ABSTRACT
Circular RNA (circRNA) forms closed loops via back-splicing in precursor mRNA, resisting exonuclease degradation. In higher eukaryotes, protein-coding genes create circRNAs through exon back-splicing. Unlike mRNAs, circRNAs possess unique production and structural traits, bestowing distinct cellular functions and biomedical potential. In this review, we explore the pivotal roles of viral circRNAs and associated RNA in various biological processes. Analysing the interactions between viral circRNA and host cellular machinery yields fresh insights into antiviral immunity, catalysing the development of potential therapeutics. Furthermore, circRNAs serve as enduring biomarkers in viral diseases due to their stable translation within specific tissues. Additionally, a deeper understanding of translational circRNA could expedite the establishment of circRNA-based expression platforms, meeting the rising demand for broad-spectrum viral vaccines. We also highlight the applications of circular RNA in biomarker studies as well as circRNA-based therapeutics. Prospectively, we expect a technological revolution in combating viral infections using circRNA.
1 Introduction to circRNAs: structure, biogenesis, known functions, and translation
Merely 5% of the human transcriptome undergoes translation [Citation1], while most cellular RNA transcripts, such as circular RNAs (circRNAs), microRNAs, small nuclear RNA (snRNA), piwi-interacting RNA (piRNA), small nucleolar RNA (snoRNA) and long-noncoding RNAs (lncRNAs), are designated as noncoding RNAs (ncRNAs) and are believed to be hardly translated [Citation2]. CircRNA is a unique subtype of ncRNA that contains covalently closed loops, but without 3’ and 5’ terminals [Citation3,Citation4], generated through back-splicing or head-to-tail splicing [Citation5–7]. Thus, the ‘back splicing junctions’ (BSJ) of circular RNAs can exclusively distinguish it from linear RNAs [Citation8]. Their unconventional structure makes them resistant to most ribonucleases, except for RNase A, RNase T1, and RNase T2 [Citation9,Citation10]. CircRNAs are found in cells from a broad range of species, including humans, with a more enriched content in specific cell or tissue types due to their longer biological half-life [Citation11]. CircRNAs not only function as post-transcriptional regulators but also act as miRNA sponges, repress viral gene expression, or function as protein decoys [Citation12].
Since their initial identification in plant viral RNA pathogens, circRNAs have been repeatedly observed in life-cycles of many viruses, including both RNA and DNA viruses, regardless they are single-stranded or double-stranded [Citation13]. This discovery has drawn elevated attention in various epidemics and pandemics. Some virus genomes even encode entire circRNA sequences, allowing for the production of large amounts of circRNAs. However, unlike the linear viral RNAs, circRNAs lose their replicative and infectious abilities as a result of the reverse splicing process that leads to the circularization of viral genes [Citation14,Citation15]. Most viral circRNAs have been found in tumour-associated viruses, such as Epstein – Barr virus (EBV), Kaposi’s sarcoma herpesvirus (KSHV), and papillomaviruses (HPVs), indicating their potential roles in tumorigenesis [Citation16–18]. Additionally, the research on viral circRNA has also explored biological functions and significances [Citation19–21]. Currently, there are two hotspots in viral circRNA research that are being closely watched, including 1) the identification of disclosing latent circRNAs as potential therapeutic targets [Citation22–24] 2) the use of tumour viruses-associated circular RNAs as long-lived tumour biomarkers.
Typical circRNAs in cells lack open reading frames, rendering them untranslated. Moreover, circRNAs are typically not translated due to the absence of the 5’ cap and 3’ poly(A) tail. In eukaryotes, the 5’ cap of m7GpppN (m7G) and 3’ poly(A) tail of mRNA are considered essential for protein synthesis. In most instances, transcripts that correspond to products with less than 100 amino acids are also neglected [Citation25]. However, some mature exonic circRNAs in the cytoplasm are still capable of recruiting the translational machinery to produce unique peptides [Citation11,Citation26]. The identification of translatable circRNAs is based on high-throughput technology, RNA sequencing (RNA-seq), polysome profiling, and a panel of circRNA-specific bioinformatics algorithms [Citation27–30]. As biological circRNAs lack a functional m7G cap, the translation initiation of circular RNA relies on the presence of other elements such as the internal ribosome entry site (IRES) [Citation27,Citation31–33] and m6A-IRES (m6A-Internal Ribosome Entry Site, MIRES) [Citation34]. Otherwise, circRNAs can also be translated via the rolling circle amplification (RCA) mechanism [Citation30]. A better understanding of noncanonical circRNA-derived proteins could facilitate the construction of effective and reliable expression platforms for the development of circRNA-based viral vaccines, providing alternative therapeutic tools in viral epidemics.
In this review, we provide an overview, as well as the recent progresses, of circRNAs in the viral life cycle, with a specific focus on virus-associated circRNAs. We highlight the latest findings on the regulatory functions of circRNA in viral infections, including their potentials as long-lived biomarkers for viral diseases. Additionally, we discuss the translation regulatory mechanism of the noncanonical circRNA-derived proteins and their implications in viral replication and host–virus interactions. Finally, we explore the potential applications of circRNA as therapeutic tools for the treatment of viral infections.
2 CircRNAs in viral infections: biogenesis, immunogenicity and functions
2.1 The biogenesis of circRNA in vivo
CircRNAs are widespread in living organisms, spanning prokaryotes, eukaryotes, and viruses, and are believed to contribute to natural evolution. Unlike linear RNAs, which are hindered from circularization due to 5’ end caps and/or 3’ end poly(A) tails, circRNAs are generally considered to be more structurally stable. Given their increased stability, circRNAs are suspected to function as the genetic information storage molecule in viroids and virus-like particles [Citation35].
The mechanisms behind the biogenesis of circular RNAs remain unclear, although two models have been proposed and widely accepted to explain their formation of circRNA and had been reviewed otherwhere [Citation36,Citation37] (). These circRNA biogenesis models include the direct back-splicing model and the lariat intermediate model [Citation38,Citation39]. Back-splicing is a unique form of splicing that differs from canonical splicing. Canonical splicing involves joining exons at an upstream 5’ splice site with a downstream 3’ splice site, releasing the lariat intron. Whereas back-splicing joins a downstream 5’ splice site with an upstream 3’ splice site in reverse order across exon(s) to generate circular RNAs (). In the lariat intermediate model, canonical splicing produces a lariat intermediate containing skipped exons, which is then back-spliced to generate a circRNA consisting of the exons that were originally skipped (). A circRNA can also be generated from an exon-skipped lariat or from an intron lariat that escapes debranching () [Citation40].
Figure 1. In vivo and in vitro generation of circRnas.
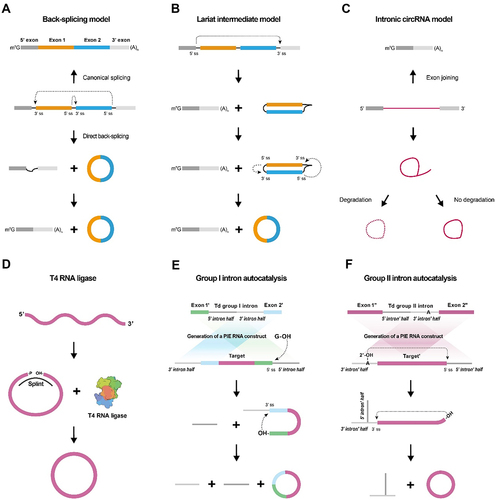
Many ‘tans’ and ‘cis’ factors regulate circRNA biogenesis. Complementary Alu repeats adjacent to circularized exons facilitate circRNA formation [Citation41]. DExH-Box Helicase 9 (DHX9), a reverse Alu helicase, has been shown to guide the circRNA formation by binding to reverse Alu elements (IRAlu) [Citation42]. In addition to analysing and forecasting circRNA formation mechanisms, IRAlu has become a practical tool for identifying and characterizing circRNAs in biological systems [Citation38]. Circular RNA biogenesis is influenced by various proteins, including nuclear factor 90 and 110 (NF90/NF110) [Citation43], heterogeneous nuclear ribonucleoprotein L (HNRNPL) [Citation44], splicing factor muscleblind (MBL) [Citation41], RNA-binding protein quaking I (QKI) [Citation45], and others [Citation39,Citation46–49].
2.2 RNA circularization strategies in vitro
Increasing interest in the growing number of identified circular RNA species in vivo has led to researchers attempting to establish in vitro circularization modes of RNA molecules [Citation50]. Generally, two approaches are now commonly used for in vitro synthesis of circRNA, T4 enzymatic ligation and modified group I intron self-splicing. As a preparation step, the primary linear RNA needs to be synthesized before circularization using in vitro transcription (IVT). The T4 enzymatic ligation approach uses bacteriophage T4 RNA ligase 1 (T4 Rnl 1) or RNA ligase 2 (T4 Rnl 2) to catalyse ligation reactions [Citation35]. To enable the enzymatic reactions, a linear RNA precursor with a 3’-OH and a 5’ monophosphate present at the acceptor site and donor site, respectively, is required [Citation51] along with a short DNA or RNA oligonucleotide (complementary to the acceptor and donor site) to splint the RNA. The activated 5’-terminus of the RNA precursor is then susceptible to T4 RNA ligase mediated nucleophilic attack, resulting in a covalent 5’,3’-phosphodiester bond that circularizes the RNA (). The group I intron self-splicing approach, also known as permuted introns and exons (PIE) [Citation52–54], utilizes self-splicing of group I introns to ligate RNA by two transesterifications at defined splice sites with GTP and Mg2+ as cofactors. Circular permutation of the group I intron precursor RNA disrupts the half-intron sequences with coupled exons [Citation53,Citation55], yielding a circular RNA exon in vitro. In the first transesterification, the free 3’-OH in GTP attacks the acceptor splice site in the 5’-half intron, releasing the 3’-terminal sequence. In the second transesterification, the newly generated 3’-splice site in exon 1 with a free 3’-OH attacks the acceptor splice site in the 3’-half intron and finally cyclizes exons 1 and 2. In practice, to study the function of a gene of interest, it can be inserted between exons 1 and 2 (). Group II introns constitute a nucleic acid catalytic self-splicing system similar to Group I introns. The process begins with the attack of the 2’-OH from the 5’ splice site, followed by the free 3’-OH attacking the acceptor splice site in the 3’−half intron. The excised Group II intron is connected through 2’−5’phosphodiester bonds, and the circularized exon is connected through 3’-5’ phosphodiester bonds, ultimately forming a complete circRNA molecule (). CircRNAs produced by Group II introns enable the full replacement of the original exonic sequence, distinguishing them from Group I introns ().
The synthesis of circular RNA is achieved both in cells and in tubes using the PIE method [Citation56], which offers broader application compared to enzymatic ligation. Unlike enzymatic ligation, the PIE method is capable of cyclizing larger linear RNA precursors, and its reaction conditions and purification methods are superior. Therefore, the PIE method is currently the most widely used approach for RNA circulation [Citation53,Citation57,Citation58].
2.3 The production and function of viral-derived circRNAs
CircRNAs are found in diverse viral families such as herpesviruses, coronaviruses and orthohepadnavirus. Among all circRNAs encoded viruses, circRNAs from Epstein–Barr Virus (EBV) are most intensively investigated (). Recently, high-throughput sequencing of RNA libraries (ribosomal RNA- and linear RNA-depleted) have revealed a repertoire of circRNAs in EBV and Kaposi’s sarcoma herpesvirus (KSHV)-infected cells [Citation22,Citation68]. Besides EBV and KSHV [Citation21,Citation68,Citation69], other DNA viruses, such as HPV16 [Citation70], hepatitis B virus (HBV) [Citation71] and Marek’s Disease Virus (MDV) [Citation72] also encode circular RNA. Recently, an RNA-sequencing study of viral infection revealed thousands of circRNAs in the life-threatening coronaviruses MERS-CoV, SARS-CoV-1 and SARS-CoV-2 [Citation73]. In Grass carp reovirus (GCRV) infected kidney cell, more than 30 specific circRNAs were identified and confirmed to engage in the viral proliferation [Citation74]. Two RNA virus-derived circRNA, circALTO1 and circALTO2, were detected in the Merkel Cell Polyomavirus (MCPyV) infected cells and patients’ tissues [Citation75] ().
Table 1. The production and function of viral-derived circRNAs.
Functionally, viral-derived circRNAs regulate viral gene expression, modulate host immune responses, and contribute to viral pathogenesis [Citation76]. One comprehensively analysed example of viral-derived circRNAs is the spectrum of circular RNAs encoded by the latency and lysis cycles of EBV as well as spanned cell lines in different latency states [Citation22]. EBV encodes more than 30 unique viral circRNAs [Citation22,Citation68]. EBV-circrpms1, circlmp2, and EBV-circbhlf1 have been associated with EBV-associated cancer [Citation23,Citation77]. Two EBV-encoded circRNAs, circRPMS1 and ebv-circLMP2A, are often overexpressed in cells and tissues infected with EBV, and both of them further play a pivotal role in the development and maintenance of cancer stem cell properties [Citation77]. Understanding the biogenesis and functions of circRNAs in viral infections provides valuable insights into the intricate interplay between viruses and hosts.
2.4 The immunogenicity of viral-derived circRNAs
Given the prevalence of circRNAs in viral genomes, it is not surprising that mammalians possess innate immunity to distinguish viral circRNAs from self-produced ones. For example, circRNAs from KSHV and hepatitis delta virus (HDV) were targeted by the host immune system [Citation69]. The ability of the host to identify and react to viral circRNAs could contribute to the host’s immune response to combat viral infections. In mammalian cells, the retinoic acid-inducible gene-I (RIG-I) mediates the recognition of exogenous circRNAs, while endogenous circRNAs modified with m6A to recognize non-self circRNAs. These specific molecular characteristics or modifications play a crucial role in identifying circRNAs that are not derived from the host organism [Citation78]. RIG-I, belonging to the pattern recognition receptors (PRRs) of the host, is essential in innate immunity. It recognizes signals from pathogen-associated molecular patterns (PAMPs) of viruses and induces non-specific reactions against viral infection [Citation59]. RIG-I distinguishes endogenous from exogenous circRNAs by discriminating the self-intronic regions from the non-self regions. This distinction is facilitated by the association of endogenous spliceosomes with exogenous RNAs, which is enabled by RNA binding proteins (RBPs). The inability of exogenous circRNAs to bind to host RBPs is speculated to trigger RIG-I-mediated innate immune responses [Citation60]. The m6A RNA modification on circRNA is also implicated in differentiating host and exogenous circRNAs [Citation55,Citation78]. The m6A reader protein binds to the m6A modified sites on the host circRNA, preventing the activation of innate immunity [Citation61]. However, the transcribed viral circRNAs within infected cells exploit the host cell machinery, evading detection as foreign circRNAs due to the presence of the same nucleotide modification patterns (e.g. m6A modification) [Citation62,Citation63].
2.5 Roles of host derived circRNAs in viral infections
Analysis of RNA-seq data obtained from cells infected with diverse viruses revealed changes in host circRNAs in response to viral infection [Citation64]. Several studies have demonstrated possible functions played by host circRNAs to counteract viral infections [Citation65–67]. Upon de novo KSHV infection, has_circ_0001400 expression was found to result in the reduction of KSHV LANA-1 and RTA transcripts significantly, without blocking viral entry into cells. These findings suggest hsa_circ_0001400 may act as an antiviral factor [Citation79].
CircRNAs that are normally found in host cells can be co-opted by viruses to enhance viral replication and pathogenesis. For instance, CircGATAD2A was found to play a role in promoting H1N1 influenza virus (IAV) virus replication, as evidenced by increased viral titre and nucleocapsid protein (NP) levels in overexpressing cells [Citation80]. Additionally, the cellular content of circGATAD2A was upregulated upon H1N1 IAV infection. Furthermore, a negative regulation of vacuolar sorting protein 34 (VPS34)-dependent autophagy by circGATAD2A was exhibited in this work, which was accomplished by inhibiting autophagy-related protein light-chain 3-II (LC3-II) formation and increasing the level of p62. VPS34-dependent autophagy is known to be essential for the removal of intracellular viruses. In hepatitis C virus (HCV), Chen et al. (2020) observed proviral activities of host circRNAs in HCV-infected cells [Citation81]. The expression of circular RNA circPSD3, which is encoded by the Pleckstrin and Sec7 gene, was upregulated during HCV infection. This resulted in the inhibition of the cellular nonsense-mediated decay (NMD) pathway, which serves as a broad-spectrum antiviral defence mechanism. Similarly, infection of lung cells with Middle East respiratory syndrome coronavirus (MERS-CoV) led to the increased expression of host circRNAs (e.g. circFNDC3B and circCNOT1) with proviral function [Citation82]. MERS-CoV viral load was significantly reduced when knocked down the host-derived circFNDC3B and circCNOT1 [Citation82].
3 Therapeutic applications of circRNAs: as biomarkers, RNA-based therapies, or expression platform
3.1 The circRNAs as diagnostic markers for viral diseases
Numerous studies investigating on implications of viral-encoded circRNAs or viral infection-induced host circRNAs have suggested possibilities for developing new diagnostic and prognostic tools, as well as new treatments, for virus-associated diseases (). For example, circRNAs have been reported to be stably detected in human peripheral whole blood and plasma, indicating their potency in detecting a variety of diseases [Citation83,Citation84]. The exceptional stability and specificity of both host and viral circRNAs according to cell type and developmental stage make them highly promising as effective diagnostic and prognostic biomarkers for diseases associated with viruses. Furthermore, the presence of viral circRNAs in transplanted organs or tissues can be used to monitor transplant recipients for the development of transplant-related complications [Citation85]. The presence of viral circRNAs indicates a greater likelihood of virus-related illnesses.
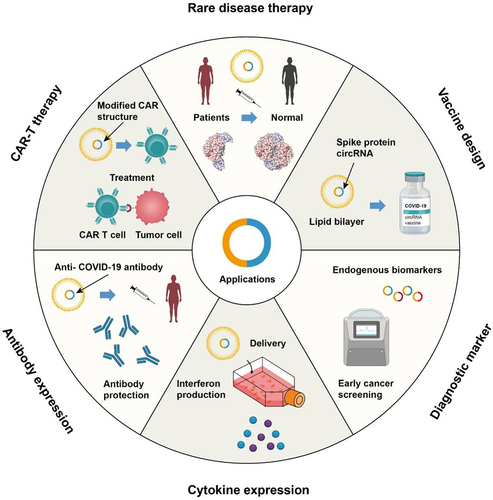
Figure 2. Perspectives of circRNA nanomedicine.
Viral circRNAs, including those derived from EBV, KSHV, and HPV, were shown to be detectable in patient samples [Citation22,Citation23,Citation68]. However, EBV circRNAs have not been compared to the clinical diagnostic marker EBV-encoded RNA transcripts (EBER) in detecting EBV in tumours with respect to specificity and sensitivity for now. Constructive information will be obtained by performing such tests. Similarly, HPV circE7 can also be compared to the operational HPV DNA testing for its potency as a diagnostic marker for cervical cancer screening [Citation86]. Additionally, the concentration of immune-related circRNAs is a potential marker for antiviral immunity [Citation87–89]. Albeit the promising clinical perspectives of circRNAs, the current understanding of them is still rough. Recent advances in high-throughput techniques have facilitated the probing of promising circRNA biomarkers that can potentially be used to validate respiratory virus diseases [Citation90]. However, compared to established clinical diagnostic methods for viral diseases, circRNA as a diagnostic marker still has a long way to go.
3.2 CircRNA-based RNA therapies
The stability of RNA molecules can significantly extend their potentials in therapeutic applications [Citation57,Citation91–93]. Antisense circRNAs and oligos targeting the 5’-untranslated region of SARS-CoV-2 viral mRNAs exhibited greater inhibition efficiency in cell-based virus proliferation compared to linear modified antisense oligos, attributed to the circRNAs’ enhanced stability [Citation94]. Some circRNAs were shown to target disease-linked miRNA and were proposed as potential cures. One example is from HCV, microRNA-122 is essential for efficient HCV replication [Citation95]. Jost et al. (2018) found that miR sponge, an artificial circRNA can disrupt the life cycle of HCV by targeting microRNA-122 [Citation96]. Similar to Miravirsen, the first anti-microRNA drug that was manifested to decrease HCV titre in patients, miR sponge exhibits the ability to capture and eliminate microRNA-122 from HCV cells. Through the utilization of advanced high-throughput RNA sequencing and cutting-edge bioinformatics tools, a myriad of circRNAs have been identified across various organisms. Notably, circMbl and circANRIL act as protein sponges [Citation12,Citation97], regulating MBL and PES1, respectively. Elevated MBL levels enhance circMbl biosynthesis, impacting mRNA splicing. Conversely, circMbl overexpression sequesters MBL impairing its functionality. CircANRIL disrupts PES1 function, affecting rRNA processing, linked to atherosclerosis. circRNAs intricately interact with proteins, uncovering a complex regulatory network with broad gene expression implications [Citation12,Citation97]. To develop safe and effective circRNA-based therapies for virus-related diseases, it is important for future research to concentrate on identifying circRNA targets and enhancing patient delivery methods.
3.3 The discovery of translated circRNA
Natural circRNAs were previously thought to be untranslated for lacking the m7G cap and poly(A) tail until the introduction of a synthetic circRNA carrying an ORF and IRES of encephalomyocarditis virus. An approximate 23 kDa protein was produced from this artificial circRNA, manifesting the translation potency of IRES-element equipped circRNAs () [Citation98]. The single-stranded circular RNA originated from the genome of the hepatitis delta virus was found to contain an open reading framework (ORF), a start codon AUG, and a stop codon, suggesting a chance to be translated [Citation99]. Plant viroids and virusoids were also revealed to comprise similar translated circular RNAs [Citation100]. Additionally, covalently closed RNAs from virusoids were also shown to produce 16 kDa proteins using overlapping initiation-termination codons (UGAUGA) and in IRES-dependent manner [Citation101]. The translation potency of circRNAs in human and Drosophila cells was also proven [Citation102]. Natural exonic circRNAs containing GFP fragments and IRES-elements were translated into functional proteins [Citation56]. Apart from the IRES-dependent translation, an alternative protein synthesis mechanism termed ‘rolling circle amplification (RCA)’ was found to be utilized by an exonic circRNA that contains an infinite ORF (iORF) and a start codon AUG [Citation103,Citation104]. Even without IRES element, 5’m7G cap, and 3’ poly(A) tail, this construct still produced a set of unexpected protein bands in rabbit reticulocyte lysate and human cells. Altogether, the feasibility of circRNAs to undergo the cap-independent translation has been demonstrated. Correspondingly, one recent study reported the detection of a large number of circRNA-coded peptides using mass spectrometry [Citation105]. Most of the translated peptides were rapidly degraded after the translation and were present in low abundance.
Figure 3. circRNA translating processes.
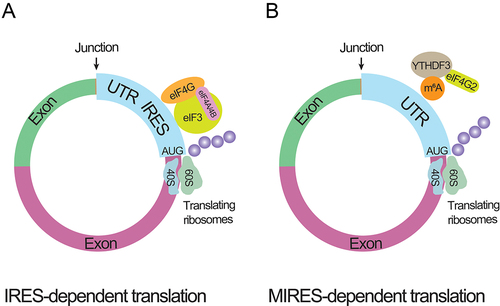
Additionally, solid evidence has already been found to confirm the polysomes involved natural circRNAs translation process [Citation27,Citation28,Citation34,Citation106]. Using ribosome footprinting (RFP) datasets, 192 circRNAs were found to be polysome-bound in rat/mouse tissues and 151 circRNAs were in Drosophila. Pamudurti et al. (2017) detected a 37.03 kDa protein with Western blotting and MS analysis at the overexpression of circMbl3, a circRNA derived from the mbl transcript [Citation28]. Similarly, using a polysome sucrose gradient fractionation assay, a circ-ZNF609-heavy polysome complex has been examined in human and mouse tissues. A UTR-directed mechanism generated an approximately 30 kDa protein from this ribo-circRNA [Citation27].
MIRES-dependent circRNA translation was also discovered. According to the m6A-RIP assay, numerous circRNAs possess m6A site density near the start codon and possess an RRACH (R: G or A; H: A, C or U) consensus motif of m6A modification. Circular RNA could be translated at one or two m6A sites, mainly through the recruitment of m6A binding proteins, like YTH domain family (YTHDF) proteins [Citation34]. Human embryonic stem cells were mapped across their genome for m6A-methylated circular RNAs, which implies that circRNA translation is dependent on MIRES [Citation106] ().
3.4 The application of translated circRNA as an expression platform
As a proof of concept, some circRNAs have demonstrated the potential for protein translation applications [Citation27,Citation28,Citation107]. In an innovative study, artificially engineered circRNAs with IRES were shown to robustly and stably express proteins in eukaryotic cells [Citation53]. To enhance the splicing, their circRNA backbone was equipped with a self-splicing intron with well-designed accessory sequences. A sequence of exogenous RNAs up to 5 kb in length was circularized into the engineered backbone in vitro. In preclinical studies, Wei’s group used circular RNA as a vaccine candidate, expressing receptor-binding domain (RBD) of SARS-CoV-2, and had demonstrated greater efficacy against SARS-CoV-2 and its emerging variants in mice and rhesus macaques [Citation108] (). These results illustrate that circRNAs hold great promise as a safe and effective platform for developing virus-targeted vaccines.
Table 2. The translational usage of circRNA.
Both mRNA and circRNA vaccines have shown great potential in expressing neutralizing antibodies, effectively inhibiting viral replication and providing robust protection [Citation108]. For instance, the administration of LNP-encapsulated SARS-CoV-2 neutralizing antibody HB27 resulted in long-term protection in mice and hamsters [Citation109], and the mRNA-based therapeutic antibody VRC01 demonstrates potential for passive immunotherapy against HIV-1 and other diseases [Citation110]. Furthermore, the development of mRNA-LNP-mediated CAR-T therapy has been established [Citation111]. CircRNAs have demonstrated impressive protein expression capabilities, making them promising candidates for various trials using antibody and cytokines, as well as CAR-T therapy. They have been proposed as potential treatments for diseases such as cancers, genetic diseases, and rare diseases [Citation112,Citation113]. Considering the superior expression capability of circRNAs, their immense potential in the field of medical applications can be predicted ().
3.5 Challenges for translational circRNA platform
Despite the success of SARS-CoV-2 vaccines, circRNA-associated nanomedicine is facing a primary challenge in sequence design to improve the protein expression efficiency. Directions to improve the expression modules include RNA modifications [Citation57,Citation78], optimization of homologous arms [Citation53,Citation114], incorporation of IRES [Citation57], utilization of aptamers [Citation115] (). Recently, a novel circRNA platform was developed with optimized vector topology, 5’ and 3’ UTRs, IRESs, and synthetic aptamers, enabling potent and durable protein production process [Citation57]. The optimized circRNA backbone from this platform increases the yield of circRNA proteins by several hundred-fold in cells ().
Figure 4. Challenges of circRNA nanomedicine.
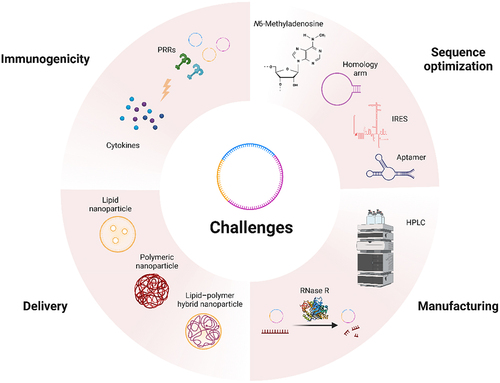
Another approach is to improve manufacturing for better expression. Purification techniques like HPLC and the use of RNase R are employed to achieve high purity and enrichment of circular RNA for downstream applications (). Anderson’s data supported this improvement direction [Citation114]. Safety is another significant concern when it comes to RNA therapy aimed at achieving protein expression, as immune responses can lead to potential toxicity. Therefore, enhancing the safety of circRNA-associated nanomedicine is crucial. Encouragingly, research from ORNA has demonstrated that circRNAs induce fewer undesirable immune responses than linear mRNAs [Citation58]. As we summarize in part 2.4, non-self circRNA is recognized by RIG-I [Citation60] and m6A modification on circRNA can evade the activation of innate immunity [Citation78]. The immune recognition of circRNA is influenced by factors like sequence, m6A modifications [Citation57,Citation78], structure, and cellular environment. The in vitro synthesized circular RNA should be more carefully checked for its immunogenicity as the impurity intermediate RNA generated during the synthetic process [Citation114].
CircRNA can be delivered using various methods, including lipid nanoparticles (LNPs), for efficient and targeted delivery. Yet, delivery efficiency and target delivery are other challenges facing circRNA (). Novel RNA delivery systems are under development, which is better reviewed elsewhere [Citation58]. Increasing evidence had suggested that several biological pathways may interfere with RNA delivery [Citation116,Citation117], yet the molecular mechanisms are unclear. Better understanding can further improve the efficacy of RNA drugs. Overall, to expedite the clinical translation of circRNA nanomedicines, additional breakthroughs and advancements are necessary to overcome the existing obstacles.
3.6 Future perspectives
CircRNA presents several advantages and promising prospects for the future. Its natural accumulation in non-dividing cells indicates enhanced stability and safety, enabling sustained expression due to its slower degradation. However, designing the sequence of circRNA poses significant challenges compared to traditional linear mRNA. The circular nature of circRNA, coupled with the lack of well-established methods for secondary structure prediction, complicates the design process. Factors such as codon usage, enzyme-cleavage motifs, translation initiation of IRES, and auxiliary sequences must be carefully considered. This demands a more sophisticated and nuanced approach.
In the production of circRNA, three major challenges need to be addressed. 1), large-scale production of circRNA remains underdeveloped within the mRNA production landscape, hindering its practical application. Robust large-scale production is necessary to achieve cost control and to facilitate widespread use. 2), the circular structure of circRNA lacks a Poly-A tail at certain design points, making conventional large-scale affinity chromatography purification methods inadequate. Thus, developing effective purification strategies for large-scale production of high-quality and pure circRNA is crucial. 3), formulating circRNA as a novel product capable of long-term and targeted expression in vivo requires further investigation to ensure its efficacy, safety, and stability within the body.
To unlock the full potential of circRNA as a revolutionary mRNA platform, research and development efforts must focus on overcoming these challenges. By addressing the issues surrounding large-scale production, purification, and in vivo expression, circRNA can fulfil its promise as a versatile and effective therapeutic platform.
4 Conclusions
CircRNAs are featured with high tissue-specificity and remarkable stability, making them promising disease markers and therapeutic targets [Citation118]. They can function as miRNA sponges [Citation119], playing crucial roles in regulating various viruses-associated cancers [Citation120] and viral infection diseases [Citation121]. Furthermore, the study of viruses-associated circRNAs has extended our understanding of viral life cycle and how viruses interact with their host [Citation64], which may expedite the development of antiviral therapy in the near future.
Compared to well-established mRNA nanomedicine, circRNA holds great promise for application in various drug fields, including cell and gene therapy, antibody drugs, and protein replacement therapy. Its potential has sparked interest in circRNA nanomedicine across a wide range of fields, driving innovation and transformative changes.
Author contributions
WW, LS, MH, TJ, and YQ drafted the manuscript. ZM and QZ finalized the paper.
Acknowledgments
This work was supported by grants from R&D Program of Guangzhou Laboratory (Grant No. YW-JCYJ0612, SRPG22-002, SRPG22-003, SRPG22-006, SRPG22-007), State Key Laboratory of Pathogen and Biosecurity (Academy of Military Medical Science, Grant No. SKLPBS2238), Natural Science Foundation of China (Grant No. 32270707), and the National Key R&D Program of China (Grant No. 2021YFF1200900, 2021YFF1200903).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study.
Additional information
Funding
References
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian Genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014
- Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24(7):344–352. doi: 10.1016/j.tig.2008.04.005
- Adelman K, Egan E. More uses for genomic junk. Nature. 2017;543(7644):183–185. doi: 10.1038/543183a
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928
- Liu X, Hu Z, Zhou J, et al. Interior circular RNA. RNA Biol. 2020;17(1):87–97. doi: 10.1080/15476286.2019.1669391
- Zhao Q, Liu J, Deng H, et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS Output. Cell. 2020;183(1):76–93.e22. doi: 10.1016/j.cell.2020.08.009
- Jiao J, Li C, Ning L, et al. Electrochemical detection of circRnas based on the combination of back-splice junction and duplex-specific nuclease. Sensors And Actuat B Chem. 2020;302:127166. doi: 10.1016/j.snb.2019.127166
- Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281(40):29769–29775. doi: 10.1074/jbc.M606744200
- Suzuki H. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34(8):e63–e63. doi: 10.1093/nar/gkl151
- Wang Y, Wu C, Du Y, et al. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol Cancer. 2022;21(1):13. doi: 10.1186/s12943-021-01484-7
- Huang A, Zheng H, Wu Z, et al. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517. doi: 10.7150/thno.42174
- Niu M, Ju Y, Lin C, et al. Characterizing viral circRnas and their application in identifying circRnas in viruses. Brief Bioinform. 2022;23(1):bbab404. doi: 10.1093/bib/bbab404
- Yan L, Chen YG. Circular RNAs in immune response and viral infection. Trends Biochem Sci. 2020;45(12):1022–1034. doi: 10.1016/j.tibs.2020.08.006
- Tagawa T, Oh D, Dremel S, et al. A virus-induced circular RNA maintains latent infection of Kaposi’s sarcoma herpesvirus. Proc Natl Acad Sci, USA. 2023;120(6):e2212864120. doi: 10.1073/pnas.2212864120
- Liu C-X, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865–880.e21. doi: 10.1016/j.cell.2019.03.046
- Gong L, Chen J, Dong M, et al. Epstein–Barr virus‐derived circular RNA lMP2A induces stemness in EBV‐associated gastric cancer. EMBO Rep. 2020;21(10):e49689. doi: 10.15252/embr.201949689
- Niu M, Zhang J, Li Y, et al. CirRNAPL: a web server for the identification of circRNA based on extreme learning machine. Computat Struct Biotechnol J. 2020;18:834–842. doi: 10.1016/j.csbj.2020.03.028
- Berman TA, Schiller JT. Human papillomavirus in cervical cancer and oropharyngeal cancer: one cause, two diseases. Cancer. 2017;123(12):2219–2229. doi: 10.1002/cncr.30588
- Torresi J, Tran BM, Christiansen D, et al. HBV-related hepatocarcinogenesis: the role of signalling pathways and innovative ex vivo research models. BMC Cancer. 2019;19(1):707. doi: 10.1186/s12885-019-5916-6
- Abere B, Li J, Zhou H, et al. Kaposi’s sarcoma-associated herpesvirus-encoded circRnas are expressed in infected tumor tissues and are incorporated into Virions. MBio. 2020;11(1):e03027–19. doi: 10.1128/mBio.03027-19
- Ungerleider N, Concha M, Lin Z, et al. The Epstein Barr virus circRnaome. PLOS Pathog. 2018;14(8):e1007206. doi: 10.1371/journal.ppat.1007206
- Huang J, Chen J, Gong L, et al. Identification of virus-encoded circular RNA. Virology. 2019;529:144–151. doi: 10.1016/j.virol.2019.01.014
- Wu F, Cheng W, Zhao F, et al. Association of N6-methyladenosine with viruses and related diseases. Virol J. 2019;16(1):133. doi: 10.1186/s12985-019-1236-3
- Tonkin J, Rosenthal N. One small step for muscle: a New micropeptide regulates performance. Cell Metab. 2015;21(4):515–516. doi: 10.1016/j.cmet.2015.03.013
- Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRnas. Biochim Biophys Acta Gene Regul Mech. 2016;1859(10):1245–1251. doi: 10.1016/j.bbagrm.2016.07.009
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Molecular Cell. 2017;66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021
- van Heesch S, Witte F, Schneider-Lunitz V, et al. The translational landscape of the human heart. Cell. 2019;178(1):242–260.e29. doi: 10.1016/j.cell.2019.05.010
- Gao X, Xia X, Li F, et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR–STAT3 signalling. Nat Cell Biol. 2021;23(3):278–291. doi: 10.1038/s41556-021-00639-4
- Liang W-C, Wong C-W, Liang P-P, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the wnt pathway. Genome Biol. 2019;20(1):84. doi: 10.1186/s13059-019-1685-4
- Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via hippo-YAP signaling. Mol Cancer. 2019;18(1):47. doi: 10.1186/s12943-019-1010-6
- Wu X, Xiao S, Zhang M, et al. A novel protein encoded by circular SMO RNA is essential for hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021;22(1):33. doi: 10.1186/s13059-020-02250-6
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31
- Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–2465. doi: 10.1093/nar/gkv045
- Chen L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. doi: 10.1038/s41580-020-0243-y
- Yang L, Wilusz JE, Chen L-L. Biogenesis and regulatory roles of circular RNAs. Annu Rev Cell Dev Biol. 2022;38(1):263–289. doi: 10.1146/annurev-cellbio-120420-125117
- Zhang X-O, Wang H-B, Zhang Y, et al. Complementary sequence-mediated Exon Circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019
- Wilusz JE. Circular RNAs: unexpected outputs of many protein-coding genes. RNA Biol. 2017;14(8):1007–1017. doi: 10.1080/15476286.2016.1227905
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
- Aktaş T, Avşar Ilık İ, Maticzka D, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544(7648):115–119. doi: 10.1038/nature21715
- Li X, Liu C-X, Xue W, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Molecular Cell. 2017;67(2):214–227.e7. doi: 10.1016/j.molcel.2017.05.023
- Fei T, Chen Y, Xiao T, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci, USA Internet. 2017 [[cited 2023 Mar 9]];114(26): doi: 10.1073/pnas.1617467114
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRnas. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014
- Stagsted LVW, O’Leary ET, Ebbesen KK, et al. The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. Elife. 2021;10:e63088. doi: 10.7554/eLife.63088
- Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8(1):14741. doi: 10.1038/ncomms14741
- Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRnps, and SR proteins. Genes Dev. 2015;29(20):2168–2182. doi: 10.1101/gad.270421.115
- Liang D, Tatomer DC, Luo Z, et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Molecular Cell. 2017;68(5):940–954.e3. doi: 10.1016/j.molcel.2017.10.034
- Chen X, Lu Y. Circular RNA: biosynthesis in vitro. Front Bioeng Biotechnol. 2021;9:787881. doi: 10.3389/fbioe.2021.787881
- Moore MJ. Joining RNA molecules with T4 DNA ligase [Internet. In RNA-Protein interaction protocols. New Jersey: Humana Press; 1999. cited 2023 Mar 9 pp. 11–19. http://link.springer.com/10.1385/1-59259-676-2:11
- Umekage S, Kikuchi Y. In vitro and in vivo production and purification of circular RNA aptamer. J Biotechnol. 2009;139(4):265–272. doi: 10.1016/j.jbiotec.2008.12.012
- Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9(1):2629. doi: 10.1038/s41467-018-05096-6
- Rausch JW, Heinz WF, Payea MJ, et al. Characterizing and circumventing sequence restrictions for synthesis of circular RNA in vitro. Science. 2021;49(6):e35–e35. doi: 10.1093/nar/gkaa1256
- Chen YG, Kim MV, Chen X, et al. Sensing self and foreign circular RNAs by intron identity. Molecular Cell. 2017;67(2):228–238.e5. doi: 10.1016/j.molcel.2017.05.022
- Meganck RM, Liu J, Hale AE, et al. Engineering highly efficient backsplicing and translation of synthetic circRnas. Mol Ther Nucleic Acids. 2021;23:821–834. doi: 10.1016/j.omtn.2021.01.003
- Chen R, Wang SK, Belk JA, et al. Author correction: engineering circular RNA for enhanced protein production. Nature Biotechnol. 2022;1–11. doi: 10.1038/s41587-022-01472-2
- Huang X, Kong N, Zhang X, et al. The landscape of mRNA nanomedicine. Nat Med. 2022;28(11):2273–2287. doi: 10.1038/s41591-022-02061-1
- Paludan SR, Pradeu T, Masters SL, et al. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat Rev Immunol. 2021;21(3):137–150. doi: 10.1038/s41577-020-0391-5
- Chen X, Yang T, Wang W, et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9(2):588–607. doi: 10.7150/thno.29678
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730
- Lu M, Zhang Z, Xue M, et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol. 2020;5(4):584–598. doi: 10.1038/s41564-019-0653-9
- Baquero-Perez B, Geers D, Díez J. From a to m6A: the emerging viral epitranscriptome. Viruses. 2021;13(6):1049. doi: 10.3390/v13061049
- Tan K, Lim Y. Viruses join the circular RNA world. FEBS J. 2021;288(15):4488–4502. doi: 10.1111/febs.15639
- Du L, Wang X, Liu J, et al. A Previously undiscovered circular RNA, circTNFAIP3, and its role in coronavirus replication. MBio. 2021;12(6):e02984–21. doi: 10.1128/mBio.02984-21
- Zhang X, Liang Z, Wang C, et al. Viral circular RNAs and their possible roles in virus-host interaction. Front Immunol. 2022;13:939768. doi: 10.3389/fimmu.2022.939768
- Role of noncoding RNAs in dengue virushost interaction. Front Biosci (Schol Ed) 2021;13:44.
- Toptan T, Abere B, Nalesnik MA, et al. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci, USA Internet. 2018 [cited 2023 Mar 9];115(37). doi:10.1073/pnas.1811728115
- Tagawa T, Gao S, Koparde VN, et al. Discovery of Kaposi’s sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci, USA. 2018;115(50):12805–12810. doi: 10.1073/pnas.1816183115
- Zhao J, Lee EE, Kim J, et al. Translation and transforming activity of a circular RNA from human papillomavirus [Internet]. Mol Biol. 2019 [cited 2023 Mar 9]. Available from: doi: 10.1101/600056
- Zhu M, Liang Z, Pan J, et al. Hepatocellular carcinoma progression mediated by hepatitis B virus-encoded circRNA HBV_circ_1 through interaction with CDK1. Mol Ther Nucleic Acids. 2021;25:668–682. doi: 10.1016/j.omtn.2021.08.011
- Chasseur AS, Trozzi G, Istasse C, et al. Marek’s disease virus virulence genes encode circular RNAs. J Virol. 2022;96(9):e00321–22. doi: 10.1128/jvi.00321-22
- Cai Z, Lu C, He J, et al. Identification and characterization of circRnas encoded by MERS-CoV, SARS-CoV-1 and SARS-CoV-2. Brief Bioinform. 2021;22(2):1297–1308. doi: 10.1093/bib/bbaa334
- Pan J, Zhang X, Zhang Y, et al. Grass carp reovirus encoding circular RNAs with antiviral activity. Aquaculture. 2021;533:736135. doi: 10.1016/j.aquaculture.2020.736135
- Yang R, Lee EE, Kim J, et al. Characterization of ALTO-encoding circular RNAs expressed by Merkel cell p olyomavirus and trichodysplasia spinulosa polyomavirus. PLOS Pathog. 2021;17(5):e1009582. doi: 10.1371/journal.ppat.1009582
- Li X, Yang L, Chen L-L. The biogenesis, functions, and challenges of circular RNAs. Molecular Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034
- Zhang J, Du Y, Gong L, et al. Ebv-circRPMS1 promotes the progression of EBV-associated gastric carcinoma via Sam68-dependent activation of METTL3. Cancer Lett. 2022;535:215646. doi: 10.1016/j.canlet.2022.215646
- Chen YG, Chen R, Ahmad S, et al. N6-methyladenosine modification controls circular RNA immunity. Molecular Cell. 2019;76(1):96–109.e9. doi: 10.1016/j.molcel.2019.07.016
- Uppal T, Banerjee S, Sun Z, et al. KSHV LANA—the master regulator of KSHV latency. Viruses. 2014;6(12):4961–4998. doi: 10.3390/v6124961
- Yu T, Ding Y, Zhang Y, et al. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet Microbiol. 2019;231:238–245. doi: 10.1016/j.vetmic.2019.03.012
- Chen T-C, Tallo-Parra M, Cao QM, et al. Host-derived circular RNAs display proviral activities in hepatitis C virus-infected cells. PLOS Pathog. 2020;16(8):e1008346. doi: 10.1371/journal.ppat.1008346
- Zhang X, Chu H, Wen L, et al. Competing endogenous RNA network profiling reveals novel host dependency factors required for MERS-CoV propagation. Emerg Microbes Infect. 2020;9(1):733–746. doi: 10.1080/22221751.2020.1738277
- Koh W, Pan W, Gawad C, et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc Natl Acad Sci, USA. 2014;111(20):7361–7366. doi: 10.1073/pnas.1405528111
- Memczak S, Papavasileiou P, Peters O, et al. Identification and characterization of circular RNAs as a New class of putative biomarkers in human blood. PLoS One. 2015;10(10):e0141214. doi: 10.1371/journal.pone.0141214
- Zebardast A, Latifi T, Shirzad M, et al. Critical involvement of circular RNAs in virus-associated cancers. Genes Dis. 2022;10(6):S2296–2305. doi: 10.1016/j.gendis.2022.04.009
- Chamseddin BH, Lee EE, Kim J, et al. Assessment of circularized E7 RNA, GLUT1, and PD-L1 in anal squamous cell carcinoma. Oncotarget. 2019;10(57):5958–5969. doi: 10.18632/oncotarget.27234
- Su H, Chu Q, Zheng W, et al. Circular RNA circPikfyve acts as a sponge of miR-21-3p to enhance antiviral immunity through regulation of MAVS in teleost fish. J Virol. 2021;95(8):e02296–20. doi: 10.1128/JVI.02296-20
- Su H, Zheng W, Pan J, et al. Circular RNA circSamd4a regulates antiviral immunity in teleost fish by Upregulating STING through sponging miR-29a-3p. J Immunol. 2021;207(11):2770–2784. doi: 10.4049/jimmunol.2100469
- Chu Q, Zheng W, Su H, et al. A highly conserved circular RNA, circRasgef1b, enhances antiviral immunity by regulating the miR-21-3p/MITA pathway in lower vertebrates. J Virol. 2021;95(7):e02145–20. doi: 10.1128/JVI.02145-20
- Yao W, Pan J, Liu Z, et al. The cellular and viral circRnaome induced by Respiratory syncytial virus infection. MBio. 2021;12(6):e03075–21. doi: 10.1128/mBio.03075-21
- Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed. 2009;48(8):1378–1398. doi: 10.1002/anie.200802092
- Setten RL, Rossi JJ, Han S. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov. 2019;18(6):421–446. doi: 10.1038/s41573-019-0017-4
- Ni S, Zhuo Z, Pan Y, et al. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl Mater Interfaces. 2021;13(8):9500–9519. doi: 10.1021/acsami.0c05750
- Pfafenrot C, Schneider T, Müller C, et al. Inhibition of SARS-CoV-2 coronavirus proliferation by designer antisense-circRnas. Nucleic Acids Res. 2021;49(21):12502–12516. doi: 10.1093/nar/gkab1096
- Henke JI, Goergen D, Zheng J, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27(24):3300–3310. doi: 10.1038/emboj.2008.244
- Jost I, Shalamova LA, Gerresheim GK, et al. Functional sequestration of microRNA-122 from hepatitis C virus by circular RNA sponges. RNA Biol. 2018;1–8. doi: 10.1080/15476286.2018.1435248
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7
- Chen C, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344
- Wang K-S, Choo Q-L, Weiner AJ, et al. Structure, sequence and expression of the hepatitis delta (δ) viral genome. Nature. 1986;323(6088):508–514. doi: 10.1038/323508a0
- Makino S, Chang M-F, Shieh C-K, et al. Molecular cloning and sequencing of a human hepatitis delta (δ) virus RNA. Nature. 1987;329(6137):343–346. doi: 10.1038/329343a0
- AbouHaidar MG, Venkataraman S, Golshani A, et al. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci, USA. 2014;111(40):14542–14547. doi: 10.1073/pnas.1402814111
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRnas. RNA. 2015;21(2):172–179. doi: 10.1261/rna.048272.114
- Abe N, Hiroshima M, Maruyama H, et al. Rolling circle amplification in a prokaryotic translation System using small circular RNA. Angew Chem Int Ed. 2013;52(27):7004–7008. doi: 10.1002/anie.201302044
- Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5(1):16435. doi: 10.1038/srep16435
- Fan X, Yang Y, Chen C, et al. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat Commun. 2022;13(1):3751. doi: 10.1038/s41467-022-31327-y
- Zhou C, Molinie B, Daneshvar K, et al. Genome-wide maps of m6A circRnas identify widespread and Cell-type-specific methylation patterns that are distinct from mRnas. Cell Rep. 2017;20(9):2262–2276. doi: 10.1016/j.celrep.2017.08.027
- Koch L. Translated circular RNAs. Nat Rev Genet. 2017;18(5):272–273. doi: 10.1038/nrg.2017.27
- Qu L, Yi Z, Shen Y, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185(10):1728–1744.e16. doi: 10.1016/j.cell.2022.03.044
- Deng Y-Q, Zhang N-N, Zhang Y-F, et al. Lipid nanoparticle-encapsulated mRNA antibody provides long-term protection against SARS-CoV-2 in mice and hamsters. Cell Res. 2022;32(4):375–382. doi: 10.1038/s41422-022-00630-0
- Pardi N, Secreto AJ, Shan X, et al. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun. 2017;8(1):14630. doi: 10.1038/ncomms14630
- Rurik JG, Tombácz I, Yadegari A, et al. CAR T cells produced in vivo to treat cardiac injury. Science. 2022;375(6576):91–96. doi: 10.1126/science.abm0594
- Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365. doi: 10.1136/jmedgenet-2016-103758
- Ye D, Gong M, Deng Y, et al. Roles and clinical application of exosomal circRnas in the diagnosis and treatment of malignant tumors. J Transl Med. 2022;20(1):161. doi: 10.1186/s12967-022-03367-x
- Wesselhoeft RA, Kowalski PS, Parker-Hale FC, et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Molecular Cell. 2019;74(3):508–520.e4. doi: 10.1016/j.molcel.2019.02.015
- Litke JL, Jaffrey SR. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat Biotechnol. 2019;37(6):667–675. doi: 10.1038/s41587-019-0090-6
- Lokugamage MP, Gan Z, Zurla C, et al. Mild innate immune activation overrides efficient nanoparticle‐mediated RNA delivery. Adv Mater. 2020;32(1):1904905. doi: 10.1002/adma.201904905
- Paunovska K, Da Silva Sanchez A, Foster MT, et al. Increased PIP3 activity blocks nanoparticle mRNA delivery. Sci Adv. 2020;6(30):eaba5672. doi: 10.1126/sciadv.aba5672
- Westholm JO, Miura P, Olson S, et al. Genome-wide Analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993
- Meng J, Chen S, Han J-X, et al. Twist1 regulates Vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78(15):4150–4162. doi: 10.1158/0008-5472.CAN-17-3009
- Xie H, Sun H, Mu R, et al. The role of circular RNAs in viral infection and related diseases. Virus res. 2021;291:198205. doi: 10.1016/j.virusres.2020.198205
