Abstract
A method of identifying fourteen mango hybrids along with their parents was designed using a DNA typing procedure employing RAPD markers. Ten of the 80 operon primers screened produced 139 usable bands. Cluster analysis based on Jaccard's coefficient of similarity using UPGMA revealed low to medium diversity among hybrids. Hybrids, which had “Neelum,” “Alphonso,” and “Mulgoa” as one of their parents, formed into distinct cluster. Hybrid “Neelgao” with “Mulgoa” as its male parent stood distinctly in the dendrogram comprising hybrids along with their parents. The study indicates the potential for use of RAPD markers in identification of parent similarity among hybrids.
KEYWORDS:
INTRODUCTION
The mango (Mangifera indica L.) is undoubtedly the most important fruit crop of India. It covers the largest production area compared to any other fruit crop in the country and thrives in almost all regions except at the altitudes above 3000 feet. From Cape Comorin to the foothills of Himalayas and from the Punjab to Assam, one can see mango trees flourishing everywhere. The mango is the prominent member of Anacardiaceae and is said to have originated in Indo-Burma region (CitationDe Candolle, 1904). CitationVavilov (1926) also felt that the mango originated in the Indo-Burma region. India probably has more commercial plantings than rest of the world (CitationOchse et al., 1961). On account of the hardy nature of the tree, low cost of maintenance, and profuse yield, it has come to be known as the poor man's fruit and thus possesses a mass appeal. However, it must be acknowledged that the real economic importance of mango at present lies in its tremendous local consumption rather than its export value, although it has great potential as an item of export both fresh and in its processed form. Mango has been cultivated in India and Southeast Asian countries over last 4,000 years (CitationDe Candolle, 1884). The planted area of this fruit is 1.23 million ha. with an annual production of 10.99 million tons accounting to 57.2% of the world's production (CitationNegi, 2000).
Its demand in export markets is ever increasing. Nevertheless, this fruit crop presents difficulties for orchardists due to its low orchard efficiency caused by the problem of biennial cropping (alternate bearing) inherent in trees. Analysis of hybrids and their parents is essential to know the contribution of each parent to their progenies, which will help with further analysis of hybridization programs. “Neelum” has been extensively used as one of the parents in hybridization programs to transfer the regular bearing habit to hybrids. It has contributed much to the evolution of many regular bearing Indian hybrid cultivars.
Mango cultivars are currently identified on the basis of morphological traits such as leaf and fruit characteristics (CitationCampbell, 1992; CitationJintanawong et al., 1992). However, this type of cultivar identification usually requires growing plants to maturity, and it often lacks decisiveness and objectivity. Furthermore, morphological traits cannot serve as unambiguous markers because they may vary with the environmental conditions (CitationTanksley et al., 1989). Recently, reliable genetic markers have been developed and introduced for mango cultivar identification. These include isozymes, random amplified polymorphic DNAs (RAPDs), and variable number tandem repeats (VNTRs).
RAPDs are DNA markers that are produced by the amplification of random DNA segments using single short primers by means of a polymerase chain reaction (PCR). It helps in identification of complete or partial nucleotide sequence homology, between the genomic DNA, and the oligonucleotide primer, at each end of the amplified product. On an average, each primer will direct the amplification of several discrete loci in the genome, making the assay an efficient way to screen for nucleotide sequence polymorphisms between individuals. The major advantage of this assay is that there is no requirement for DNA sequence information. The protocol is also relatively quick and easy to perform and uses fluorescence in lieu of radioactivity (Williams et al., 1992).
MATERIALS AND METHODS
Plant Material and DNA Extraction
Fourteen economically significant hybrids and their parents were selected for the study as shown in . The leaf samples were obtained from the orchard of University of Agricultural Sciences, Gandhi Krishi Vignana Kendra, Bangalore and Agricultural Research Station, Chintamani (Karnataka), India. Only the recently matured leaves were collected. Later in the laboratory, they were oven dried at 40°C for 48 hours, ground to fine powder using a “Remi” mixer, filtered, used for DNA extraction.
TABLE 1 Mango Hybrids and Their Parents Examined in the Study
DNA Isolation
Total DNA was extracted as standardized earlier (CitationKumar et al., 2001). 500 mg powdered leaf tissue and preheated (65°C) 20 ml of extraction buffer containing 3% CTAB, 100 mM Tris, 20 mM EDTA, 1.4 M NaCl, 2% PVP and 1% β-Mercaptoethanol were incubated for one hour with intermittent shaking at 65°C. The centrifuge tube was brought to room temperature before adding 6 ml of chloroform and isoamyl alcohol (24:1). The contents were mixed well by inverting the tube gently 25–30 times and spun at 6,000 rpm for 15 minutes. The supernatant was transferred to a fresh tube and this clean-up step was repeated until a clear supernatant was obtained. Supernatant was kept overnight at 4°C to precipitate DNA by adding half a volume of 5 M NaCl and 1 volume of isopropanol. Centrifuging at 10,000 rpm for 20 minutes pelleted the DNA, and the pellet was washed with 70% ethanol. The dried DNA pellet was resuspended in 1 ml of TE [Tris-EDTA] buffer. Contaminating RNA was removed by digestion with 10 μg RNase for 30 minutes at 37°C. The DNA was further purified by extracting twice with an equal volume of phenol followed by an equal volume of phenol to chloroform (1:1) and finally with an equal volume of chloroform. The DNA was precipitated by the addition of 1 volume of isopropanol and spun at 5000 rpm for 5 minutes. The final pellet was dissolved in 0.5 ml TE. The DNA concentration was determined using “Hoefer's Dynaquant” (procedure as given by manufacturer) and the quality was verified by electrophoresis on a 2% agarose gel.
RAPD Assay
The basic protocol reported by CitationWilliams et al. (1990) for PCR was followed with slight modifications to suit a mango crop (CitationKumar et al., 2001). A single decamer of arbitrary sequence was used in each PCR reaction. Amplification reactions were carried out in 25 μl reaction mixture containing template DNA (25 ng), 5 pmols of primer (Operon USA, Inc.), 2 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl, 0.1% Triton X-100, 1 unit of Taq DNA polymerase (Bangalore Genei), and 200 μM of each dNTP (Finnzymes). The mixture was overlaid with a drop of mineral oil to prevent evaporation of the reaction mixture. Amplification was performed in a thermal cycler (MJ Research, PTC 100) for 45 cycles after an initial denaturation at 94°C for 2 minutes. In each cycle, denaturation for one minute at 94°C, annealing for one minute at 35°C and extension for two minutes at 72°C was programmed with a final extension step at 72°C for five minutes after the 45th cycle. Negative control was used initially to check the fidelity of the PCR reaction. For further reactions negative controls were not used.
DNA Electrophoresis
The amplified DNA fragments were separated out on 2% agarose gel (polysaccharide derivative of agar) stained with ethidium bromide. A running buffer containing Tris-base, Boric acid and EDTA (pH-8.0) was used for electrophoresis and for preparing gels. Wells were loaded with 25 μl of reaction volume and 5 μl of loading buffer (Sucrose and Bromo-cresol green dye) together. Electrophoresis was conducted at 40–75 volts for 5–8 hours, and the gel was photographed under UV light by using a gel doc system (Herolab, Germany).
Dendrogram Construction
GelCompar, Ver 4.1 (Applied Maths, Kortrijik, Belgium), a computer application was used to generate Jaccard's coefficient of similarity matrices, which was later used to construct a dendrogram by UPGMA (Unweighted Pair Group Method with Arithmetical Averages). Coefficient of Jaccard's (SJ) = nAB / nA+nB = nAB, where nAB is the number of bands common for samples A and B, nA is the total number of bands in sample A and nB is the total number of bands in sample B. Jaccard's coefficient considers only the presence of band as similarity and hence, is more conservative in declaring genetic relatedness.
RESULTS
The recently matured leaf samples of mango hybrids and their parents were collected and dried in the hot air oven at a temperature of 40°C for 48 hours by spreading them out evenly. Drying for 48 hours was found to be optimum, with prolonged drying of the samples resulting in degraded DNA. Powdering around 5 g of leaf tissue yielded 1.5 g of fine powder when filtered through 60-mesh sieve.
DNA yield was 12–150 μg/ml for every g of powdered leaf sample. The quality of DNA obtained was of high quality and easily amplifiable. A Spectrophotometer reading of 1.8 to 2.0 (260 nm/280 nm) and electrophoresis confirmed the quality of DNA, which was above 50 Kb.
The basic protocol described by CitationWilliams et al. (1990) and Welsh and Mc Clelland (1990) for PCR was optimized as described in Materials and Methods. A template DNA of 25 ng, MgCl2 of 2.0 mM and dNTPs of 200 μM concentration was found to be optimum for high quality amplification and obtaining intense repeatable banding patterns. Large changes in template DNA concentrations, lead to reduction in amplification of fainter bands, while too much DNA produced a smear effect, emphasizing the importance of precise quantification of the DNA for clear amplification.
In this study 80 operon random ten-base long single stranded primers (OPA to OPD with 20 primers in each group) were screened using the few genotypes that on average gave 6 bands. In each reaction a control (water) was used to check contamination during initial reactions. At least one product was amplified in all the primers screened. The selected ten primers yielded an average of 13 bands per primer of sizes ranging from 200 to 3000 bp (.). All the 139 bands amplified were either monomorphic or polymorphic which were considered for the precise calculation of genetic diversity.
TABLE 2 Synthetic Deoxyribonucleotides Used as Primers for Amplification of Mango DNA
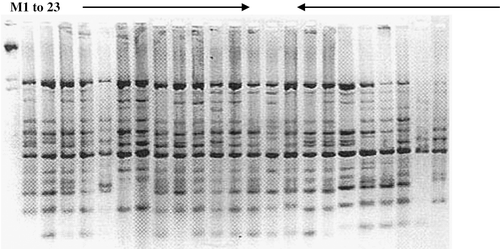
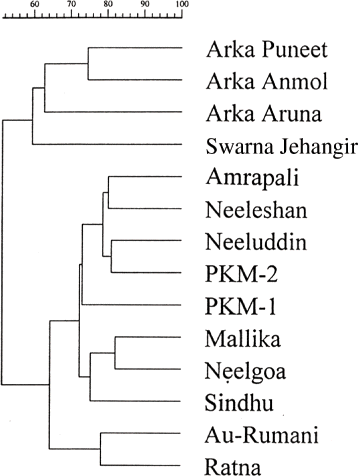
A representative of the PCR amplification product of 14 hybrids and their parents is shown in , which yielded sufficient polymorphisms to distinguish hybrids with their parents. UPGMA cluster analysis of 14 hybrids and their progenies using Jaccard's coefficient of similarity revealed 52% similarity in two major clusters. Hybrids “Neeleshan,” “Neeluddin,” “Ratna,” “Mallika,” “Amrapali,” “Neelgoa,” “Sindhu” and “PKM-1” shared “Neelum” as one of their parents formed a separate cluster. They were also regular bearers. Hybrids “Arka Puneet” and “Arka Anmol, sharing “Alphonso” as female parent, formed another cluster.
FIGURE 1 A gel profile of the mango hybrids with their corresponding parents using Operon primer D1. M—Lambda marker. Lane 1-23: Au-Rumani, Neelum, Baneshan, Ratna, Mulgoa, Dashehri, Alphonso, Suvarnarekha, PKM-1, Rumani, Sindhu, Mallika, Amrapali, Neeleshan, Neelgoa, Neeluddin, PKM-2, Himayuddin, Jehangir, Swarna Jehangir, Arka Aruna, Arka Puneet, and Arka Anmol.
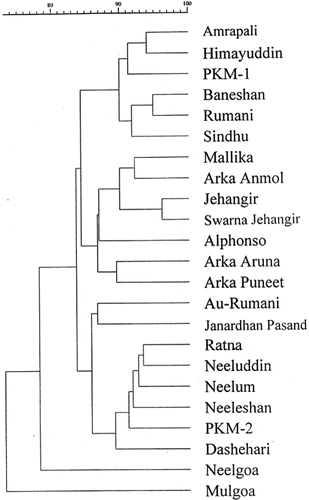
In another cluster analysis where hybrids, along with their parents, were used it formed four distinct clusters. The hybrids were distributed in the dendrogram and most of them had “Neelum” as one of their parents. The hybrid “Neelgoa,” which was clearly distinct in the dendrogram had “Mulgoa” as its male parent and was placed close to “Mulgoa.” “Mulgoa” was placed distinctly in the RAPD dendrogram drawn using commercial cultivars in our previous study (CitationKumar et al., 2001) in that it is totally different from other cultivars in several characters.
DISCUSSION
Mango (2n = 40) is reported to have a DNA content of around 439 million bases (CitationArmuganathan and Earle, 1991). Our analysis of relatedness was based on an average of one marker per 300 million bases, which was realistic to represent the whole genetic makeup of the mango and to make meaningful statements about relatedness among mango hybrids, although there could be gaps of 10 to 15 million bases which were not covered in the genome.
The analysis of mango hybrids suggested that the differences among the hybrids are low, as most of the hybrids had same parent. The generated dendrogram was based on Jaccard's coefficient of similarity, which considers only the presence of bands among hybrids as similarity and not the absence of bands (CitationJaccard, 1908). Our results revealed a minimum similarity of 52% (). The hybrids “Arka Aruna,” “Arka Puneet” and “Arka Anmol” were placed closely in the dendrogram, which was comprised of “Alphonso” and “Banganpalli” as one of their parents. The similarity of morphological characters compared in the dendrogram revealed that the hybrids “Arka Puneet” and “Arka Anmol” were placed close in the cluster as both have “Alphonso” as a parent, fruits of a medium size, with excellent quality, and both hybrids are mid season, which strengthens the close relationship. Other hybrids were placed in a separate cluster, as they had “Neelum” as one of their parents. Further, among these hybrids “Mallika,” “Neelgoa” and “Sindhu” were placed much closer as both “Mallika” and “Neelgoa” are high yielding and late bearing hybrids. “Mallika” and “Sindhu” produce good quality fruits and are late bearing. “Sindhu” is seedless and in “Mallika” seedlessness is noticed to some extent. There was a close relationship between “Au-Rumani” and “Ratna.” They both are high yielding, have large sized fruits with broad shoulders, and are late hybrids (June harvesting).
FIGURE 2 Association among mango hybrids revealed by UPGMA cluster analysis according to Jaccard's genetic similarity coefficients calculated from RAPD data generated by ten primers.
The dendrogram representing hybrids, along with their parents, showed distinct clusters (), but hybrids sharing “Neelum” as one of the parents were distributed all along the dendrogram. The comparison of morphological characters and the obtained dendrogram revealed that the hybrids grouped in the first cluster viz, “Amrapali,” “Himayuddin” and “PKM-1” have exceptionally good quality fruits with high TSS (>24°B). In this group of hybrids “PKM-1” and “Amrapali” are regular bearers and high yielders, but “Himayuddin” is a shy and alternate bearer. Cultivars “Baneshan” and “Rumani” were placed close, as both are very hardy and most suitable for dry regions. In the second cluster the relationship between the hybrids “Mallika” and “Arka Anmol” may be due to their late bearing ability, elongated fruits with slight similarity in beak, and the development of cadmium yellow skin color upon ripening. The hybrid “Swarna Jehangir” which had “Jehangir” as one of the parents was obviously placed next to “Jehangir.” “Alphonso,” being one of the parents for “Arka Aruna,” “Arka Puneet,” and “Arka Anmol,” topped the cluster and was placed close to “Alphonso.” The whole group had a similar pulp texture to that of “Alphonso.” The hybrids “Arka Aruna” and “Arka Puneet” were placed close as parentage of both these hybrids is the same, it is a reciprocal cross of each other, and both are regular bearers. In the third cluster hybrids “Ratna,” “Neeluddin,” and “Neeleshan” formed a cluster and were close to “Neelum” as they had “Neelum” as one of their parents and are also regular bearers like “Neelum” in addition to having similar fruit shape. Close association of hybrids “Ratna” and “Neeluddin” may be attributed to similar characters such as late bearing, excellent fruit quality, regular bearing ability, and large sized fruits. The hybrid “PKM-2” was placed close to “Dashehari,” as “Dashehari” is one of the parents for “PKM-2.” In the fourth cluster hybrid “Neelgoa” was clearly distinct when compared to other hybrids, because “Mulgoa” is the male parent of this hybrid, showing major contribution of “Mulgoa” to the hybrid. “Mulgoa” occupied a distinct position on the dendrogram as it is distinctly shy bearer than rest of the cultivars, producing spherical shaped fruits with broad basal cavity and fruits will have green patches even after ripening. Most of the hybrids are regular bearers from South India. There was little difference among hybrids as most of them have at least one of their parents in common.
FIGURE 3 Association among mango hybrids and their corresponding parents revealed by UPGMA cluster analysis according to Jaccard's genetic similarity coefficients calculated from RAPD data generated by ten primers.
Although analysis on relatedness of RAPD markers was based on non-coded regions of DNA, it is expected to give a measure of difference among hybrids even in the coded regions of DNA. This is primarily due to the fact that the differences in genomes are random. Hence, a measure of similarity based on RAPD markers (noncoded regions) also offers a relative measure of similarity in coding regions, which are responsible for phenotype. Therefore, grouping of individuals based on RAPD patterns also indicates similarity in the coded region of the genome and hence common morphological traits.
In conclusion, RAPD analysis revealed a low degree of difference among the hybrids examined in the study. RAPD technique is less restricting than other molecular techniques. RAPD markers in the cluster were closely linked with important traits like regular or alternate bearing habit, high yielding ability, fruit shape, and color of fruit etc. This kind of association could contribute to the efficient selection and hybridization work in mango.
CONCLUSIONS
A rapid DNA fingerprinting technique for identifying fourteen mango hybrids along with their parents was developed. It involved identification of amplicons from genomic DNA of an isolate, produced according to RAPD markers through conventional sequence tagged site PCR using oligonucleotides as primers of disclosed sequences. Out of the 80 Operon primers screened 10 produced 139 usable bands. Cluster analysis of the data based on Jaccard's coefficient of similarity using UPGMA revealed low to medium diversity among hybrids as most of them had at least one of their parents in common. The hybrids, which had “Neelum,” “Alphonso” and “Mulgoa” as one of their parents formed into a distinct cluster. The dendrogram comprising hybrids and their corresponding parents showed that hybrid “Neelgoa” stood distinctly different as compared to others due to the presence of “Mulgoa.” The study indicated the efficiency of RAPD markers for the identification of parent similarities among hybrids and their analysis.
The author is extremely grateful to the Department of Science and Technology, New Delhi, for providing financial support and to the University of Agricultural Sciences, Bangalore, for the rest of the facilities.
LITERATURE CITED
- Armuganathan , K. and Earle , E.D. 1991 . Nuclear DNA content of some important plant species . Plant Molecular Biol. Rpt. , 9 ( 3 ) : 208 – 218 .
- Campbell , R.J. 1992 . Mangos: A guide to mango in Florida , Edited by: Campbell , R.J. 227 Miami, Florida : Fairchild Tropical Garden .
- De Candolle , A. 1884 . Origin of cultivated plants , Kegan Paul : Trench, London .
- De Candolle , A. 1904 . Origin of cultivated plants , Kegan Paul : Trench, London .
- Jaccard , P. 1908 . Nouvelles recherches sur la distribution florale . Bul. Soc. Vaud. Sci. Nat. , 44 : 223 – 270 .
- Jintanawong , S.H. , Hiranpradit , P. , Polprasid , K. and Duangpikul , P. 1992 . Group characterization of Thai mango, Mangifera indica L . Acta Hort. , 321 : 254 – 261 .
- Kumar , H.N. , Narayanaswamy , V.P. , Prasad , D.T. , Mukunda , G.K. and Sondur , S.N. 2001 . Estimation of genetic diversity of commercial mango (Mangifera indica L.) cultivars using RAPD markers . J. Hort. Sci. Biotech. , 76 ( 5 ) : 529 – 533 .
- Negi , S.S. 2000 . Mango production in India . Acta Hort. , 509 : 69 – 77 .
- Ochse , J.J. 1961 . Tropical and subtropical agriculture. I , New York : MacMillan .
- Tanksley , S.N. , Young , A. , Paterson , B. and Bonierbale , M. 1989 . RFLP mapping in plant breeding; new tools for an old science . Bio/Technol. , 7 : 257 – 264 .
- Vavilov , N.I. 1926 . The origin, variation, immunity and breeding of cultivated plants . Chronica Botanica , 13 ( 1/6 ) : 1949 – 1950 .
- Welsh , J. and McClelland , M. 1990 . Fingerprinting genomes using PCR with arbitrary primers . Nucleic Acids Res. , 18 : 7213 – 7218 .
- Williams , J.G.K. , Kubelik , A.R. , Livak , K.J. , Rafalski , J.A. and Tingey , S.V. 1990 . DNA polymorphisms amplified by arbitrary primers are useful as genetic markers . Nucleic Acids Res. , 18 : 6531 – 6535 .