Abstract
Diesel exhaust particles (DEP) were reported to have adverse effects on the immune system of laboratory animals and to induce thymic involution, particularly when exposure occurred during the fetal or lactational period. DEP consist of a carbon core to which many organic compounds are adsorbed, including polyaromatic hydrocarbons (PAHs) and their derivatives (e.g., dioxins and quinones). Although it has been suggested that these organic compounds were responsible for mediating the effects of DEP through their regulation of gene expression, the molecular mechanism of action of DEP has not been fully elucidated. In this study, we examined the direct effect of DEP extracts and their constituents on gene expression and phenotype in the fetal thymus. Fetal thymuses from C57BL/6 mice were exposed to DEP extracts for 24 hrs, after which their gene expression was analyzed using an Affymetrix GeneChip system. DEP extracts up-regulated several genes known as arylhydrocarbon receptor (AhR)-target genes, including cytochrome P450 1a1 (Cyp1a1), 1b1 (Cyp1b1), TCDD-inducible poly(ADP-ribose) polymerase (Tiparp), and scinderin (Scin). Similarly, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and benzo[a]pyrene (B[a]P), which are AhR ligands, induced remarkably similar changes in gene expression compared to DEP extracts. In addition, our data showed little contribution of quinones to DEP extracts-induced changes in gene expression in fetal thymus through oxidative stress responses. These changes in gene expression were also confirmed by semi-quantitative RT-PCR. Furthermore, DEP extracts skewed thymic T-cell differentiation in favor of the production of CD8 T-cells, which was also observed when exposed to AhR ligands. Our results suggest that organic compounds adsorbed onto DEP alter thymic gene expression and directly affect thymocyte development by activating the AhR.
INTRODUCTION
Diesel exhaust particles (DEP) are the main air pollutants in urban areas and have been reported to induce the adverse effects on immune systems in adult mice (Heo et al., Citation2001). DEP also have been reported to affect immune development in the fetus (Watanabe and Ohsawa, Citation2002). This report showed that DEP promoted thymic involution, which was most sensitive during the fetal and lactational periods (Watanabe and Ohsawa, Citation2002). DEP contain a variety of organic chemicals, such as polyaromatic hydrocarbons (PAHs), dioxins, and oxygenated-PAHs (Miyabara et al., Citation1999; Cho et al., Citation2004; Masaki et al., Citation2005). In addition, these chemicals have been reported to play a pivotal role in mediating DEP's biological effects (Ichinose et al., Citation1995; Bonvallot et al., Citation2001). Therefore, it is suspected that these organic chemicals adsorbed onto DEP penetrate from blood capillary in alveolus, followed by transferred from mother to fetuses via placenta or to neonates via breast-feeding. However, whether organic chemicals adsorbed onto DEP directly affect immune development has not been clarified.
Of the organic chemicals known to adsorb onto DEP, several act as a ligand for the arylhydrocarbon receptor (AhR). It has been reported that DEP adsorb 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and benzo[a]pyrene (B[a]P) (Miyabara et al., Citation1999; Masaki et al., Citation2005), both of which have been well studied as environmental contaminants and were reported to exert their toxic effects in an AhR-dependent manner (Fernandez-Salguero et al., Citation1996; Shimizu et al., 2000; Vorderstrasse et al., Citation2001). The AhR is a ligand-dependent transcriptional factor that is ubiquitously expressed in various tissues (Whitlock, Citation1999). Ligand-activated AhR enhances the expression of a number of genes, most notably the cytochrome P450 1a1 (Cyp1a1) gene (Whitlock, Citation1999; Mimura and Fujii-Kuriyama, Citation2003), by binding to the xenobiotics responsive element (XRE) in their enhancer regions. Previous reports showed that organic extracts of DEP were able to induce the expression of the Cyp1a1 gene in several cell types (Bonvallot et al., Citation2001; Baulig et al., Citation2003). In addition to AhR ligands, it has also been reported that quinone compounds are involved in DEP-induced toxicities (Sagai et al., Citation1993; Kumagai et al., Citation1997). DEP have several quinone compounds, such as 1,2-naphthoquinone (NPQ) and 9,10-phenanthraquinone (PHQ) (Cho et al., Citation2004). These quinones are known to cause oxidative stress responses through their redox cycling (Doherty et al., Citation1984; Kumagai et al., Citation2002). Since previous studies showed that DEP induce oxidative stress responses in lung (Sagai et al., Citation1993; Kumagai et al., Citation1997) and the expression of stress response genes including heme oxygenase 1 (Hmox1) and NAD(P)H:quinone oxidoreductase (Nqo1) in alveolar epithelial cells (Baulig et al., Citation2003; Koike et al., Citation2004), it is strongly suggested that quinones play a pivotal role in DEP-induced toxicities through oxidative stress. The foregoing notwithstanding, it remains to be clarified which organic compounds are involved in DEP-induced toxicity of thymus.
In the present study, we examined the effects of DEP extracts and their constituents on thymic gene expression and phenotype, and determined which kind of their organic compounds mediated these effects. The model system that we used was the fetal thymus organ culture (FTOC). In the thymus, CD4−CD8− (DN) thymocytes, the most immature T-cells, transiently develop into CD4+CD8+ (DP) thymocytes, which then terminally differentiate into mature CD4+CD8− (CD4 T) or CD4−CD8+ (CD8 T)-cells. These differentiation processes of immature T-cells normally proceed in these thymic explants (Kingston et al., Citation1985), thereby providing us with the ability to examine the direct effects of DEP on thymus without influence of endogenous hormones.
MATERIALS AND METHODS
Animals
Female and male C57BL/6J mice (8 weeks old) were purchased from Clea Japan (Tokyo, Japan) and acclimated to their housing environment for 1 week prior to use. They were given access to food and distilled water ad libitum and their room was maintained under controlled conditions with a temperature of 24 ± 1°C, humidity of 50 ± 10%, and a 12/12-hr light/dark cycle. The mice were handled in a humane manner according to National Institute for Environmental Studies guidelines.
DEP Extracts and Chemicals
DEP extracts were prepared as previously described (Koike et al., Citation2004). Briefly, DEP were collected in the dilution tunnel of diesel inhalation facility and extracted with methylene chloride. The extracts were concentrated by evaporation and the residue was dissolved in dimethyl sulfoxide (DMSO) (500 mg/ml). DEP extracts were finally diluted to 10 mg/ml with DMSO and stored at −80°C. TCDD was purchased from Cambridge Isotope Laboratories (Andover, MA) and 10 μM TCDD was prepared by diluting it with DMSO. B[a]P was purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan) and dissolved to a final concentration of 1 mM in DMSO. Stock solutions of TCDD and B[a]P were stored at −30°C. 10 mM NPQ (Tokyo Kasei Kogyo Co., Ltd., Tokyo, Japan) and 1 mM PHQ (Wako) were freshly prepared by dissolving them in DMSO. The stock solutions of DEP extracts and these chemicals were diluted 1000-fold with culture medium just prior to exposure of tissues.
Fetal Thymus Organ Culture (FTOC)
FTOC was performed as previously described (Tsukumo et al., Citation2002). Briefly, thymic lobes were randomly collected from 16.5 day-old C57BL/6J fetuses. Each lobe was cultured on a nitrocellulose filter in RPMI 1640 media (Sigma, St. Louis, MO) supplementedwith 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY), 10 mM HEPES (pH 7.1), 1 mM pyruvate, and 50 μM 2-mercaptoethanol in the presence of either 0.1% DMSO, 10 μg/ml of DEP extracts, 10 nM TCDD, 1 μM B[a]P, 10 μM NPQ, or 1 μM PHQ at 37°C in 5% CO2 for the indicated times.
Affymetrix GeneChip Analysis
Affymetrix GeneChip analysis was performed as recommended in the Affymetrix expression analysis technical manual (Affymetrix, Santa Clara, CA). After the end of the culture period, total RNA was isolated from 5 thymic lobes/each treatment groups using an RNeasy Mini Kit (Qiagen, Chatsworth, CA). Total RNA was quantitated using a NanoDrop spectrometer (NanoDrop Technologies, Rockland, DE) and its purity was confirmed by determining the absorbance ratio A260nm/A280nm. All RNA samples yielded ratios of 1.8 or greater. The quality of the total RNA was also confirmed by visualizing 18 and 28S ribosomal RNA bands in an agarose gel. Double-stranded cDNA (ds-cDNA) was synthesizedfrom 5 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) and T7 oligo (dT)24 primer (Affymetrix). The double-strandedcDNA was purified using the phenol/chloroform extraction method, which was followed by ethanol precipitation. The in vitro transcription reactionwas performed using a BioArray high yield RNA transcriptlabeling kit (Enzo Diagnostics, Farmingdale, NY).
The amount of synthesized cRNA was measured using a NanoDrop spectrometer and the quality was confirmed on an Agilent BioAnalyzer 2100 (Agilent Technologies, Palo Alto, CA) and agarose gel electrophoresis. Fifteen μg of the biotin-labeled cRNA were fragmented and hybridized to a Murine Genome U74A.v2 array (Affymetrix). The hybridized probe array was washed, stained, and scanned. Criteria for an acceptable hybridization (noise, background, and 3′/5′ ratio of housekeeping genes) were checked. The data obtained from two independent experiments were analyzed using Affymetrix GCOS 1.2 software. Comparison analysis was performed, followed by analysis using the statistical algorithms as described on the web site 〈http://www.affymetrix.com/support/technical/technotes/statistical_referen ce_guide.pdf〉. Up-regulated transcripts were defined only when identified as “Present” in experiment arrays by the detection algorithm, “Increase” by the change algorithm, and signal log ratio of 1.0 or greater by the signal log ratio algorithm in both of two independent experiments. On the other hand, down-regulated transcripts were defined only when identified as “Present” in Baseline arrays by the detection algorithm, “Decrease” by the change algorithm, and signal log ratio of −1.0 or lesser by the signal log ratio algorithm in both of two independent experiments. For hierarchical clustering, analysis was performed using GeneSpring 7.0 (Silicon Genetics, Redwood City, CA).
Semi-Quantitative RT-PCR
Ds-cDNA of each gene was amplified using an RNA PCR kit (AMV) Ver.2.1 (TaKaRa Biomedicals, Kyoto, Japan). Primers used in this study were designed using PRIMER3 〈frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi〉 based on mouse sequences published in the NCBI database. Primer sequences, PCR cycle numbers, and the annealing temperatures for each gene are shown in . Semi-quantitative RT-PCR was carried out with cycle numbers that had been tested to be in the linear range of product amplification. The amplification was carried out at an initial incubation at 94°C for 2 min, followed by 18-28 cycles of 94°C for 30 sec, 60 or 64°C for 30 sec, and 72°C for 30 sec, and a final extension at 72°C for 7 minutes. The PCR products were separated in a 1.2% SYNER gel (Diversified Biotech, Boston, MA) containing 0.5 μg/ml ethidium bromide and visualized using Kodak EDAS 290.
TABLE 1 List of primers used for semi-quantitative RT-PCR
Flow Cytometric Analysis
The percentages of CD4 and CD8 fetal thymocytes were determined as previously described (Tsukumo et al., Citation2002). After 4 d in culture, a single cell suspension of thymocytes was prepared by gently homogenizing the thymus lobes using slide glasses. Isolated thymocytes were then incubated with titrated amounts of phycoerythrin-labeled anti-mouse CD4 (GK1.5) and fluorescein isothiocyanate-labeled anti-mouse CD8 (53-6.7) antibodies. After washing, the cells were treated with 7-aminoactinomycin D (5 μg/ml) and were analyzed using FACSCalibur (BD Biosciences, San Diego, CA). All antibodies were purchased from PharMingen (San Diego, CA).
Statistical Analysis
Statistical analysis was performed by the Mann–Whitney, 2-tailed test using the GraphPad Prism 4 (GraphPad Software Inc., San Diego, CA).
RESULTS
Identification of DEP Extracts-Induced Gene Expression Changes by Affymetrix GeneChip Analysis
In this study, we used fetal thymuses from GD16.5 fetuses. Following a culture period of 6 hrs, DN cells comprised the most abundant subset of thymocytes in our fetal thymuses (approx. 60%), followed by DP thymocytes (approximately 25%). After 24 hrs in culture, the percentage of DN thymocytes decreased (approximately 15%) while the DP percentage increased to approximately 80%. When cultured for ≥4 days, the thymuses were observed to contain small numbers of CD4 T- and CD8 T-cells (approximately 7% and 2%, respectively), which is in accordance with our previous findings (Tsukumo et al., Citation2002).
To explore genes that are directly affected by organic compounds adsorbed onto DEP, we analyzed changes in gene expression in thymocytes 6 and 24 hrs following exposure. DEP extracts were used at a concentration of 10 μg/ml, since our previous study showed that 10 μg/ml of DEP extracts induced substantial amount of several genes, such as Hmox1, in alveolar macrophages (Koike et al., Citation2002). At these exposure time points and this dose, DEP extracts showed no cytotoxicity and no changes in the percentages of T-cell subpopulations in the fetal thymus (data not shown). lists the genes that showed at least a 2-fold change in expression following exposure of the fetal thymus to DEP extracts, in both of two independent experiments. A 6-hr exposure to DEP extracts resulted in the up-regulation of Cyp1a1, TCDD-inducible poly(ADP-ribose) polymerase (Tiparp), cytochrome P450 1b1 (Cyp1b1), galectin 3 (Lgals3), scinderin (Scin), and matrix metalloproteinase 13 (Mmp13) genes. A 24 hr exposure up-regulated the expression of lymphocyte antigen 6 complex, locus A (Ly6a), and chemokine (C-X-C motif) ligand 9 (Cxcl9), in addition to the 6 genes that were up-regulated at 6 hrs. No down-regulation in gene expression was found at 6 hrs, and only 1 gene i.e., arginase 1 (Arg1) was down-regulated at 24 hr. The gene that showed the greatest change was Cyp1a1, which is an AhR target gene. Cyp1b1, Tiparp, and Scin were also reported to be induced in an AhR-dependent manner (Ma et al., Citation2001; Svensson et al., Citation2002).
TBALE 2 Up- and down-regulated gene in mouse fetal thymus exposed to DEP extracts
Comparison of Changes in Gene Expression Induced by DEP Extracts and AhR Ligands
The finding that DEP extracts up-regulated the expression of several AhR target genes prompted us to examine the role played by AhR ligands in mediating this effect. Therefore, we compared changes in gene expression levels induced by DEP extracts with those caused by TCDD and B[a]P, which are well known ligands for the AhR. showed the Venn diagram of genes that were increased at least a 2-fold following a 24 hr exposure to DEP extracts, TCDD, or B[a]P. TCDD up-regulated the expression of 6 of the 8 genes that were up-regulated by a 24 hr exposure to DEP extracts; these 6 genes were also up-regulated following a 6 hr exposure to DEP extracts (). Similarly, B[a]P up-regulated the expression of 5 genes that were also regulated by DEP extracts and TCDD. On the other hand, neither TCDD nor B[a]P changed expression of Cxcl9 and Ly6a. We checked the changes in expression of these common genes that were regulated in both DEP extracts and AhR ligands by semi-quantitative RT-PCR. As shown in , our data obtained from semi-quantitative RT-PCR were well consistent with those from Affymetrix GeneChip analysis.
FIG. 1 Venn diagram of DEP extracts and AhR ligand (TCDD and B[a]P)-regulated genes. Fetal thymus was exposed to DEP extracts, TCDD, or B[a]P for 24 hrs. Total RNA was isolated and gene expression was analyzed using an Affymetrix GeneChip. These genes showed at least a 2-fold change in expression in both of two independent experiments, compared to controls.
![FIG. 1 Venn diagram of DEP extracts and AhR ligand (TCDD and B[a]P)-regulated genes. Fetal thymus was exposed to DEP extracts, TCDD, or B[a]P for 24 hrs. Total RNA was isolated and gene expression was analyzed using an Affymetrix GeneChip. These genes showed at least a 2-fold change in expression in both of two independent experiments, compared to controls.](/cms/asset/92cb733f-c292-4f45-af7f-99bea13b313d/iimt_a_149611_uf0001_b.gif)
FIG. 2 Confirmation of gene chip data in fetal thymuses exposed to DEP extracts, TCDD, and B[a]P using semi-quantitative RT-PCR. Fetal thymus was exposed to DEP extracts, TCDD, or B[a]P for 24 hrs, and ds-cDNAs obtained in two independent experiments were subjected to semi-quantitative PCR using each specific primer pair (). PCR products were separated in a 1.2% SYNER gel containing 0.5 μg/ml ethidium bromide.
![FIG. 2 Confirmation of gene chip data in fetal thymuses exposed to DEP extracts, TCDD, and B[a]P using semi-quantitative RT-PCR. Fetal thymus was exposed to DEP extracts, TCDD, or B[a]P for 24 hrs, and ds-cDNAs obtained in two independent experiments were subjected to semi-quantitative PCR using each specific primer pair (Table 1). PCR products were separated in a 1.2% SYNER gel containing 0.5 μg/ml ethidium bromide.](/cms/asset/f90ada9c-fc1e-41d2-bf88-1b79595dddb3/iimt_a_149611_uf0002_b.gif)
In this study, TCDD and B[a]P were used at concentrations of 10 nM and 1 μM, respectively. At these doses, TCDD and B[a]P induced the significant levels of expression of Cyp1a1, which were shown by Affymetrix GeneChip analysis (55.9- and 12.1-fold, respectively) and semi-quantitative RT-PCR (). In addition, they showed no cytotoxicity and no change of the percentages of T-cell subpopulations in the fetal thymuses when exposed for 24 hrs at these doses, although 10 μM B[a]P exerted cytotoxicity in fetal thymus (data not shown).
Comparison of Changes in Gene Expression Induced by DEP Extracts and the Quinones Contained in the Extracts
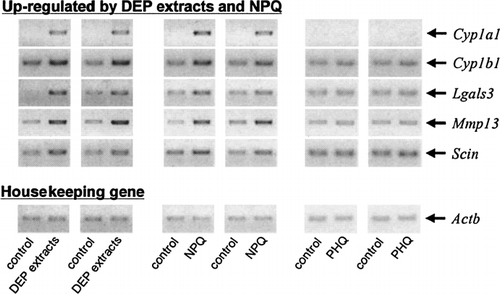
In order to determine whether quinones may have played a role in the regulation of gene expression, particularly through stress response, we compared the changes in gene expression induced by DEP extracts to those obtained with NPQ and PHQ, which are quinones contained in DEP extracts. showed the Venn diagram of genes that were increased at least a 2-fold following a 24 hr exposure to DEP extracts, NPQ, or PHQ, when analyzed using Affymetrix GeneChip system. Unexpectedly, exposure to NPQ induced changes in expression of Cyp1a1, Cyp1b1, Lgals3, Mmp13, and Scin that were also up-regulated by DEP extracts and AhR ligands. On the other hand, PHQ induced up-regulation of only 1 gene (Ddx3y). We checked the changes in expression of these common genes that were regulated in both DEP extracts and quinones by semi-quantitative RT-PCR (). In consistence with data obtained from Affymetrix GeneChip analysis, NPQ, but not PHQ, increased expression of genes that were up-regulated by both DEP extracts and NPQ. In this study, NPQ and PHQ were used at concentrations of 10 μM and 1 μM, respectively. These doses of NPQ and PHQ showed no cytotoxicity and no change of the percentages of T-cell subpopulations in fetal thymus exposed for 24 hrs, while exposure to them at higher doses caused cytotoxicity (data not shown).
FIG. 3 Venn diagram of DEP extracts and quinones (NPQ and PHQ)-regulated genes. Fetal thymus was exposed to DEP extracts, NPQ, or PHQ for 24 hours. Total RNA was isolated and gene expression was analyzed using an Affymetrix GeneChip. These genes showed at least a 2-fold change in expression in both of two independent experiments, compared to controls.
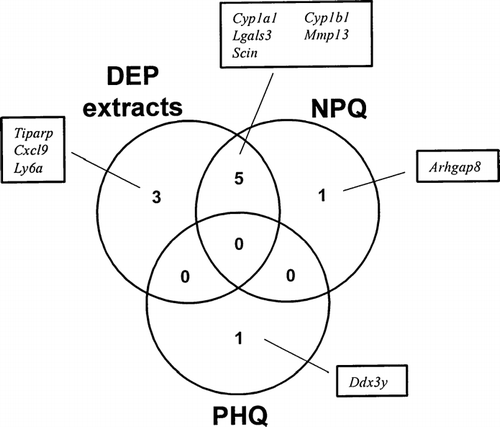
FIG. 4 Confirmation of gene chip data in fetal thymuses exposed to DEP extracts, NPQ, and PHQ using semi-quantitative RT-PCR. Fetal thymus was exposed to DEP extracts, NPQ, or PHQ for 24 hrs, and ds-cDNAs obtained in two independent experiments were subjected to semi-quantitative PCR using each specific primer pair (). PCR products were separated in a 1.2% SYNER gel containing 0.5 μg/ml ethidium bromide.
Although DEP extracts are known to induce the expression of stress response genes such as Hmox1 and Nqo1 in several cell types, we did not detect any up-regulation in these stress response genes in thymic explants cultured with DEP-extracts, and even with quinones, using Affymetrix GeneChip MG-U74Av.2. Similarly, we found no changes in mRNA expression levels for these two genes by semi-quantitative RT-PCR in exposed fetal thymuses ().
FIG. 5 Measurement of the mRNA expression of stress response genes using semi-quantitative RT-PCR. Ds-cDNAs of stress response genes (Nqo1 and Hmox1) were amplified using semi-quantitative PCR and visualized using ethidium bromide. Ds-cDNAs were identical to the samples used in the Affymetrix GeneChip analysis ( and ).

Hierarchical Clustering
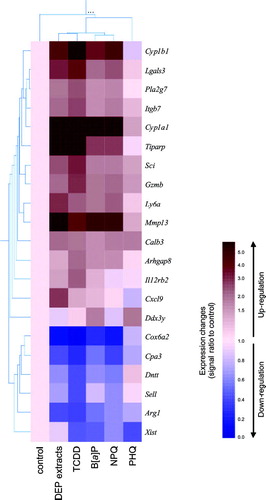
We used hierarchical clustering to facilitate data interpretation (). Up and down-regulated genes in either exposure to DEP extracts, TCDD, B[a]P, NPQ, or PHQ were collected from and , and the mean signal intensities in two independent experiments were normalized using the mean of control fetal thymuses. Hierarchical clustering analysis revealed that patterns of changes in gene expression were highly similar among DEP extracts, TCDD, B[a]P, and NPQ, while PHQ showed little similarity to any chemicals. Cxcl9, which was only up-regulated over 2-fold by DEP extracts, was nearly unaffected by any exposure of TCDD, B[a]P, NPQ, and PHQ.
FIG. 6 Hierarchical clustering analysis of the genes that were either up- or down-regulated by a 24 hr exposure to DEP extracts or their constituents. Values obtained from Affymetrix GeneChip analysis were normalized using the mean of the vehicle-treated controls (cream color), and hierarchical clustering was performed using GeneSpring. The results are presented as the mean of ratio of the signal intensities from two independent experiments. Up-regulated genes appear red while down-regulated genes appear blue.
DEP Extracts Induced Changes in Thymocyte Subsets That Were Similar to Those Induced by AhR Ligands
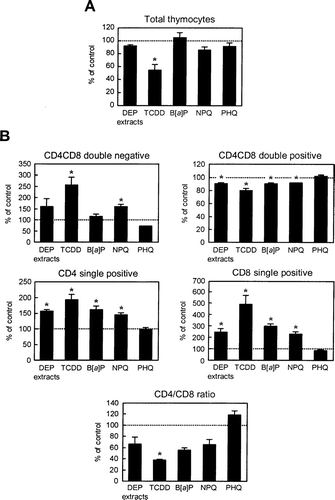
The data in and strongly suggested that DEP extracts altered gene expression by activating AhR. AhR activation is known to induce thymic involution, including loss of DP thymocytes and skewing of thymocyte maturation toward the CD8 lineage (Lai et al., Citation1998; Tsukumo et al., Citation2002). In consistence with previous reports, 4-day exposure to TCDD significantly reduced thymocyte cell numbers (). In addition, TCDD significantly increased the percentages of DN, CD4 T-, and CD8 T-cells and decreased the percentage of DP cells (). The CD4/CD8 ratio was also dramatically reduced by TCDD (). As in the case of TCDD, DEP extracts, B[a]P, and NPQ increased the percentages of DN, CD4 T-, and CD8 T-cells, decreased the percentage of DP cells and CD4/CD8 ratio (), although the effects of DEP extracts, B[a]P, and NPQ on thymic cell number were less notable (). The magnitude of these effects of DEP extracts, B[a]P, and NPQ was less than for TCDD. On the other hand, PHQ did not induce any changes in thymocyte cell number and subsets.
FIG. 7 Effects of DEP extracts or their constituents on thymocyte cellularity. Fetal thymus was exposed to DEP extracts or the described chemicals for 4 days and thymocyte cell numbers (A) and subset populations (B) were examined. The data are expressed as the mean percent of control ± SEM (n = 3), compared to control. Asterisk indicates statistically significant difference (p < 0.05) from the corresponding control.
DISCUSSION
The objective of our study was to determine the effects of DEP extracts and their constituents on gene expression and phenotype in cultured fetal thymuses, and to clarify which kind of organic compounds in these extracts were involved in mediating these effects. Using the Affymetrix GeneChip system, we found that DEP extracts up-regulated several AhR target genes i.e., Cyp1a1, Cyp1b1, Tiparp, and Scin. Hierarchical clustering analysis revealed that the patterns of changes in gene expression were highly similar among DEP extracts and AhR ligands. These results were derived from analyses of genes that showed 2- or more fold increases or decreases in expression. Analyses of genes with 1.5- or more fold changes in expression also draw the same conclusion that DEP extracts up-regulate genes that are commonly up-regulated by AhR ligands. We also found that DEP extracts skewed the differentiation of thymocytes towards the CD8 lineage, an effect that is known to occur following AhR activation (Lai et al., Citation1998; Tsukumo et al., Citation2002). Our results strongly suggest that organic chemicals adsorbed onto DEP directly regulate gene expression in the fetal thymus and influence thymocyte differentiation through their activation of the AhR.
In order to determine the role of quinones in DEP extracts-induced changes in gene expression, we examined the effects of NPQ and PHQ. Our results demonstrated that NPQ also up-regulated AhR target genes. To our knowledge, this is the first report strongly suggesting that NPQ acts as a ligand for the AhR. On the other hand, PHQ did not change expression of any genes that were up-regulated by DEP extracts (). We used PHQ at a concentration one-tenth of NPQ, because exposure to 10 μM PHQ for 24 hrs exerted cytotoxicity in fetal thymus (data not shown). Since 1 μM PHQ is still considerably higher than the concentration in DEP extracts (Cho et al., Citation2004), it seems that PHQ is not involved in DEP extracts-induced changes in gene expression in the fetal thymus observed in the present study. With regard to the involvement of oxidative stress responses, exposure of the fetal thymus to DEP extracts did not change the expression levels of Hmox1 and Nqo1, which are stress response genes known to be up-regulated by DEP extracts in alveolar macrophages and epithelial cells (Li et al., Citation2000; Baulig et al., Citation2003; Koike et al., Citation2004) (). Discrepancies in these results may be a reflection of cell- and tissue-specific responses to oxidative stress. We examined expression levels of Hmox1 and Nqo1 in Hepa-1c1c7, a mouse hepatoma cell line, exposed to DEP extracts and these quinones at the concentrations used in this study, and found that DEP extracts and NPQ, but not PHQ, up-regulated expression of these genes (data not shown). Therefore, fetal thymus may be insensitive to quinones-induced oxidative stress responses, unlikely alveolar macrophages, epithelial cells, and liver. Taken together, these results suggested little contribution of oxidative stress responses by quinones to DEP extracts-induced changes in gene expression in fetal thymus.
A previous study has reported the contents of dioxins (PCDDs and PCDFs) in the DEP samples from which the DEP extracts we used in the present study were prepared (Miyabara et al., Citation1999). The amounts of B[a]P and quinones in DEP were also reported by previous studies (Bonvallot et al., Citation2001; Cho et al., Citation2004; Masaki et al., Citation2005). Estimated from these studies, the concentrations of TCDD, B[a]P, NPQ and PHQ in 10 μg/ml DEP extracts are about 0.1 fM, 0.04 pM, 1 nM, and 1 nM, respectively. Thus, the concentrations of organic compounds used in this study are much higher than those contained in 10 μg/ml DEP extracts. On the other hand, our results from Cyp1a1 measurement by GeneChip analyses and RT-PCR showed that 10 μg/ml DEP extracts have AhR-ligand activity approximately comparative to 1 μM B[a]P and 10 μM NPQ, and about half by 10 nM TCDD. In consistence with our results, Baulig et al. (Citation2003) have reported that 10 μg/ml DEP extracts induced a comparative level of expression of Cyp1a1 when compared with 3 μM B[a]P. These findings suggest that DEP extracts contain, in addition to compounds investigated in the present study, many constituents that activate the AhR, possibly in a cooperative manner. To further demonstrate roles of their constituents in DEP's toxicities, it may be necessary to perform combined exposure of representative compounds at their lower doses. In addition, dose-response relationship of toxicants would need to be considered.
Although exposure of fetal thymus to DEP extracts induced the changes in gene expression and phenotype that were highly similar to AhR ligands, opposite effects of DEP extracts and AhR ligand on immune systems have been also reported. Heo et al. (Citation2001) have reported that exposure to DEP extracts enhanced the production of antigen-specific IgG1 and IgE in mice, while exposure to TCDD suppressed it, even though the immune cells, such as mature T-cells and B-cells, have the functional AhR (Doi et al., Citation2003). As in the case of specific response to oxidative stress, the involvement of AhR pathway in DEP-induced immunotoxicity may differ in their target cells.
Although our findings suggest that the effects of DEP extracts on the fetal thymus are dependent on AhR activation, the precise mechanism by which AhR activation affects thymocyte phenotypes has not been elucidated. A genome-wide approach using gene chip analysis allowed us to broadly search for genes that were activated by the AhR and played a role in mediating the effects of DEP in the thymus. Our data showed that DEP extracts and AhR ligands induced the expression of Cyp1a1, Cyp1b1, Tiparp, and Scin, which confirmed the results of previous studies (Lai et al., Citation1998; Ma et al., Citation2001; Svensson et al., Citation2002; Tsukumo et al., Citation2002). These genes are known to have some XRE sequences in their enhancer region. In addition to these genes, we found that the 5′ up-stream region of Lgals3 has a consensus sequence of XRE (5′-TNGCGTG-3′). Lgals3 is widely distributed in tissues, including the thymus, as well as in other immune cells, and galectin 3 protein coded by Lgals3 has several physiological functions (Hsu et al., Citation2000). We previously showed that the extracellular signal-regulated kinase (ERK) pathway was involved in the TCDD-induced differentiation of thymocytes towards the CD8 lineage in FTOC (Tsukumo et al., Citation2002). Interestingly, galectin 3 has been reported to activate ERK1/2 through phosphorylation (Maeda et al., Citation2003). These data suggest that galectin 3 may play a role in enhancing CD8 cell differentiation in the DEP extracts-treated fetal thymus.
In conclusion, we studied the direct effect of organic chemicals adsorbed onto DEP on gene expression in the fetal thymus by using FTOC and Affymetrix GeneChip analysis. We provided data showing that various AhR ligands contained in DEP extracts directly alter the development of fetal thymus. Our approach in the present study would be useful in assessing the effects of various immunotoxicants on immune development.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Ministry of Environment. The authors wish to thank Drs. M. Kubo, R. Abe (Science University of Tokyo) and H. Takano (National Institute for Environmental Studies) for their useful discussions. We also thank Ms. M. Matsumoto for her excellent technical assistance.
Haruko Nagai is also affiliated with the Research Institute for Biological Sciences, Science University of Tokyo, Noda, Chiba, Japan.
REFERENCES
- Baulig A., Garlatti M., Bonvallot V., Marchand A., Barouki R., Marano F., Baeza-Squiban A. Involvement of reactive oxygen species in the metabolic pathways triggered by diesel exhaust particles in human airway epithelial cells. Am. J. Physiol. 2003; 285: L671–L679, [CSA]
- Bonvallot V., Baeza-Squiban A., Baulig A., Brulant S., Boland S., Muzeau F., Barouki R., Marano F. Organic compounds from diesel exhaust particles elicit a proinflammatory response in human airway epithelial cells and induce cytochrome p450 1A1 expression. Am. J. Respir. Cell Mol. Biol. 2001; 25: 515–521, [PUBMED], [INFOTRIEVE], [CSA]
- Cho A. K., Stefano E. D., You Y., Rodriguez C. E., Schmitz D. A., Kumagai Y., Miguel A. H., Eiguren-Fernandez A., Kobayashi T., Avol E., Froines J. R. Determination of four quinones in diesel exhaust particles, SRM 1649a, and atmospheric PM2.5. Aerosol Sci. Technol. 2004; 38: 1–14, [CSA], [CROSSREF]
- Doherty M. D., Cohen G. M., Smith M. T. Mechanisms of toxic injury to isolated hepatocytes by 1-naphthol. Biochem. Pharmacol. 1984; 33: 543–549, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Doi H., Baba T., Tohyama C., Nohara K. Functional activation of arylhydrocarbon receptor (AhR) in primary T-cells by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicology 2003; 52: 655–662, [CSA]
- Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., Gonzalez F. J. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 1996; 140: 173–179, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Heo Y., Saxon A., Hankinson O. Effect of diesel exhaust particles and their components on the allergen-specific IgE and IgG1 response in mice. Toxicology 2001; 159: 143–158, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Hsu D. K., Yang R. Y., Pan Z., Yu L., Salomon D. R., Fung-Leung W. P., Liu F. T. Targeted distribution of the galectin-3 gene results in attenuated peritoneal inflammatory responses. Am. J. Pathol. 2000; 156: 1073–7083, [PUBMED], [INFOTRIEVE], [CSA]
- Ichinose T., Furuyama A., Sagai M. Biological effects of diesel exhaust particles (DEP). II. Acute toxicity of DEP introduced into lung by intratracheal instillation. Toxicology 1995; 99: 153–167, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Kingston R., Jenkinson E. J., Owen J. T. A single stem cell can recolonize an embryonic thymus, producing phenotypically distinct T-cell populations. Nature 1985; 317: 811–813, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Koike E., Hirano S., Furuyama A., Kobayashi T. cDNA microarray analysis of rat alveolar epithelial cells following exposure to organic extract of diesel exhaust particles. Toxicol. Appl. Pharmacol. 2004; 201: 178–185, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Koike E., Hirano S., Simojo N., Kobayashi T. cDNA microarray analysis of gene expression in rat alveolar macrophages in response to organic extract of diesel exhaust particles. Toxicol. Sci. 2002; 67: 241–246, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Kumagai Y., Arimoto T., Shinyashiki M., Shimojo N., Nakai Y., Yoshikawa T., Sagai M. Generation of reactive oxygen species during interaction of diesel exhaust particle components with NADPH-cytochrome P450 reductase and involvement of the bioactivation in the DNA damage. Free Radic. Biol. Med. 1997; 22: 479–487, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Kumagai Y., Koide S., Taguchi K., Endo A., Nakai Y., Yoshikawa T., Shimojo N. Oxidation of proximal protein sulfhydryls by phenanthraquinone, a component of diesel exhaust particles. Chem. Res. Toxicol. 2002; 15: 483–489, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Lai Z. W., Fiore N. C., Gasiewicz T. A., Silverstone A. E. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and diethylstilbestrol affect thymocytes at different stages of development in fetal thymus organ culture. Toxicol. Appl. Pharmacol. 1998; 149: 167–177, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Li N., Venkatesan M. I., Miguel A., Kaplan R., Gujuluva C., Alam J., Nel A. Induction of heme oxygenase-1 expression in macrophages by diesel exhaust particle chemicals and quinones via the antioxidant-responsive element. J. Immunol. 2000; 165: 3393–3401, [PUBMED], [INFOTRIEVE], [CSA]
- Ma Q., Baldwin K. T., Renzelli A. J., McDaniel A., Dong L. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 2001; 289: 499–506, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Maeda N., Kawada N., Seki S., Arakawa T., Ikeda K., Iwao H., Okuyama H., Hirabayashi J., Kasai K., Yoshizato K. Stimulation of proliferation of rat hepatic stellate cells by galectin-1 and galectin-3 through different intracellular signaling pathways. J. Biol. Chem. 2003; 278: 18938–18944, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Masaki H., Susaki H., Korenaga T. Development of a micro dual beam fluorometric detector specific for microchip analysis of benzo[a]pyrene and benzo[k]fluoranthene in diesel exhaust particulate samples. Analyst 2005; 130: 1253–1257, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Mimura J., Fujii-Kuriyama Y. Functional role of AhR in the expression of toxic effects by TCDD. Biochim. Biophys. Acta 2003; 1619: 263–268, [PUBMED], [INFOTRIEVE], [CSA]
- Miyabara Y., Hashimoto S., Sagai M., Morita M. PCDDs and PCDFs in vehicle exhaust particles in Japan. Chemosphere 1999; 39: 143–150, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Sagai M., Saito H., Ichinose T., Kodama M., Mori Y. Biological effects of diesel exhaust particles. I. In vitro production of superoxide and in vivo toxicity in mice. Free Radic. Biol. Med. 1993; 14: 37–43, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Snimizu Y., Nakatsuru Y., Ichinose M., Takahashi Y., Kume H., Mimura J., Fujii-Kuriyama Y., Ishikawa T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2000; 97: 779–782, [CSA], [CROSSREF]
- Svensson C., Silverstone A. E., Lai Z. W., Lundberg K. Dioxin-induced adseverin expression in the mouse thymus is strictly regulated and dependent on the aryl hydrocarbon receptor. Biochem. Biophys. Res. Commun. 2002; 291: 1194–1200, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Tsukumo S., Iwata M., Tohyama C., Nohara K. Skewed differentiation of thymocytes toward CD8 T cells by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires activation of the extracellular signal-related kinase pathway. Arch. Toxicol. 2002; 76: 335–343, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Vorderstrasse B. A., Steppan L. B., Silverstone A. E., Kerkvliet N. I. Aryl hydrocarbon receptor-deficient mice generate normal immune responses to model antigens and are resistant to TCDD-induced immune suppression. Toxicol. Appl. Pharmacol. 2001; 171: 157–164, [PUBMED], [INFOTRIEVE], [CSA], [CROSSREF]
- Watanabe N., Ohsawa M. Elevated serum immunoglobulin E to Cryptomeria japonica pollen in rats exposed to diesel exhaust during fetal and neonatal periods. BMC Pregnancy Childbirth 2002; 2: 1–9, [CSA], [CROSSREF]
- Whitlock J. P., Jr. Induction of cytochrome P4501A1. Ann. Rev. Pharmacol. Toxicol. 1999; 39: 103–125, [CSA], [CROSSREF]