ABSTRACT
An increased appreciation of the role of RNA dynamics in governing RNA function is ushering in a new wave of dynamic RNA synthetic biology. Here, we review recent advances in engineering dynamic RNA systems across the molecular, circuit and cellular scales for important societal-scale applications in environmental and human health, and bioproduction. For each scale, we introduce the core concepts of dynamic RNA folding and function at that scale, and then discuss technologies incorporating these concepts, covering new approaches to engineering riboswitches, ribozymes, RNA origami, RNA strand displacement circuits, biomaterials, biomolecular condensates, extracellular vesicles and synthetic cells. Considering the dynamic nature of RNA within the engineering design process promises to spark the next wave of innovation that will expand the scope and impact of RNA biotechnologies.
Introduction
RNAs play central roles in biological systems [Citation1,Citation2]. These roles range from messenger RNA sequences that encode proteins, to the folding of non-coding RNAs into structures that perform many functional roles including regulating and coordinating gene expression and performing catalysis central to life [Citation2]. Because of these features, engineered RNAs have been long sought after components of the synthetic biologists’ toolbox [Citation3], where the promise of programming the RNA sequence-structure-function relationship has great potential to contribute solutions to society’s most pressing challenges in environmental health [Citation4], human health [Citation5] and sustainable manufacturing [Citation6]. Decades of work towards this promise has resulted in powerful new technologies such as CRISPR gene editing [Citation7], mRNA vaccines [Citation5] and point-of-care biosensor diagnostics [Citation8,Citation9] with far-reaching impacts. Yet these technologies only scratch the surface of RNA’s natural abilities, suggesting that this is the beginning of an era of RNA synthetic biology.
One of the most exciting developments in RNA synthetic biology is an appreciation of how dynamic RNA behaviour influences RNA function. A renewed interest in understanding how the dynamic, out-of-equilibrium environment created by cellular processes affects how RNAs fold in the cell, and how their function is governed by these folds, is adding new layers to understanding the RNA sequence-structure-function relationship [Citation10]. This understanding is in turn teaching us new principles by which RNAs act as sensors, how RNAs control gene expression processes, and how RNAs drive the formation of higher-order structures inside cells. These principles are forming the basis of a new approach to RNA design that uses dynamic RNA properties to control the formation of functional RNA structures that can be used to program biological systems in a range of applications from diagnostics to therapeutics.
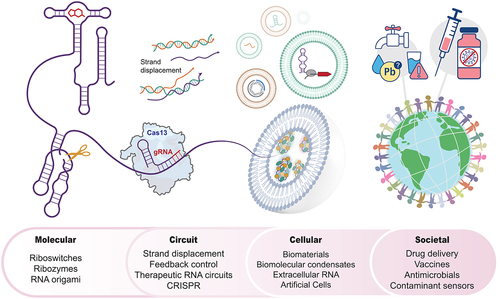
Here, we highlight recent advances and opportunities in dynamic RNA synthetic biology, organized along the spatial scales of biological systems where RNA engineering is finding the most impact (). We start with the molecular scale, where new principles for programming RNA folding dynamics are giving rise to new approaches to program riboswitches as biosensors, ribozymes as genetic control elements, and RNA origami as scaffold systems. Next, we discuss the circuit scale, where advances in controlling interactions of multiple RNAs are giving rise to engineering new dynamical systems including RNA logic circuits and feedback loops. Finally, we highlight RNA engineering at the cellular scale, where higher-order RNA assemblies are being engineered into new classes of biomaterials, into biomolecular condensates that control localization and function of cellular components, and RNAs that act as core components of extracellular vesicle signalling systems and synthetic cells. In each case, we introduce the core concepts of the scales and phenomena, and then discuss applications of technologies being developed to solve important societal-scale challenges.
Figure 1. Engineering dynamic RNA folding and function can play key roles across multiple spatiotemporal scales within engineered biological systems. From left to right: Riboswitches and ribozymes are examples of molecular-level RNA tools that control gene expression processes in synthetic biology applications. These and other RNAs can act dynamically together at the circuits scale, for example within CRISPR-Cas13-mediated and toehold-mediated strand displacement circuits to allow signal propagation, logic, timing control and other circuit-level function. At the cellular scale, engineering of RNA components can help control the function of biomolecular condensates, extracellular vesicles and synthetic cells. Finally, designing RNAs to function across the molecular, circuit and cellular scales can have societal-scale impacts through the development and deployment of medical and environmental biotechnologies.
Molecular scale
The molecular-scale phenomenon of RNA folding is intimately related to the myriad roles that non-coding RNAs play across the cell, including controlling transcription [Citation11–13], translation [Citation14–16], transcript stability [Citation15–17], epigenetic modification [Citation18,Citation19], splicing [Citation20–22], polyadenylation [Citation23] and scaffolding [Citation24]. As a single-stranded polynucleotide, RNA can flexibly fold to form diverse structures, from simple hairpins that can block transcription [Citation11] and translation [Citation14], to three-dimensional structures that bind ligands [Citation25] or perform catalysis [Citation26]. Recently, there has been renewed focus on the dynamic nature of RNA folding [Citation10], which is leading to new concepts in programming RNA folding to optimize or engineer new functions within biotechnologies [Citation27–30]. Here, we discuss several classes of RNAs where these concepts are being developed with exciting results ().
Figure 2. Recent advances in understanding and utilizing molecular-scale interactions to drive dynamic RNA folding and function. A. Riboswitches are composed of a ligand-binding aptamer domain connected to an expression platform that can dynamically change its fold to control gene expression [Citation35] B. These and other dynamic RNA folding pathways can be implemented and controlled through internal strand displacement, a process which involves exchanging base-pair complementarity between different RNA strands. Dynamic RNA switches utilize cotranscriptional strand displacement processes [Citation37]. C. Kinetic barriers to such internal strand displacement can affect riboswitch properties such as dynamic range [Citation31]. D. Engineered riboswitches have been used for field-deployable biosensing of ions and small molecules [Citation30]. E. Recent work has focused on utilizing self-cleaving and self-splicing ribozymes to control genetic processes in cellular systems in two exciting ways: F. through the automated selection of ribozymes that can sense a range of ligands in the De novo Rapid In Vitro Evolution of RNA biosensors (DRIVER) platform [Citation48] and G. through the split-ribozyme Ribozyme-ENabled Detection of RNA (RENDR) platform that acts as an RNA sensor [Citation49]. H. Beyond individual RNA molecules and switches, RNA origami is being built inside cells through the assembly of individual tiles that are cotranscriptionally folded [Citation69]. I. Fusing CRISPR guide RNAs to the RNA origamis is allowing the development and optimization of modular transcriptional regulators that function in cells [Citation73].
![Figure 2. Recent advances in understanding and utilizing molecular-scale interactions to drive dynamic RNA folding and function. A. Riboswitches are composed of a ligand-binding aptamer domain connected to an expression platform that can dynamically change its fold to control gene expression [Citation35] B. These and other dynamic RNA folding pathways can be implemented and controlled through internal strand displacement, a process which involves exchanging base-pair complementarity between different RNA strands. Dynamic RNA switches utilize cotranscriptional strand displacement processes [Citation37]. C. Kinetic barriers to such internal strand displacement can affect riboswitch properties such as dynamic range [Citation31]. D. Engineered riboswitches have been used for field-deployable biosensing of ions and small molecules [Citation30]. E. Recent work has focused on utilizing self-cleaving and self-splicing ribozymes to control genetic processes in cellular systems in two exciting ways: F. through the automated selection of ribozymes that can sense a range of ligands in the De novo Rapid In Vitro Evolution of RNA biosensors (DRIVER) platform [Citation48] and G. through the split-ribozyme Ribozyme-ENabled Detection of RNA (RENDR) platform that acts as an RNA sensor [Citation49]. H. Beyond individual RNA molecules and switches, RNA origami is being built inside cells through the assembly of individual tiles that are cotranscriptionally folded [Citation69]. I. Fusing CRISPR guide RNAs to the RNA origamis is allowing the development and optimization of modular transcriptional regulators that function in cells [Citation73].](/cms/asset/79571e66-687c-4ebe-bef4-82e6dad22078/krnb_a_2269508_f0002_oc.jpg)
Riboswitches
As the RNA chain elongates and exits the RNA polymerase during transcription, intermolecular interactions among the nascent RNA’s nucleotides and with interacting molecules in its proximity drive RNA-folding changes [Citation31–33]. The importance of such intermolecular interactions in driving dynamic RNA folding is particularly evident in riboswitches – non-coding RNA molecules that can rearrange in response to ligand binding to control gene expression at multiple levels, including transcription, translation and splicing [Citation34].
Riboswitches consist of two domains – an aptamer domain that, when folded, creates a binding pocket for a specific ligand, and an expression platform that controls a specific gene expression process through its structure [Citation35] (). For example, an expression platform that folds into an intrinsic terminator hairpin will regulate transcription elongation [Citation11], while an expression platform hairpin that sequesters a ribosome binding site will block translation initiation [Citation36]. In many cases, the aptamer and expression platform folds are mutually exclusive, allowing ligand binding to ‘flip’ the RNA structure into different conformations, leading to genetic decision-making [Citation35].
Recent studies on transcriptional riboswitches demonstrate the importance of RNA-folding dynamics in regulating transcription. Specifically, multiple transcriptional riboswitch classes that sense different ligands have all been shown to utilize internal strand displacement, where base-pairing complementarity is exchanged between regions of the growing RNA transcript (), driving large-scale changes in expression platform folding within the millisecond time scales of transcription [Citation37]. In these mechanisms, aptamer domains have been found to fold first, enabling ligand binding early in the folding pathway [Citation37]. In the absence of ligand, synthesis of additional nucleotides triggers strand displacement, leading to a specific expression platform fold. However, ligand-binding blocks this strand displacement, leading to a ligand-dependent switch [Citation33,Citation37].
Fascinatingly, more detailed studies of riboswitch strand displacement are revealing links to lessons learned from studies of DNA nanotechnology. Molecular-scale features such as base-pair mismatches, bulges, deletions or wobbles appear to slow the process of internal strand displacement [Citation31], much like how these features slow strand displacement in DNA circuits [Citation38]. In the riboswitch context, recent work shows that tuning the kinetics of strand displacement in the expression platform fold can lead to functional changes such as tuning dynamic range (), without having to alter the aptamer ligand-binding interactions [Citation31]. Furthermore, sequences can be added to compete with expression platform strand displacement folding, causing larger-scale rearrangements that flip riboswitch logic [Citation31].
Viewing riboswitches through the lens of strand displacement is establishing a new foundation for engineering riboswitch-based biosensors (). Knowledge of strand displacement from DNA nanotechnology [Citation38] creates new approaches for programming riboswitch dynamic range and sensitivity to meet application needs. Exciting opportunities also exist to leverage transcription factors that can modulate transcription kinetics by competing with ligands to bind co-transcriptionally folding RNA [Citation32]. A deeper understanding of how RNA folding dynamics direct riboswitch decisions is moving us closer to the vision of using directed evolution to generate new ligand-binding aptamers [Citation39] and rational expression platform design to make programmable biosensor switches.
Outside of bacteria, exciting new approaches are illuminating the existence and possible roles of RNA switches in human cells to treat genetic diseases by controlling gene expression by shifting the balance between RNA conformations [Citation40]. We speculate that further exploration of such RNA switches in humans will enhance our understanding of the molecular basis of gene regulation and enable the identification of novel therapeutic targets.
Ribozymes
The importance of molecular-scale interactions in driving dynamic RNA behaviour is also highlighted in ribozymes – catalytically active RNAs with diverse functions such as cleavage [Citation41,Citation42] and splicing [Citation43] (). Self-cleaving and self-splicing ribozymes were first discovered decades ago and have been used extensively to elucidate the impacts of dynamic RNA folding conditions on their ability to fold into functional forms [Citation44]. For such ribozymes, molecular-scale interactions including intra-chain base-pair complementarity and interactions with ions or small molecules in solution influence progression through different folding pathways [Citation45–47], thereby inducing transitions between active and inactive conformations and thus cleavage or splicing activity [Citation47].
Self-cleaving and self-splicing ribozymes remain at the forefront of RNA research and are enabling exciting new RNA technologies. For example, in the De novo Rapid In Vitro Evolution of RNA biosensors (DRIVER) platform, the influence of interacting molecules on folding pathway progression and consequently cleavage activity has been exploited for high-throughput biosensor development, where ribozymes that self-cleave in their native conformation are selected against cleavage in the presence of a desired ligand. The selected ribozymes can then be used to yield a translatable RNA output in a ligand-dependent fashion [Citation48] (). Base-pair complementarity has also been exploited to construct systems where RNA inputs are used to template trans-acting self-splicing ribozymes. For example, in the Ribozyme-ENabled Detection of RNA (RENDR) platform, these interactions are used to conditionally control splicing activity – in the presence of a specific input RNA, the transcript is spliced to yield an mRNA that encodes for a downstream protein output that can be used in a range of applications including detection of antibiotic resistance in microbes [Citation49] ().
Although many ribozymes are well characterized, a mechanistic understanding of their kinetics and the dynamic equilibrium that exists among their active and inactive conformations remains challenging [Citation47,Citation50]. An understanding of the rules that govern ribozyme folding pathway progression and kinetics, especially in the presence of ligands of interest, would pave the way for engineering ribozymes that avoid misfolded states and thus achieve greater kinetic functionality in applications such as sensing or controlling gene expression.
Dynamic RNA Origami
Nucleic acid origami involves DNA and RNA strands that programmatically self-assemble into two – and three-dimensional structures with potential applications in therapeutics delivery [Citation51–54], tissue engineering [Citation55,Citation56] and gene therapy [Citation57–59]. Historically, focus has been on DNA origami, where controlled annealing of high-purity DNA strands is used to achieve desired structures [Citation60,Citation61]. However, a growing interest in utilizing origami systems to control cellular processes has begun to shift focus towards RNA [Citation62,Citation63].
RNA origami provides advantages for cellular applications because RNA can be generated via transcription. However, challenges arise because RNA origami designs must consider the isothermal, out-of-equilibrium cotranscriptional folding environments of cells [Citation64–67], which are starkly different from the controlled annealing environments used for DNA origami. While complex, exciting progress is being made in RNA origami including in vivo folded double and tetra-squares [Citation68] and Z-tiles that cotranscriptionally fold and oligomerize via intramolecular kissing-loop interactions [Citation69] (). Novel characterization techniques with higher structural and temporal resolution are also enhancing our understanding of cellular RNA origami design. For example, atomistic models derived from Cryo-EM have been used to identify kinetic traps in RNA origamis that can take hours to escape in a cotranscriptional folding regime [Citation70].
A growing understanding of RNA origami design rules is enabling the development of applications such as apta-FRET systems that use fluorescent RNA aptamers scaffolded into responsive RNA origami structures [Citation71] and translation regulators in E. coli that use protein-binding RNA origami scaffolds [Citation72]. In eukaryotic cells, RNA origami scaffolds have been combined with protein-binding RNA aptamers, regulatory proteins and guide RNAs to engineer CRISPR-based systems that achieve precise and modular activation of transcription [Citation73] (). We anticipate that the expansion of cellular RNA origami will enhance our ability to engineer processes using nanomachines that can sense and respond to their cellular environment and execute precise tuning control of metabolism and gene expression, with applications in biomanufacturing and biomedicine [Citation74–77].
Circuit scale
Programming RNA interactions with other nucleic acids and proteins can be used to build molecular circuits that expand the capabilities of bioproduction strains, therapeutics and diagnostics. Recent studies have shown that synthetic circuits can interface with endogenous gene networks to control apoptosis [Citation78], differentiation [Citation79], cell proliferation [Citation79] and cell–cell communication [Citation79]. In application settings, RNA circuits are being used to control metabolic pathway flux [Citation80], to improve the efficacy and safety of ‘smart’ therapeutics [Citation81] and in field-deployable diagnostic platforms where RNA circuits can tune the specificity and sensitivity of transcription factor-based biosensors [Citation82]. A particularly powerful feature of RNA circuits is the ability to tune circuit reaction kinetics using engineering of RNA–DNA interactions [Citation83]. Here, we discuss several types of RNA-mediated circuits and their use in application contexts ().
Figure 3. Advances in dynamic RNA circuits that can be programmed to process information in a range of contexts. A. Toehold-mediated strand displacement (TMSD). TMSD gates are formed by annealing substrate and incumbent DNA or RNA strands to leave a double-stranded region and a single-stranded toehold overhang. An invading DNA or RNA strand, complementary to the substrate, can bind to the toehold and strand displace the gate, releasing the incumbent. The released strand can interact with additional downstream gates not shown, allowing gates to be configured into a range of circuit architectures [Citation83]. B. Example of TMSD OR gate. Two different gates are made with the same incumbent sequence but different toehold sequences. When two different input invaders are present, these gates release the same incumbent, which can then interact with a downstream signal gate labelled with fluorophore and quencher. When the quencher strand is released, a fluorescent signal is generated [Citation83]. C. Genelets use TMSD circuits to control the activation of transcription. Genelets consist of an incomplete T7 RNAP transcription template that is specifically lacking a strand that is complementary to the promoter. In the absence of an input, this incomplete T7 RNAP promoter genelet is in a transcription OFF state. Addition of a designed ssDNA activator that complements the promoter enables transcription activation. The outputs of genelet transcription can then act as activators for downstream genelets, or can displace existing activators via TMSD to repress downstream genelets [Citation87]. D. Cotranscriptionally encoded RNA strand displacement (ctRSD) gates allow TMSD gates to be synthesized on the fly, enabling gate designs to be encoded by easier to purify DNA templates that are activated when needed. In this design, an RNA transcript has complementary regions and folds into a double-stranded gate as it is transcribed. Inclusion of a self-cleaving ribozyme exposes a toehold region in the gate for TMSD reactions to occur, including activating fluorophore-quencher ‘signal’ gates [Citation90].
![Figure 3. Advances in dynamic RNA circuits that can be programmed to process information in a range of contexts. A. Toehold-mediated strand displacement (TMSD). TMSD gates are formed by annealing substrate and incumbent DNA or RNA strands to leave a double-stranded region and a single-stranded toehold overhang. An invading DNA or RNA strand, complementary to the substrate, can bind to the toehold and strand displace the gate, releasing the incumbent. The released strand can interact with additional downstream gates not shown, allowing gates to be configured into a range of circuit architectures [Citation83]. B. Example of TMSD OR gate. Two different gates are made with the same incumbent sequence but different toehold sequences. When two different input invaders are present, these gates release the same incumbent, which can then interact with a downstream signal gate labelled with fluorophore and quencher. When the quencher strand is released, a fluorescent signal is generated [Citation83]. C. Genelets use TMSD circuits to control the activation of transcription. Genelets consist of an incomplete T7 RNAP transcription template that is specifically lacking a strand that is complementary to the promoter. In the absence of an input, this incomplete T7 RNAP promoter genelet is in a transcription OFF state. Addition of a designed ssDNA activator that complements the promoter enables transcription activation. The outputs of genelet transcription can then act as activators for downstream genelets, or can displace existing activators via TMSD to repress downstream genelets [Citation87]. D. Cotranscriptionally encoded RNA strand displacement (ctRSD) gates allow TMSD gates to be synthesized on the fly, enabling gate designs to be encoded by easier to purify DNA templates that are activated when needed. In this design, an RNA transcript has complementary regions and folds into a double-stranded gate as it is transcribed. Inclusion of a self-cleaving ribozyme exposes a toehold region in the gate for TMSD reactions to occur, including activating fluorophore-quencher ‘signal’ gates [Citation90].](/cms/asset/8535a133-21e8-40c2-8fd1-8a2dcd08bc58/krnb_a_2269508_f0003_oc.jpg)
RNA toehold-mediated strand displacement circuits
One particularly powerful class of nucleic acid circuits is the toehold-mediated strand displacement (TMSD) circuit [Citation83]. TMSD circuits are composed of a series of nucleic acid ‘gates’ that contain a double-stranded region and a single-stranded ‘toehold’ region (). Gates are formed when two different lengths of complementary strands, denoted ‘substrate’ and ‘incumbent’, are annealed to each other [Citation84]. The sequence of the gates determines whether they respond to an input ‘invader’ strand – if the invader is complementary to the substrate, it can bind to the toehold and strand displace the incumbent. This results in the release of the incumbent which can serve as an input to another gate. In this way, networks of gates with different sequence complementary patterns can be created to process information by programmed cascades of strand displacement interactions. For example, an OR gate can be created by having two gates that have the same incumbent sequence, but different toehold regions for invaders to release the incumbent and allow it to, for example, generate signal by strand displacing a fluorophore-quencher labelled ‘signal’ gate [Citation83] ().
Cell-free biosensors can also be interfaced with in vitro transcribed TMSD circuits to improve performance and expand functionality. Allosteric transcription factors can be configured to regulate the transcription of RNAs designed to interact with signal processing TMSD gates to perform information processing such as logic computation (). Since input RNAs are only generated when the transcription factors detect specific ligands, this combination in effect creates an interface between chemical sensing and TMSD signal processing. Importantly, these circuits leverage the powerful ability to program TMSD kinetics within gate design: increasing toehold length increases TMSD circuit speed, while adding mismatches in TMSD gates slows kinetics [Citation85,Citation86]. This feature can be used to create advanced circuit architectures, for example, an analogue-to-digital conversion circuit that provides a semi-quantitative read-out of input chemical concentration [Citation83]. In general, the interface of transcription factor sensing and TMSD gates creates exciting opportunities to perform complex logic on chemical inputs.
While TMSD gates have been traditionally been implemented with DNA, progress has been made in creating nucleic-acid-based TMSD gates that can interface with input strands generated by transcription. One example is the genelet [Citation87] (). A genelet consists of a T7 RNAP transcription template where the T7 promoter region is missing a portion of a complementary DNA strand, rendering it non-functional and in a transcriptional OFF state [Citation87]. The addition of a complementary strand (activator) can complete the promoter, leading to a transcriptional ON state. Similarly, genelets can be designed to be turned OFF by leaving a toehold on the genelet construct to allow an incoming nucleic acid strand to bind, displace and disassemble the promoter. In addition, genelets can be layered, with the RNA transcribed by one genelet interacting with a downstream genelet template, which has been used to build nucleic acid implementations of bistable circuits, feedforward and feedback circuits, as well as pulse generating circuits [Citation87,Citation88]. Genelet circuits show exciting potentials to increase the modularity and scale-up of transcriptional circuits to regulate and functionalize biological systems.
Traditionally TMSD gates have been prepared by annealing DNA strands, which often requires careful purification to isolate correctly assembled gates [Citation89]. This has begun to change with a recent advance to encode TMSD gates in transcribable RNA sequences. Called cotranscriptionally encoded RNA strand displacement (ctRSD) gates [Citation90,Citation91], these systems consist of a single RNA molecule that can fold back on itself to form a double-stranded region with a toehold overhang (). Incorporation of a self-cleaving ribozyme sequence cleaves the single strand into two, thus completing the TMSD gate architecture. With this approach, ctRSD circuits have been shown to perform logic computation and multilayer cascades [Citation90]. ctRSD gates can be encoded on transcription templates which are far simpler to synthesize and purify than traditional TMSD gates [Citation91]. ctRSD circuits also have the powerful potential to be genetically encoded and continuously operated inside living cells [Citation90], thus opening new circuit design strategies to leverage gates that can be continuously produced and assembled.
Strand displacement within CRISPR technologies
The CRISPR/Cas mechanism depends on dynamic RNA interactions that influence CRISPR complex formation and target recognition. Specifically, gRNA length, secondary structure and structural stability can all affect the activity of Cas proteins [Citation92]. Exciting recent progress has leveraged this feature with the power of strand displacement to conditionally render gRNAs active or inactive, providing an additional level of control over Cas-9 genome editing activity [Citation93]. In this system, gRNAs are engineered to interact with different mRNA inputs via strand displacement to control CRISPR/Cas9 response outputs and perform logic. Programmability can also be added to Cas12a-based systems by introducing strand displacement-based circuits that control gRNA loading into Cas12a [Citation94]. In this system, the gRNA is originally unable to bind to Cas12a due to a designed secondary structure that occludes the Cas12a binding region. This interference can then be removed by a trigger RNA sequence that unfolds the gRNA via a TMSD reaction, facilitating Cas12a binding. Orthogonal trigger RNA sequences can be designed for different gRNAs to enable multi-input logic gates with digital behaviours. These TMSD gRNA circuits can be implemented in bacterial systems for logic gated transcriptional control of gene expression in E. coli [Citation94]. Finally, positive feedback loop circuits have been created in the Cas12a diagnostic system. In these systems, activation of indiscriminate Cas12a cleavage upon target recognition can be used to cleave inactive or ‘caged’ gRNAs into active gRNAs, which recruits and activates additional Cas enzymes to improve diagnostic sensitivity [Citation95]. Overall, integrating strand displacement circuits with CRISPR systems can enhance control of biological systems and improve diagnostic tools.
RNA feedback control circuits
Positive and negative feedback control circuits help the performance of complicated tasks in biological systems, tune circuit dynamics and increase system robustness [Citation83,Citation96]. However, the majority of synthetic biology feedback circuits rely on protein-based regulation to control circuit output [Citation97,Citation98]. Recent work shows that RNA-based feedback circuits can also be designed to improve system performance, including by improving the sensitivity of cell-free biosensors. For example, in the RNA Output Sensors Activated by Ligands INDuction (ROSALIND) system, allosteric transcription factors activate transcription of fluorescent RNA aptamers to produce signals in response to chemical inputs [Citation82]. Typically, the sensitivity is determined by the properties of the transcription factor. However, a negative feedback loop can be created by simultaneously activating transcription of an RNA aptamer that binds to the allosteric transcription factor and inhibits its function to allow more transcription, leading to enhanced sensitivity.
Negative feedback circuits are also useful for compensating for unpredictable disturbances. Multiple negative feedback circuits can be layered to construct combinatorial regulation, activating cascade and nested autoregulatory motifs in E. coli cells [Citation99]. Recent work has used an RNA negative feedback loop in mammalian cells to build robust control systems [Citation96]. In this case, there are two genes that are encoded on two separate plasmids. The first plasmid (activator) expresses a synthetic transcription factor that activates the expression of an antisense RNA encoded on a separate plasmid. The antisense RNA hybridizes with the activator mRNA in a sequestration reaction, thus closing the negative feedback loop. The closed-loop feedback control allows the system to robustly deal with disturbances and adjust to initial setpoints, which can be adjusted by varying different ratios of the plasmids. Prospective application of this feedback circuit in mammalian system includes exogenous insulin therapy for type 1 diabetes mellitus, as well as other applications in biotechnology and biomedicine that require robust controllers to restore homoeostasis and deliver precise drug doses [Citation96].
RNA circuits are also shown to be important components of new classes of therapeutics based on circular RNA (circRNA), which has shown promise due to increased stability and persistent gene expression in cells compared to non-circularized RNAs [Citation100,Citation101]. However, controlling translation of circRNAs has been challenging in cells due to a lack of mechanisms to sense intracellular conditions and autonomously control translation level. Recent work showed that circRNA protein expression can be controlled by a circuit with miRNA or protein inputs. In this circuit design, miRNA-binding and protein-binding sequences are inserted into the untranslated regions of circRNAs so gene expression can be turned off in the presence of these inputs, either through mRNA degradation for a miRNA input, or in cap-dependent translation repression for a protein input. Since the activity of miRNAs differs among cell types, miRNA levels can be used to precisely control circRNA expression between different cell types [Citation102].
Cellular scale
An exciting new trend in synthetic biology is designing RNAs to accomplish cellular-scale engineering. These include RNAs designed to be incorporated into biomaterials and synthetic biomolecular condensates, as components of extracellular RNA-based signalling and delivery platforms, and as key components of artificial cells. New challenges arise at the cellular scale, including those related to larger-scale phenomena such as transport and structural mechanics, as well as those associated with the complex in vivo environments where many of these systems are applied. Recent work is addressing these challenges, creating cellular scale engineered RNA systems that address therapeutic applications with exciting new developments that leverage the dynamic properties of RNA ().
Figure 4. Cellular-scale dynamic RNA design has many biotechnological applications. A. RNA can be designed to form nanowires regulated by the blue light-responsive protein [Citation108]. This enables functions such as dynamic RNA release that can be used in therapeutic applications. B. RNA can also be designed to form artificial condensates or to modulate natural condensates. Altering the balance of charge between anionic RNA and cationic proteins can be used to regulate condensate formation and dissolution [Citation119–121]. C. Extracellular vesicles secreted from cells contain many types of RNA that could be detected to diagnose diseases [Citation125–127]. D. Artificial cells can also be engineered to form biosensors by encapsulating transcription/translation machinery [Citation107]. For example, this has been applied to build an encapsulated fluoride riboswitch followed by a reporter gene to detect fluoride in water [Citation144].
![Figure 4. Cellular-scale dynamic RNA design has many biotechnological applications. A. RNA can be designed to form nanowires regulated by the blue light-responsive protein [Citation108]. This enables functions such as dynamic RNA release that can be used in therapeutic applications. B. RNA can also be designed to form artificial condensates or to modulate natural condensates. Altering the balance of charge between anionic RNA and cationic proteins can be used to regulate condensate formation and dissolution [Citation119–121]. C. Extracellular vesicles secreted from cells contain many types of RNA that could be detected to diagnose diseases [Citation125–127]. D. Artificial cells can also be engineered to form biosensors by encapsulating transcription/translation machinery [Citation107]. For example, this has been applied to build an encapsulated fluoride riboswitch followed by a reporter gene to detect fluoride in water [Citation144].](/cms/asset/5b8ad558-3adb-4723-a734-9f11f0f9fc41/krnb_a_2269508_f0004_oc.jpg)
Biomaterials
RNA biomaterials are ordered structural networks that span multiple-length scales, from nanostructures to larger mesoscale materials [Citation103]. While much of the work in nucleic acid nanomaterials originated with DNA, recently researchers have recognized the potential for RNA’s wider range of structural motifs to expand nanomaterial functionality [Citation103], such as metabolic pathway scaffolding [Citation104].
Some of the earliest work in RNA material design engineered squares of RNA called tectosquares to assemble into many different patterns and sizes [Citation105]. Interestingly, RNA-based nanostructures can be tuned to have ‘rubber-like’ properties, meaning that they can bend and return to their original shape [Citation106]. RNA nanostructures can also act as scaffolds to incorporate protein components, providing additional functionality. For example, nanowires have been engineered from RNA dimers that can bind to peptides fused with proteins, forming a wire structure in which the length could be regulated by the blue light-responsive protein LOV2 [Citation108]. For soft material engineering, nucleic acids can be engineered to form hydrogels, consisting of a network of cross-links that are formed from base pairing interactions (). In DNA hydrogels, strand displacement reactions have been used to break or re-form hydrogel crosslinks, causing hydrogels to swell or propagate waves of autocatalytic amplification reactions in response to strand-displacing DNA or chemical stimuli [Citation109,Citation110]. Hydrogel engineering can also leverage higher-order RNA structures and dynamic RNA design features, such as using RNA G-quadruplexes for phase separation and sub-localization of translation [Citation111].
In application spaces, nucleic acid-based biomaterials are being pursued for therapeutics, where they are used in drug and vaccine delivery, tissue engineering and other applications [Citation112]. The complexity of in vivo environments, however, presents challenges with stability and toxicity [Citation103]. RNA modifications, such as fluorination of the 2’ ribose position, can improve resistance to nucleases while retaining RNA structure and biological activity [Citation113]. There are opportunities to use dynamic RNA design principles to further enhance RNA material function. For example, size, shape and sequence of RNA nanoparticles can be designed to optimize immunogenicity [Citation114]. Programming these nanostructures to self-assemble at the in vivo target site [Citation68] could further alleviate challenges with immunogenic structures. RNA structures are also transient, enabling delivery of, for example, a hepatocellular carcinoma treatment that is then cleared quickly from the system after treatment [Citation115]. Hydrogels have been engineered out of RNA utilizing structures such as the triple helix, a strategy that is showing promise in potential breast cancer treatments [Citation116–118]. The dynamic nature of RNA has been shown to be useful for regulating soft material properties and optimizing RNA therapeutics kinetics. Full leverage of dynamic RNA properties in biomaterials has the potential to create highly programmable biocompatible therapeutics.
Biomolecular condensates
At the cellular scale, molecular function – including enzymatic reaction kinetics, gene regulation and macromolecular assembly – can be regulated by controlling localization [Citation119,Citation120]. While membrane-bound organelles have long been appreciated to achieve this, there has been exciting recent progress in our understanding of how biomolecular condensates can achieve ‘membrane-less organelle’ function [Citation121]. Biomolecular condensates are cellular coacervates, formed when negatively charged RNAs and tens to hundreds of different charged proteins interact with each other to form micron-scale structures through a phase separation process () [Citation121]. Condensate formation is governed by charge balance – lower RNA concentrations promote condensate formation through weak, multivalent interactions between proteins and RNA [Citation122], while higher concentrations of RNA disfavour the formation because of the large negative charge [Citation123]. Biomolecular condensates have many functions, including gene regulation, signal transduction, RNA metabolism, and translation regulation [Citation121,Citation122].
In addition to modifying existing condensates, artificial condensates have also been developed that can add additional functionalities, such as giving optogenetic control over phase transition behaviour [Citation124]. While most condensate engineering has focused on the protein components, there is a great opportunity in the future to engineer dynamic properties of the RNA components to control the timing of formation, location and regulatory function to expand the tools available in this area.
Extracellular vesicles
While RNA is mostly recognized for its cellular roles, it can also play important extracellular roles as core components of membrane-bound extracellular vesicles (EVs) (). EVs consist of membrane-bound coding and non-coding RNAs, which can be present alongside proteins, lipids, carbohydrates and DNA [Citation125–127]. RNA-binding proteins can regulate RNA enrichment in EVs through RNA sequence motif recognition [Citation128]. Membrane encapsulation of RNA protects them from extracellular nucleases present in tissues, allowing RNAs to be transported between cells [Citation127]. In this way, EVs can play important roles in intercellular communication [Citation129–131].
Because extracellular RNAs within EVs have been associated with many types of cancer, a number of diagnostic approaches focus on capturing and analysing EVs [Citation132]. For example, a colorimetric assay was developed to detect RNA in cancer exosomes [Citation133]. Vesicles with RNAs are also released by pathogens such as Epstein–Barr virus and uropathogenic E. coli [Citation134,Citation135], suggesting that those vesicles could be a target for nucleic acid-based pathogen detection.
Because of their natural role in delivering RNA components to cells, applications of engineered EVs have mostly focused on solving therapeutic delivery challenges. In one study, red blood cell-based vesicles were used to transport therapeutic antisense oligonucleotides to leukaemia and breast cancer cells to inhibit a miRNA [Citation136]. To improve targeting, T-cell-targeting single-chain variable fragments were displayed on an EV surface to target the CRISPR-Cas9 cargo to T-cells [Citation137]. Dynamic properties of RNA can be used to enable complex circuitry in cargo delivery to expand the functions of EVs.
Synthetic cells
Engineered RNAs are also emerging as important components of synthetic cells. There has been a long-standing interest in both top-down and bottom-up approaches to building synthetic cells [Citation138,Citation139]. Recent work has been expanding the capabilities of synthetic cells to include intercellular communication [Citation140,Citation141] and artificial membrane-less organelle compartmentalization [Citation142,Citation143].
Many of the principles of molecular and network-scale engineering of RNA systems discussed above can be directly applied in a synthetic cell context. For example, encapsulated genetic networks and riboswitch biosensors have been explored that offer protection from environmental matrix effects through having a barrier, which can be designed to be selective to the molecule of interest [Citation144–146]. In this way, fluoride biosensors have been created by encapsulating a fluoride riboswitch-regulated reporter in a membrane that was able to sense environmental fluoride while preventing RNase-mediated degradation from the environment () [Citation146]. In another example, a histamine-sensing riboswitch was encapsulated to trigger a histamine-responsive release of small molecules or cleavage of the phospholipid membrane in an artificial cell context [Citation147]. There is great promise for adding additional RNA-level functionalities, such as dynamic regulatory circuitry and assembly of higher-order structures, to enhance the function of synthetic cells in applications, and even help uncover principles of the origins of natural cells [Citation148,Citation149]. By leveraging encapsulating technologies that are based on dynamic RNA such as genetic networks and riboswitch-based biosensors, the capabilities of synthetic cells have been expanded and can perform higher-order functions.
Discussion
The dynamic nature of RNA folding and function is giving rise to new concepts, tools and approaches that are being applied across the molecular, circuit and cellular scales to create exciting new RNA-based technologies that address societal-scale challenges in environmental health, sustainable biomanufacturing and human health. However, we are likely only beginning to understand how dynamic RNA behaviour manifests itself across scales, and how this behaviour can be leveraged to engineer RNA systems. New fundamental approaches to understanding RNA cotranscriptional folding and processing [Citation10], assembly of RNA systems [Citation87] and the control of biomolecular condensates [Citation119] promise to accelerate the discovery of new dynamic RNA design principles that can be used to program RNA function. At the same time, the continual merger of concepts from DNA nanotechnology [Citation38] into RNA synthetic biology [Citation84] promises to introduce powerful new design motifs into engineered RNA systems that interface more naturally with other biological functions. These two trends, combined with recent efforts in computational modelling of RNA folding pathways [Citation150] and breakthroughs in AI-directed RNA 3D structure prediction [Citation151], promise to dramatically accelerate RNA design. Although we are still far from fully grasping the nuances and potential of dynamic RNA behaviour, a mindset that views RNA through the lens of dynamics [Citation152] will serve the field especially well in the pursuit of a comprehensive understanding of RNA behaviour and its potential applications.
Author contributions
All authors conceptualized, wrote, reviewed, and edited the manuscript.
Data availability
All relevant data is contained within the manuscript.
Acknowledgments
We thank Edric Choi for contributions to figures. We also thank Nestor Sampedro Valina for insightful conversations on RNA origami.
Disclosure statement
J.B.L has submitted national and international patent applications related to cell-free biosensing technologies. J.B.L. is a founder and has financial interests in Stemloop, Inc. which is commercializing cell-free biosensing technologies. These interests are reviewed and managed by Northwestern University in according with their conflict-of-interest policies. Y. L., A. A., T. L. and R. A. R. declare no competing interests.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Sharp PA. The centrality of RNA. Cell. 2009;136(4):577–580.
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94.
- Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24(5):545–554.
- Thavarajah W, Hertz LM, Bushhouse DZ, et al. RNA engineering for public health: innovations in RNA-based diagnostics and therapeutics. Annu Rev Chem Biomol Eng. 2021;12(1):263–286. doi: 10.1146/annurev-chembioeng-101420-014055.
- Pardi N, Hogan MJ, Porter FW, et al. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243.
- Lo J, Humphreys JR, Jack J, et al. The metabolism of clostridium ljungdahlii in phosphotransacetylase negative strains and development of an ethanologenic strain. Front Bioeng Biotechnol. 2020;8:560726.
- Jinek M, East A, Cheng A, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471–e00471.
- Kellner MJ, Koob JG, Gootenberg JS, et al. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2.
- Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245.
- Bushhouse DZ, Choi EK, Hertz LM, et al. How does RNA fold dynamically? J Mol Biol. 2022;434(18):167665. doi: 10.1016/j.jmb.2022.167665.
- Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284(5414):611–615.
- Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes to cells: devoted to molecular & cellular mechanisms. Genes to Cells: Devoted to Molecular & Cellular Mechanisms. 2002;7(8):755–768.
- Larson MH, Greenleaf WJ, Landick R, et al. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132(6):971–982. doi: 10.1016/j.cell.2008.01.027.
- de Smit MH, van Duin J. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol. 2003;331(4):737–743.
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3(12):a003798.
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379.
- Alifano P, Bruni CB, Carlomagno MS. Control of mRNA processing and decay in prokaryotes. Genetica. 1994;94(2–3):157–172.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641.
- Wei J-W, Huang K, Yang C, et al. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9. doi: 10.3892/or.2016.5236.
- Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21(4):366–372.
- Saldi T, Cortazar MA, Sheridan RM, et al. Coupling of RNA polymerase ii transcription elongation with pre-mRNA splicing. J Mol Biol. 2016;428(12):2623–2635. doi: 10.1016/j.jmb.2016.04.017.
- Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24(24):10505–10514.
- Wu X, Bartel DP. Widespread influence of 3’-end structures on mammalian mRNA processing and stability. Cell. 2017;169(5):905–917.e11.
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145–166.
- Ren A, Rajashankar KR, Patel DJ. Global RNA fold and molecular recognition for a pfl riboswitch bound to ZMP, a master regulator of one-carbon metabolism. Structure. 2015;23(8):1375–1381.
- Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280(4):597–609.
- Holtkamp S, Kreiter S, Selmi A, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108(13):4009–4017.
- Zhou J, Lazar D, Li H, et al. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics. 2018;8(6):1575–1590. doi: 10.7150/thno.23085.
- Wheeler LA, Vrbanac V, Trifonova R, et al. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol ther. 2013;21(7):1378–1389. doi: 10.1038/mt.2013.77.
- Thavarajah W, Silverman AD, Verosloff MS, et al. Point-of-use detection of environmental fluoride via a cell-free riboswitch-based biosensor. ACS Synth Biol. 2020;9(1):10–18. doi: 10.1021/acssynbio.9b00347.
- Bushhouse DZ, Lucks JB. Tuning strand displacement kinetics enables programmable ZTP riboswitch dynamic range in vivo. Nucleic Acids Res. 2023;51(6):2891–2903.
- Chauvier A, Ajmera P, Yadav R, et al. Dynamic competition between a ligand and transcription factor NusA governs riboswitch-mediated transcription regulation. Proc Natl Acad Sci U S A. 2021;118(47):e2109026118. doi: 10.1073/pnas.2109026118.
- Cheng L, White EN, Brandt NL, et al. Cotranscriptional RNA strand exchange underlies the gene regulation mechanism in a purine-sensing transcriptional riboswitch. Nucleic Acids Res. 2022;50(21):12001–12018. doi: 10.1093/nar/gkac102.
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5(6):451–463.
- Landgraf T, Völklein AE, Fürtig B, et al. The cotranscriptional folding landscape for two cyclic di-nucleotide-sensing riboswitches with highly homologous aptamer domains acting either as ON- or OFF-switches. Nucleic Acids Res. 2022;50(12):6639–6655. doi: 10.1093/nar/gkac514.
- de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990;87(19):7668–7672.
- Watters KE, Strobel EJ, Yu AM, et al. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol. 2016;23(12):1124–1131. doi: 10.1038/nsmb.3316.
- Machinek RRF, Ouldridge TE, Haley NEC, et al. Programmable energy landscapes for kinetic control of DNA strand displacement. Nat Commun. 2014;5(1):5324. doi: 10.1038/ncomms6324.
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822.
- Khoroshkin M, Asarnow D, Navickas A, et al. A systematic search for RNA structural switches across the human transcriptome. bioRxiv. 2023;2023.03.11.532161.
- Roth A, Weinberg Z, Chen AG, et al. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol. 2014;10(1):56–60. doi: 10.1038/nchembio.1386.
- Zaug AJ, Been MD, Cech TR. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986;324(6096):429–433.
- Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59(1):543–568.
- Heilman-Miller SL, Woodson SA. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9(6):722–733.
- Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308(5):839–851.
- Das R, Kwok LW, Millett IS, et al. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol. 2003;332(2):311–319. doi: 10.1016/s0022-2836(03)00854-4.
- Korman A, Sun H, Hua B, et al. Light-controlled twister ribozyme with single-molecule detection resolves RNA function in time and space. Proc Natl Acad Sci U S A. 2020;117(22):12080–12086. doi: 10.1073/pnas.2003425117.
- Townshend B, Xiang JS, Manzanarez G, et al. A multiplexed, automated evolution pipeline enables scalable discovery and characterization of biosensors. Nat Commun. 2021;12(1):1437. doi: 10.1038/s41467-021-21716-0.
- Gambill L, Staubus A, Mo KW, et al. A split ribozyme that links detection of a native RNA to orthogonal protein outputs. Nat Commun. 2023;14(1):543. doi: 10.1038/s41467-023-36073-3.
- Li S, Palo MZ, Zhang X, et al. Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM. Nat Commun. 2023;14(1):1294. doi: 10.1038/s41467-023-36724-5.
- Qi X, Liu X, Matiski L, et al. RNA origami nanostructures for potent and safe anticancer immunotherapy. ACS nano. 2020;14(4):4727–4740. doi: 10.1021/acsnano.0c00602.
- Oktay E, Alem F, Hernandez K, et al. DNA origami presenting the receptor binding domain of SARS-CoV-2 elicit robust protective immune response. Commun Biol. 2023;6(1):308. doi: 10.1038/s42003-023-04689-2.
- Du RR, Cedrone E, Romanov A, et al. Innate Immune stimulation using 3D wireframe DNA origami. ACS nano. 2022;16(12):20340–20352. doi: 10.1021/acsnano.2c06275.
- Wamhoff EC, Ronsard L, Feldman J, et al. Enhancing antibody responses by multivalent antigen display on thymus-independent DNA origami scaffolds. bioRxiv:Preprint Serv Biol. 2023. 2022.08.16.504128. doi: 10.1101/2022.08.16.504128
- Li S, Liu Y, Tian T, et al. Bioswitchable delivery of microRNA by framework nucleic acids: application to bone regeneration. Small. 2021;17(47):e2104359–e2104359. doi: 10.1002/smll.202104359.
- Kim F, Chen T, Burgess T, et al. Functionalized DNA nanostructures as scaffolds for guided mineralization. Chem Sci. 2019;10(45):10537–10542. doi: 10.1039/C9SC02811K.
- Wu X, Liu Q, Liu F, et al. An RNA/DNA hybrid origami-based nanoplatform for efficient gene therapy. Nanoscale. 2021;13(30):12848–12853. doi: 10.1039/D1NR00517K.
- Liu J, Song L, Liu S, et al. A tailored DNA nanoplatform for synergistic RNAi-/chemotherapy of multidrug-resistant tumors. Angew Chem Int Ed Engl. 2018;57(47):15486–15490. doi: 10.1002/anie.201809452.
- Wu X, Yang C, Wang H, et al. Genetically encoded DNA origami for gene therapy in vivo. J Am Chem Soc. 2023;145(16):9343–9353. doi: 10.1021/jacs.3c02756.
- Jun H, Shepherd TR, Zhang K, et al. Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges. ACS nano. 2019;13(2):2083–2093. doi: 10.1021/acsnano.8b08671.
- Julin S, Keller A, Linko V. Dynamics of DNA Origami Lattices. Bioconjug Chem. 2023;34(1):18–29.
- Zhou L, Chandrasekaran AR, Yan M, et al. A mini DNA-RNA hybrid origami nanobrick. Nanoscale Adv. 2021;3(14):4048–4051. doi: 10.1039/D1NA00026H.
- Delebecque CJ, Lindner AB, Silver PA, et al. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333(6041):470–474. doi: 10.1126/science.1206938.
- Torelli E, Kozyra J, Shirt-Ediss B, et al. Cotranscriptional folding of a bio-orthogonal fluorescent scaffolded RNA origami. ACS Synth Biol. 2020;9(7):1682–1692. doi: 10.1021/acssynbio.0c00009.
- Geary C, Rothemund PWK, Andersen ES. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science. 2014;345(6198):799–804.
- Geary C, Grossi G, McRae EK, et al. RNA origami design tools enable cotranscriptional folding of kilobase-sized nanoscaffolds. Nat Chem. 2021;13(6):549–558. doi: 10.1038/s41557-021-00679-1.
- Chopra A, Sagredo S, Grossi G, et al. Out-of-plane aptamer functionalization of RNA three-helix tiles. Nanomaterials. 2019;9(4):507.
- Li M, Zheng M, Wu S, et al. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat Commun. 2018;9(1):2196. doi: 10.1038/s41467-018-04652-4.
- Liu D, Geary CW, Chen G, et al. Branched kissing loops for the construction of diverse RNA homooligomeric nanostructures. Nat Chem. 2020;12(3):249–259. doi: 10.1038/s41557-019-0406-7.
- McRae EKS, McRae EK, Rasmussen HØ, et al. Structure, folding and flexibility of co-transcriptional RNA origami. Nat Nanotechnol. 2023;18:10.1038/s41565-023-01321–6.
- Jepsen MDE, Sparvath SM, Nielsen TB, et al. Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat Commun. 2018;9(1):18. doi: 10.1038/s41467-017-02435-x.
- Nguyen MTA, Pothoulakis G, Andersen ES. Synthetic translational regulation by protein-binding RNA origami scaffolds. ACS Synth Biol. 2022;11(5):1710–1718.
- Pothoulakis G, Nguyen MTA, Andersen ES. Utilizing RNA origami scaffolds in Saccharomyces cerevisiae for dCas9-mediated transcriptional control. Nucleic Acids Res. 2022;50(12):7176–7187.
- Jasinski D, Haque F, Binzel DW, et al. Advancement of the emerging field of RNA nanotechnology. ACS nano. 2017;11(2):1142–1164. doi: 10.1021/acsnano.6b05737.
- Zhu G, Song P, Wu J, et al. Application of nucleic acid frameworks in the construction of nanostructures and cascade biocatalysts: recent progress and perspective. Front Bioeng Biotechnol. 2022;9:792489.
- Hu M, Feng C, Yuan Q, et al. Lantern-shaped flexible RNA origami for Smad4 mRNA delivery and growth suppression of colorectal cancer. Nat Commun. 2023;14(1):1307. doi: 10.1038/s41467-023-37020-y.
- Krissanaprasit A, Key CM, Froehlich K, et al. Multivalent aptamer-functionalized single-strand RNA origami as effective, target-specific anticoagulants with corresponding reversal agents. Adv Healthc Mater. 2021;10(11):e2001826–e2001826. doi: 10.1002/adhm.202001826.
- Sedlmayer F, Aubel D, Fussenegger M. Synthetic gene circuits for the detection, elimination and prevention of disease. Nat Biomed Eng. 2018;2(6):399–415.
- Wong RS, Chen YY, Smolke CD. Regulation of T cell proliferation with drug-responsive microRNA switches. Nucleic Acids Res. 2018;46(3):1541–1552.
- Stevens JT, Carothers JM. Designing RNA-based genetic control systems for efficient production from engineered metabolic pathways. ACS Synth Biol. 2015;4(2):107–115.
- Li Y, Su Z, Zhao W, et al. Multifunctional oncolytic nanoparticles deliver self-replicating IL-12 RNA to eliminate established tumors and prime systemic immunity. Nat Cancer. 2020;1(9):882–893. doi: 10.1038/s43018-020-0095-6.
- Jung JK, Alam KK, Verosloff MS, et al. Cell-free biosensors for rapid detection of water contaminants. Nat Biotechnol. 2020;38(12):1451–1459. doi: 10.1038/s41587-020-0571-7.
- Jung JK, Archuleta CM, Alam KK, et al. Programming cell-free biosensors with DNA strand displacement circuits. Nat Chem Biol. 2022;18(4):385–393. doi: 10.1038/s41589-021-00962-9.
- Hong F, Šulc P. An emergent understanding of strand displacement in RNA biology. J Struct Biol. 2019;207(3):241–249.
- Haley NEC, Ouldridge TE, Mullor Ruiz I, et al. Design of hidden thermodynamic driving for non-equilibrium systems via mismatch elimination during DNA strand displacement. Nat Commun. 2020;11(1):2562. doi: 10.1038/s41467-020-16353-y.
- Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131(47):17303–17314.
- Schaffter SW, Schulman R. Building in vitro transcriptional regulatory networks by successively integrating multiple functional circuit modules. Nat Chem. 2019;11(9):829–838.
- Schaffter SW, Chen K-L, O’Brien J, et al. Standardized excitable elements for scalable engineering of far-from-equilibrium chemical networks. Nat Chem. 2022;14(11):1224–1232. doi: 10.1038/s41557-022-01001-3.
- Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332(6034):1196–1201.
- Schaffter SW, Strychalski EA. Cotranscriptionally encoded RNA strand displacement circuits. Sci Adv. 2022;8(12):eabl4354–eabl4354.
- Schaffter SW, Wintenberg ME, Murphy TM, et al. Design approaches to expand the toolkit for building cotranscriptionally encoded RNA strand displacement circuits. ACS Synth Biol. 2023;12(5):1546–1561. doi: 10.1021/acssynbio.3c00079.
- Konstantakos V, Nentidis A, Krithara A, et al. CRISPR-Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning. Nucleic Acids Res. 2022;50(7):3616–3637. doi: 10.1093/nar/gkac192.
- Li Y, Teng X, Zhang K, et al. RNA strand displacement responsive CRISPR/cas9 system for mRNA sensing. Anal Chem. 2019;91(6):3989–3996. doi: 10.1021/acs.analchem.8b05238.
- Oesinghaus L, Simmel FC. Switching the activity of Cas12a using guide RNA strand displacement circuits. Nat Commun. 2019;10(1):2092.
- Shi K, Xie S, Tian R, et al. A CRISPR-Cas autocatalysis-driven feedback amplification network for supersensitive DNA diagnostics. Sci Adv. 2021;7(5):eabc7802. doi: 10.1126/sciadv.abc7802.
- Frei T, Chang C-H, Filo M, et al. A genetic mammalian proportional-integral feedback control circuit for robust and precise gene regulation. Proc Natl Acad Sci U S A. 2022;119(24):e2122132119–e2122132119. doi: 10.1073/pnas.2122132119.
- Gao XJ, Chong LS, Kim MS, et al. Programmable protein circuits in living cells. Science. 2018;361(6408):1252–1258. doi: 10.1126/science.aat5062.
- Chen Z, Elowitz MB. Programmable protein circuit design. Cell. 2021;184(9):2284–2301.
- Hu CY, Murray RM. Layered feedback control overcomes performance trade-off in synthetic biomolecular networks. Nat Commun. 2022;13(1):5393.
- Chen R, Wang SK, Belk JA, et al. Engineering circular RNA for enhanced protein production. Nat Biotechnol. 2023;41(2):262–272. doi: 10.1038/s41587-022-01393-0.
- He AT, Liu J, Li F, et al. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6(1):185. doi: 10.1038/s41392-021-00569-5.
- Kameda S, Ohno H, Saito H. Synthetic circular RNA switches and circuits that control protein expression in mammalian cells. Nucleic Acids Res. 2023;51(4):e24–e24.
- Langlois NI, Ma KY, Clark HA. Nucleic acid nanostructures for in vivo applications: the influence of morphology on biological fate. Appl Phys Rev. 2023;10(1):011304.
- Sachdeva G, Garg A, Godding D, et al. In vivo co-localization of enzymes on RNA scaffolds increases metabolic production in a geometrically dependent manner. Nucleic Acids Res. 2014;42(14):9493–9503. doi: 10.1093/nar/gku617.
- Chworos A, Severcan I, Koyfman AY, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306(5704):2068–2072. doi: 10.1126/science.1104686.
- Ghimire C, Wang H, Li H, et al. RNA nanoparticles as rubber for compelling vessel extravasation to enhance tumor targeting and for fast renal excretion to reduce toxicity. ACS nano. 2020;14(10):13180–13191. doi: 10.1021/acsnano.0c04863.
- Li J, Zhao H, Zheng L, et al. Advances in synthetic biology and biosafety governance. Front Bioeng Biotechnol. 2021;9:598087.
- Younas T, Liu C, Struwe WB, et al. Engineer RNA-protein nanowires as light-responsive biomaterials. Small. 2023;19(12):e2206513–e2206513. doi: 10.1002/smll.202206513.
- Fern J, Schulman R. Modular DNA strand-displacement controllers for directing material expansion. Nat Commun. 2018;9(1):3766.
- Dorsey PJ, Scalise D, Schulman R. A model of spatio-temporal regulation within biomaterials using DNA reaction-diffusion waveguides. R Soc Open Sci. 2022;9(8):220200.
- Ahn SY, Kim J, Vellampatti S, et al. Protein-encoding free-standing RNA hydrogel for sub-compartmentalized translation. Adv Mater. 2022;34(18):e2110424–e2110424. doi: 10.1002/adma.202110424.
- Gačanin J, Synatschke CV, Weil T. Biomedical applications of DNA-based hydrogels. Adv Funct Mater. 2020;30(4):1906253.
- Liu J, Guo S, Cinier M, et al. Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS nano. 2011;5(1):237–246. doi: 10.1021/nn1024658.
- Guo S, Li H, Ma M, et al. Size, shape, and sequence-dependent immunogenicity of RNA nanoparticles. Mol Ther Nucleic Acids. 2017;9:399–408.
- Wang H, Ellipilli S, Lee W-J, et al. Multivalent rubber-like RNA nanoparticles for targeted co-delivery of paclitaxel and MiRNA to silence the drug efflux transporter and liver cancer drug resistance. J Control Release. 2021;330:173–184.
- Wang W, Liu X, Ding L, et al. RNA hydrogel combined with MnO(2) nanoparticles as a nano-vaccine to treat triple negative breast cancer. Front Chem. 2021;9:797094.
- Shinde SS, Ahmed S, Malik JA, et al. Therapeutic delivery of tumor suppressor miRNAs for breast cancer treatment. Biology (Basel). 2023;12(3):467. doi: 10.3390/biology12030467.
- Ding L, Li J, Wu C, et al. A self-assembled RNA-triple helix hydrogel drug delivery system targeting triple-negative breast cancer. J Mat Chem B. 2020;8(16):3527–3533. doi: 10.1039/C9TB01610D.
- Ryder PV, Lerit DA. RNA localization regulates diverse and dynamic cellular processes. Traffic. 2018;19(7):496–502.
- Kojima T, Takayama S. Membraneless compartmentalization facilitates enzymatic cascade reactions and reduces substrate inhibition. ACS Appl Mater Interfaces. 2018;10(38):32782–32791.
- Banani SF, Lee HO, Hyman AA, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285–298. doi: 10.1038/nrm.2017.7.
- Boeynaems S, Chong S, Gsponer J, et al. Phase separation in biology and disease; current perspectives and open questions. J Mol Biol. 2023;435(5):167971. doi: 10.1016/j.jmb.2023.167971.
- Henninger JE, Oksuz O, Shrinivas K, et al. RNA-mediated feedback control of transcriptional condensates. Cell. 2021;184(1):207–225.e24. doi: 10.1016/j.cell.2020.11.030.
- Wei M-T, Chang Y-C, Shimobayashi SF, et al. Nucleated transcriptional condensates amplify gene expression. Nat Cell Biol. 2020;22(10):1187–1196. doi: 10.1038/s41556-020-00578-6.
- O’Grady T, Njock M-S, Lion M, et al. Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biol. 2022;20(1):72. doi: 10.1186/s12915-022-01277-4.
- Henderson MC, Azorsa DO. The genomic and proteomic content of cancer cell-derived exosomes. Front Oncol. 2012;2:38.
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579.
- Fabbiano F, Corsi J, Gurrieri E, et al. RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J Extracell Vesicles. 2020;10(2):e12043–e12043. doi: 10.1002/jev2.12043.
- Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43–51.
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383.
- Wu D, Tao T, Eshraghian EA, et al. Extracellular RNA as a kind of communication molecule and emerging cancer biomarker. Front Oncol. 2022;12:960072.
- Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78.
- Yan E, Goyal G, Yildiz UH, et al. Colorimetric assaying of exosomal metabolic biomarkers. Molecules. 2023;28(4):1909.
- Cone AS, York SB, Meckes DG Jr. Extracellular vesicles in Epstein-Barr virus pathogenesis. Curr Clinic Microbiol Rep. 2019;6(3):121–131.
- Blenkiron C, Simonov D, Muthukaruppan A, et al. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PloS one. 2016;11(8):e0160440–e0160440. doi: 10.1371/journal.pone.0160440.
- Usman WM, Pham TC, Kwok YY, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9(1):2359. doi: 10.1038/s41467-018-04791-8.
- Stranford DM, Simons LM, Berman KE, et al. Bioengineering multifunctional extracellular vesicles for targeted delivery of biologics to T cells. bioRxiv. 2022;2022.05.14.491879.
- Wang C, Yang J, Lu Y. Modularize and unite: toward creating a functional artificial cell. Front Mol Biosci. 2021;8:781986.
- Mann S. Systems of creation: the emergence of life from nonliving matter. Acc Chem Res. 2012;45(12):2131–2141.
- Buddingh BC, Elzinga J, van Hest JCM. Intercellular communication between artificial cells by allosteric amplification of a molecular signal. Nat Commun. 2020;11(1):1652.
- Heili JM, Stokes K, Gaut NJ, et al. Controlled exchange of protein and nucleic acid signals from and between synthetic minimal cells. bioRxiv. 2022;2022.01.03.474826.
- Robinson AO, Lee J, Cameron A, et al. Cell-free expressed membraneless organelles sequester RNA in synthetic cells. bioRxiv. 2023. 2023.04.03.535479. doi: 10.1101/2023.04.03.535479
- Ghosh B. Artificial cell design: reconstructing biology for life science applications. Emerging Topics Life Sci. 2022;6(6):619–627.
- Boyd MA, Kamat NP. Designing artificial cells towards a new generation of biosensors. Trends Biotechnol. 2021;39(9):927–939.
- Ding Y, Contreras-Llano LE, Morris E, et al. Minimizing context dependency of gene networks using artificial cells. ACS Appl Mater Interfaces. 2018;10(36):30137–30146. doi: 10.1021/acsami.8b10029.
- Boyd MA, Thavarajah W, Lucks JB, et al. Robust and tunable performance of a cell-free biosensor encapsulated in lipid vesicles. Sci Adv. 2023;9(1):eadd6605–eadd6605. doi: 10.1126/sciadv.add6605.
- Dwidar M, Seike Y, Kobori S, et al. Programmable artificial cells using histamine-responsive synthetic riboswitch. J Am Chem Soc. 2019;141(28):11103–11114. doi: 10.1021/jacs.9b03300.
- O’Flaherty DK, Kamat NP, Mirza FN, et al. Copying of mixed-sequence RNA templates inside model protocells. J Am Chem Soc. 2018;140(15):5171–5178. doi: 10.1021/jacs.8b00639.
- O’Flaherty DK, Zhou L, Szostak JW. Nonenzymatic template-directed synthesis of mixed-sequence 3’-NP-DNA up to 25 nucleotides long inside model protocells. J Am Chem Soc. 2019;141(26):10481–10488.
- Zhang J, Fei Y, Sun L, et al. Advances and opportunities in RNA structure experimental determination and computational modeling. Nat Methods. 2022;19(10):1193–1207. doi: 10.1038/s41592-022-01623-y.
- Townshend RJL, Eismann S, Watkins AM, et al. Geometric deep learning of RNA structure. Science. 2021;373(6558):1047–1051. doi: 10.1126/science.abe5650.
- Ganser LR, Kelly ML, Herschlag D, et al. The roles of structural dynamics in the cellular functions of RNAs. Nat Rev Mol Cell Biol. 2019;20(8):474–489. doi: 10.1038/s41580-019-0136-0.
