sequences are listed in . Real-time PCR was performed using iTaq Universal SYBR Green Supermix (Catalog # 1725124, Biorad, Hercules, CA, USA) on a QuantStudio 6 Real-Time PCR System (Thermo Fischer, Waltham, MA, USA). The following thermal temperatures were used: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 65°C for 30 s. β-Actin (ACTB) was used as the endogenous reference gene. The comparative ΔCt method of calculating the relative gene expression was performed using the equation 2−ΔCt. Droplet digital PCR was performed by Q×200AutoDG droplet digital PCR system (Bio-Rad, Hercules, CA) as previously described [Citation46].
Table 1. List of real-time PCR primers for human genes.
Western blotting
Western blot analysis was performed as described previously [Citation46]. The primary and secondary antibodies were used in the dilutions as follows: rabbit anti-RABL2A, 1:1,000 dilution (Catalog # 17816–1-AP, Thermo Scientific, Rockford, IL, USA) and goat anti-rabbit HRP-conjugated secondary antibody, 1:1,000 dilution (Catalog # 111-035-003, Jackson Immuno Research Laboratories, West Grove, PA, USA). Proteins were visualized using Super Signal West Pico Chemiluminescent Substrate (Thermo Fischer, Waltham, MA, USA), and images were taken with an Amersham Imager 600 (GE healthcare system, Pittsburgh, PA, USA).
Construction of the overexpression vector
A Kapa Hotstart HiFi PCR kit (Roche, Basel, Switzerland) was used to amplify SNHG15 isoform 1 (NCBI Reference Sequence: NR_003697.2) from cDNAs of A549 cells and RABL2A from the RABL2A pLX304 plasmid (Catalog No. HsCD00440413, DNASU Plasmid Repository, Tempe, AZ, USA) with the following primers: SNHG15 forward primer, 5’- TTTCCGCTCGAGGCGCGGCGTCAGGCTTGGCT − 3’, SNHG15 reverse primer, 5’ – TTTCCGGAATTCATTTAAATCCATATTTATTAAATATCCC − 3’; and RABL2A forward primer, 5’ – TATGCGGCCGCCACCATGGCAGAAGACAAAACCAAAC- 3’ and RABL2A reverse primer, 5’- TATGAATTCTTCCTCCTCTGATGGGGTCTC- 3’. The thermal temperatures were 95°C for 3 min, followed by 35 cycles of 98°C for 20 s, 60°C for 15 s, 72°C for 60 s and a final extension at 72°C for 1 min. The PCR products were inserted into a modified lentiviral pLVX vector (Clontech, Mountain View, CA, USA) downstream of its green fluorescent protein (GFP) at XhoI and EcoRI for SNHG15 and at NotI and XhoI for RABL2A, and the inserts were confirmed by sequencing.
Construction of the shRNA vector
Using BLOCK-iT™ RNAi Designer, shRNA sequences were designed targeting exon 4 of all SNHG15 isoforms. shRNA sequences were inserted into the hCMV promoter-driven lentiviral miRZip vector (System Biosciences, Palo Alto, CA, USA) downstream of GFP at BamH1 and EcoR1. The shRNA sequences for SNHG15 are as follows: forward oligo 1, 5’-GATCCGCAGTCTTTGTCCATGAAACTTTCAAGAGAAGTTTCATGGACAAAGACTGCTTTTTG −3’ and reverse oligo 1, 5’-AATTCAAAAAGCAGTCTTTGTCCATGAAACTTCTCTTGAAAGTTTCATGGACAAAGACTGCG −3’; forward oligo 2, GATCCGCACAAGAGTGCCTGCCATCCTTCAAGAGAGGATGGCAGGCACTCTTGTGCTTTTTG −3’ and reverse oligo 2, 5’- AATTCAAAAAGCACAAGAGTGCCTGCCATCCTCTCTTGAAGGATGGCAGGCACTCTTGTGCG −3’.
Luciferase reporter assay
HEK293T-ACE2 cells were seeded at a density of 2.5 × 104 cells/well in a type I collagen-coated 96-well plate and were transfected with vector control (VC), SNHG15 overexpression plasmid, RABL2A overexpression, or shRNA plasmid (100 ng) using Lipofectamine 3000 reagent (Catalog # L3000015, Thermo Fischer, Waltham, MA, USA) for 24 h or with 2 nM siGENOME Non-Targeting siRNA Control Pool #2 (Catalog # D-001206-14-20, Dharmacon, Lafayette, CO, USA) or siGENOME Human RABL2A siRNA SMART Pool (Catalog # M-013620-00-002, Dharmacon) using Lipofectamine RNAi Max reagent for 48 h. Then, the cells were infected with SARS-CoV-2 pseudovirus at various doses and time points as indicated. Cells were lysed using 50 µl of 1× passive lysis buffer, and luciferase activity was measured using a dual luciferase assay kit (Catalog # E1910, Promega, Madison, WI, USA) according to the manufacturer’s instructions. SARS-CoV-2 entry is presented as relative luciferase units (RLUs).
RNA immunoprecipitation (RIP) assay
RIP was performed as previously described [Citation34] using rabbit IgG control and RABL2A antibodies (Catalog # 17816–1-AP, Proteintech, Rosemont, IL, USA) and A549 cell lysate. SNHG15 levels were determined by real-time PCR with cDNA prepared from 500 ng of input and total RNA from immunoprecipitated samples and were calculated using the formula 2−ct.
Authors’ contributions
S.P. designed and performed the experiments, analysed data, and wrote the manuscript; C.G. and K.V. produced and titrated the SARS-CoV-2 pseudovirus; Y.L. performed droplet digital PCR. L.L. conceived the studies, designed the experiments, interpreted data and wrote the manuscript.
Acknowledgments
The following reagents were obtained from BEI Resources, NIAID, NIH: Human Embryonic Kidney Cells (HEK-293T) Expressing Human Angiotensin-Converting Enzyme 2 (HEK-293T-hACE2, NR-52511); SARS-Related Coronavirus 2, Wuhan-Hu-1 Spike-Pseudotyped Lentiviral Kit (NR-52948).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data are available within the article or its supplementary materials.
Additional information
Funding
References
- Ghebreyesus TA. WHO director-general’s opening remarks at the media briefing on COVID-19. WHO Director General’s Speeches. 2020 [11 March 2020];4. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-11-march-2020
- Hu B, Guo H, Zhou P, et al. Characteristics of SARS-CoV-2 and COVID-19. Nature Rev Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017
- WHO Coronavirus (COVID-19) dashboard | WHO coronavirus (COVID-19) dashboard with vaccination data. https://covid19.who.int/.
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8
- V’kovski P, Kratzel A, Steiner S, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6
- Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;271(1):119–129. 2016. doi: 10.1038/cr.2016.152
- Huang Y, Yang C, Xu X, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;419(41):1141–1149. doi: 10.1038/s41401-020-0485-4
- Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3(9):e202000786. doi: 10.26508/lsa.202000786
- Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science (80). 2020;368(6491):630–633. doi: 10.1126/science.abb7269
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271. doi: 10.1016/j.cell.2020.02.052
- Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117
- D W, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCov spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507
- Evans JP, Liu S-L. Role of host factors in SARS-CoV-2 entry. J Biol Chem. 2021;297(1):100847. doi: 10.1016/j.jbc.2021.100847
- Daly JL, Simonetti B, Klein K, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science (80-). 2020;370(6518):861–865. doi: 10.1126/science.abd3072
- Cantuti-Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science (80-). 2020;370(6518):856–860. doi: 10.1126/science.abd2985
- Clausen TM, Sandoval DR, Spliid CB, et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183(4):1043–1057.e15. doi: 10.1016/j.cell.2020.09.033
- Zhang Q, Chen CZ, Swaroop M, et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020;61(1):1–14. doi: 10.1038/s41421-020-00222-5
- Wang K, Chen W, Zhang Z, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;51(5):1–10.
- Wang S, Qiu Z, Hou Y, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;312(2):126–140. doi: 10.1038/s41422-020-00460-y
- Bayati A, Kumar R, Francis V, et al. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J Biol Chem. 2021;296:100306. doi: 10.1016/j.jbc.2021.100306
- Klein S, Cortese M, Winter SL, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;111(1):1–10. doi: 10.1038/s41467-020-19619-7
- Kesheh MM, Mahmoudvand S, Shokri S. Long noncoding RNAs in respiratory viruses: A review. Rev Med Virol. 2022;32(2):e2275. doi: 10.1002/rmv.2275
- Melissari MT, Grote P. Roles for long non-coding RNAs in physiology and disease. Pflugers Archiv Eur J Physiol. 2016;468(6):945–958. doi: 10.1007/s00424-016-1804-y
- Ding Y-Z, Zhang Z-W, Liu Y-L, et al. Relationship of long noncoding RNA and viruses. Genomics. 2016;107(4):150–154. doi: 10.1016/j.ygeno.2016.01.007
- Bamunuarachchi G, Pushparaj S, Liu L. Interplay between host non-coding RNAs and influenza viruses. RNA Biol. 2021;18(5):767–784. doi: 10.1080/15476286.2021.1872170
- More S, Zhu Z, Lin K, et al. Long non-coding RNA PSMB8-AS1 regulates influenza virus replication. RNA Biol. 2019;16(3):340–353. doi: 10.1080/15476286.2019.1572448
- Vishnubalaji R, Shaath H, Alajez NM. Protein coding and long noncoding RNA (lncRNA) transcriptional landscape in SARS-CoV-2 infected bronchial epithelial cells highlight a role for interferon and inflammatory response. Genes (Basel). 2020;11(7):760. 11, 760. doi: 10.3390/genes11070760
- Wu Y, Zhao T, Deng R, et al. A study of differential circRNA and lncRNA expressions in COVID-19-infected peripheral blood. Sci Rep. 2021;11(1):1–14. doi: 10.1038/s41598-021-86134-0
- Mukherjee S, Banerjee B, Karasik D, et al. mRNA-lncRNA co-expression network analysis reveals the role of lncRNAs in immune dysfunction during severe SARS-CoV-2 infection. Viruses. 2021;13(3):402. doi: 10.3390/v13030402
- Turjya RR, Khan MAAK, Mir Md Khademul Islam AB. Perversely expressed long noncoding RNAs can alter host response and viral proliferation in SARS-CoV-2 infection. Future Virol. 2020;15(9):577–593. doi: 10.2217/fvl-2020-0188
- Yang Q, Lin F, Wang Y, et al. Long noncoding RNAs as emerging regulators of COVID-19. Front Immunol. 2021;12. doi: 10.3389/fimmu.2021.700184
- Lamers MM, van der Vaart J, Knoops K, et al. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. Embo J. 2021;40(5):e105912. doi: 10.15252/embj.2020105912
- Saeinasab M, Bahrami AR, González J, et al. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J Exp Clin Cancer Res. 2019;381(1):1–16. doi: 10.1186/s13046-019-1169-0
- Tong J, Ma X, Yu H, et al. SNHG15: a promising cancer-related long noncoding RNA. Cancer Manag Res. 2019;11:5961–5969.
- Shuai Y, Ma Z, Lu J, et al. LncRNA SNHG15: A new budding star in human cancers. Cell Proliferation. 2020;53(1). doi: 10.1111/cpr.12716
- Ma Z, Huang H, Wang J, et al. Long non-coding RNA SNHG15 inhibits P15 and KLF2 expression to promote pancreatic cancer proliferation through EZH2-mediated H3K27me3. Oncotarget. 2017;8(48):84153–84167. doi: 10.18632/oncotarget.20359
- Spearman P. Viral interactions with host cell Rab GTPases. Small GTPases. 2018;9(1–2):192. doi: 10.1080/21541248.2017.1346552
- Agola J, Jim P, Ward H, et al. Rab GTPases as regulators of endocytosis, targets of disease and therapeutic opportunities. Clin Genet. 2011;80(4):305. doi: 10.1111/j.1399-0004.2011.01724.x
- Nakayama K, Katoh Y. Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J Biochem. 2018;163(3):155–164. doi: 10.1093/jb/mvx087
- Nishijima Y, Hagiya Y, Kubo T, et al. RABL2 interacts with the intraflagellar transport-B complex and CEP19 and participates in ciliary assembly. Mol Biol Cell. 2017;28(12):1652–1666. doi: 10.1091/mbc.e17-01-0017
- Kanie T, Abbott KL, Mooney NA, et al. The CEP19-RABL2 GTPase complex binds IFT-B to initiate intraflagellar transport at the ciliary base. Dev Cell. 2017;42(1):22–36.e12. doi: 10.1016/j.devcel.2017.05.016
- Silva-Ayala D, López T, Gutiérrez M, et al. Genome-wide RNAi screen reveals a role for the ESCRT complex in rotavirus cell entry. Proc Natl Acad Sci. 2013;110(25):10270–10275. doi: 10.1073/pnas.1304932110
- Crawford KHD, Eguia R, Dingens AS, et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5):513. doi: 10.3390/v12050513
- Senavirathna LK, Huang C, Pushparaj S, et al. Hypoxia and transforming growth factor β1 regulation of long non-coding RNA transcriptomes in human pulmonary fibroblasts. Physiol Rep. 2020;8(1). doi: 10.14814/phy2.14343
- Huang CH, Liang Y, Zeng X, et al. Long noncoding RNA FENDRR exhibits antifibrotic activity in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2020;62(4):440–453. doi: 10.1165/rcmb.2018-0293OC
