Abstract
The population of grape (Vitis vinifera L), commonly cultivated in India, shows a wide range of ripening periods and fruit quality and is an unexploited resource for breeding programs. The main purpose of this study was to fingerprint these accessions and to construct a molecular database including the cultivars commonly grown in India. A total of 42 genotypes were analyzed using seven microsatellite simple sequence repeat (SSR) markers and their main morphological and agronomic characteristics were compared. A total of 45 alleles in all 42 genotypes were obtained with 7 primers with an average number of 6.4 alleles per locus. The dissimilarity matrix showed that a maximum of 110 units was obtained between the genotypes “Convent Large Black” and “Arka Hans,” while a minimum dissimilarity if 37 units were obtained between the genotypes “Anab-e-Shahi” and “Dilkhush.” In the dendrogram the microsatellite segregated the genotypes into two clusters (A and B) at 476 units with two subclusters each. The subclusters grouped genotypes predominantly as A1 with seeded fruits, A2 with seedless fruits, B1 with pigmented and seedless fruits, and B2 with colorless and seeded fruits. The use of seven polymorphic microsatellite markers and the level of genetic variability detected within Indian grapevine germplasm suggested that this is a reliable, efficient, and effective marker system that can be used for diversity analysis and subsequently in crop improvement programs.
INTRODUCTION
Grape cultivars (Vitis vinifera L) have a long history of domestication. The world's vineyards occupy about 8.7 million hectares. More than 9600 grape cultivars exist around the world (CitationGalet, 2000) and almost 16,000 prime names appear in the Vitis International Variety Catalogue (CitationMaul and Eibach, 2003). Some of these are not easily distinguishable by morphology and many cultivars appear to be synonyms, having been distributed around the world and acquiring new names in the process . Moreover, the wide distribution and long history of cultivation have led to the development of numerous cultivars with many synonyms, resulting in complexity among germplasm collections (CitationGalet, 1990). Grape in India are reported to have been introduced in 620 BC. (CitationOlmo, 1976) and commercial cultivation was started in the beginning of the 20th century. Presently, grapes are successfully grown in India over an area of 60,000 ha with a production of approximately 1.67 million MT (CitationAnonymous, 2005), primarily for use as fresh fruit.
Grape breeding had mainly relied on selection among naturally occurring spontaneous crosses for ages and to a lesser extent, due to conventional breeding during the last century (CitationAdam-Blondon et al., 2004). The varieties currently available are the results of a selection process by human and ecogeographical conditions (CitationBisson, 1995). Information on genetic diversity among plant species is important for efficient utilization of genetic resources. The existence of close genetic relationships among cultivars grown in the same region or under similar climatic influence could lead to dilution of genetic resources. Hence, studies on grape have been carried out to characterize the commercially important germplasm available in India.
Morphological characterization has been attempted among several grape cultivars for identification purposes. However, superiority of molecular markers over morphological characterization in grape cultivars is well established. The usefulness of molecular testing for grapevine identification is widely accepted (CitationBowers et al., 1996; CitationBowers and Meredith, 1997; CitationThomas et al., 1994). In the grape germplasm, characterization of endangered cultivar (CitationBocccacci et al., 2005), parentage analysis (CitationSefc et al., 1997), identification of the clones (CitationYe et al., 1998), analysis of genetic diversity (CitationMerdinoglu et al., 2000), and molecular mapping (CitationDoligez et al., 2006) is been carried out. Molecular markers like RFLP (CitationBourquín et al., 1993), RAPD (CitationGrando et al., 1995; Jean Jaques et al., 1993; CitationTamhankar et al., 2001; CitationVidal et al., 1999), microsatellite or SSR (CitationBowers et al., 1993, 1999b; CitationPellerone et al., 2001; CitationScott et al., 2000; CitationThomas and Scott, 1993) and AFLP (CitationCervera et al., 2000; CitationLabra et al., 1999; CitationMartinez et al., 2003) are successfully used to characterize grape germplasm. In this study, a combination of morphological and microsatellite amplification with appropriate statistical analysis has enabled us to identify the relationships among 42 genotypes.
Microsatellite technology has been extensively used in grapevine biology and genetics. The number of microsatellite loci available has greatly increased in the last few years largely through the establishment of the International Vitis Microsatellite Consortium, leading to the discovery of more than 350 new loci. Microsatellite markers, being abundant, multiallelic, and highly polymorphic, provide an efficient and accurate means of detecting genetic polymorphism. Most importantly, their codominant nature makes them the markers of choice for population genetic analysis to assess genetic organization in germplasm collections. Microsatellites have been used to determine parent-progeny relationships in grape (CitationBowers and Meredith, 1997; CitationBowers et al., 1999a), to develop a database of DNA profiles for use in cultivar identification (CitationBowers et al., 1996; CitationLamboy and Alpha, 1998), and as markers for mapping genetic linkage (CitationRiaz et al., 2004). The present study evaluates the genetic diversity, structure and differentiation in a grape germplasm collection using polymorphisms revealed by seven microsatellite loci. In addition, we examined the relationships between morphological and molecular characters.
MATERIALS AND METHODS
Plant Materials
Plant material from 42 grape genotypes was collected from Department of Horticulture, University of Agricultural Sciences, Bangalore and Indian Institute of Horticultural Research, Bangalore. Approximately, 50 g of recently matured leaves (15–20 d old) were collected, washed using distilled water, wiped with 70% (v/v) ethanol, then air dried prior to storage in sealed plastic bags at 4°C.
Morphological Data
Morphological characterization of each genotype with respect to their vegetative and reproductive characters was done in triplicate plants according to IPGRI (International Plant Genetic Resources Institute, Rome) descriptors for grape (CitationAnonymous, 1995). The dendrogram was constructed based on the descriptors value for all genotypes using SPSS for windows, version 11.5.0 (CitationAnonymous 2002).
DNA Isolation
DNA was extracted from the stored leaves of grapevine using a cetyl trimethyl ammonium bromide (CTAB) method (CitationSimon et al., 2007). 2 g of leaf sample were powdered in liquid nitrogen to extract the DNA. The powder was mixed with 10 ml extraction buffer, preheated to 65°C, containing 100 mM Tris-HCl, pH 8.0, 20 mM EDTA, 1.4 M NaCl, 3% (w/v) CTAB, 2% polyvinylpyrrolidone and 1% (v/v) ¾β-mercaptoethanol, then incubated at 65°C for 1 h. The mixture was cooled to room temperature, 10 ml cold 24:1 (v/v) chloroform:isoamylalcohol was added, and the contents were mixed well. After centrifugation at 7,500 × g for 12 min at 4°C, the supernatant was transferred to a fresh tube and the chloroform:isoamylalcohol step was repeated until a clear supernatant was obtained. 5 M NaCl was added to the supernatant [0.5 (v/v)] and mixed gently, followed by addition of 1 volume of cold isopropanol to precipitate the DNA. The mixture was incubated overnight at 4°C, then centrifuged at 6,500 × g for 5 min. The resulting pellet was washed with 70% (v/v) ethanol, air-dried, and dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). 2 μg RNase (bovine pancreatic ribonuclease; Bangalore Genei, Bangalore, India) was added to each sample which was incubated for 3 h at 37°C, mixed with an equal volume of phenol, and centrifuged at 7,500 × g for 12 min at room temperature. This step was followed by washing with an equal volume of 1:1 (v/v) phenol:chloroform, then with chloroform alone. The DNA was precipitated overnight at 4°C with 0.5 (v/v) 5 M NaCl and 1 volume of cold isopropanol. The resulting pellet obtained after centrifugation was dissolved in TE buffer, analyzed in an agarose gel and quantified using a spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Microsatellite Analyses
A total of seven SSR primers characterized in previous studies were used. The primers were VVMD 14, VVMD 25, VVMD 27, VVMD 28, VVMD 31, VVMD 36 (CitationBowers et al., 1996 and 1999b), and VMC 7f2 (CitationPellerone et al., 2001). These microsatellites were selected as they were the same core set in the screening programme used to access the target grapevine collections. Primer pairs were synthesized from MWG Biotech, Bangalore, India based on their published gene sequence. PCR was performed in 96-well plates in MJ Research PTC100 thermocyclers (Bio-Rad Laboratories, Bangalore, India). PCR reactions were carried out in 25 μl reactions containing 50 ng of DNA, 5 pmoles of each primer, 10x of Taq polymerase buffer (50 mM KCl, 10 mM Tris-HCl, pH 9.0, 0.05% (v/v) NP40, and 0.05% (v/v) Triton X-100), 1.5 mM of MgCl2, 0.5 mM of dNTPs (Finzymes Pvt. Ltd., India), and 1 U of Taq polymerase (Sigma-Aldrich Pvt. Ltd., India). The final volume was adjusted with sterile distilled water. The PCR amplifications were carried out with respect to the protocols for primer sets published in CitationBowers et al., 1996 and 1999b; CitationPellerone et al., 2001; and CitationThomas and Scott, 1993. Amplification was confirmed with agarose gels, and alleles were separated by running on 6% polyacrilamide denaturing gels and electrophoresed in 1 × TBE at 55 W for 2 h. The amplified products were visualized with silver staining previously described (CitationBowers et al., 1996).
Statistical Analysis
Amplified fragments from each SSR primer set were scored manually for their presence (1) or absence (0). The profiles of 42 accessions of grapevine using 7 primer pairs were assembled for statistical analysis. The sizes of the fragments were estimated using 50 bp standard DNA markers (Bangalore Genei Pvt. Ltd., India), coelectrophoresized with the amplified products. A genetic dissimilarity matrix was developed using Euclidean Distances, which estimates all pairwise differences in the amplification products (CitationSokal and Sneath, 1973). A cluster analysis was based on Ward's method using a minimum variance algorithm (CitationWard, 1963). Principle Component Analysis was used to make a multivariate statistical analysis of the binary data (CitationSokal and Sneath, 1973). Identity 1.0 software (CitationWagner and Sefc, 1999) was used to calculate expected and observed heterozygosity, null allele frequency, and the probability of finding 2 identical genotypes (probability of identity). The expected heterozygosity (H E) was calculated as H E = 1 – Σ pi2 (CitationNei, 1973), where Pi represents the frequency of allele i among the varietal set. The observed heterozygosity (H O) was obtained by direct calculation. The null allele frequency was calculated as r = (H E – H O)/(1 + H E) (CitationBrookfield 1996). The probability of identity was calculated as PI = 1 Σ pi4 + ΣΣ (2pipj)2 (CitationPaetkau et al., 1995), where Pi and Pj represent the frequency of alleles i and j, respectively.
RESULTS AND DISCUSSION
The objective of this study was to estimate the extent of the genetic diversity among Indian grapevine lines using SSR markers. Information on genetic diversity in crop plant species is important for efficient utilization of plant genetic resources. The traditional method of identifying species by morphological characters is now accompanied by DNA profiling that is more reliable on proteins, largely because of several limitations of their morphological data (CitationNayak et al., 2003). Therefore, when investigating organisms with a high tendency for morphological differentiation, studies considering both molecular and morphological characters are highly relevant (CitationBartish et al., 2000).
A constructed dendrogram based on 27 morphological characters clustered the grape genotypes into two major clusters (I and II) at 96 units (). Cluster I consisted of 23 genotypes grouped into two subclusters (A and B) linked at a distance of 33 units. Subcluster A clustered at a distance of 21 units and consisted of 8 genotypes. The seedless genotypes “Thompson Seedless” and “Flame Seedless” were clustered together, but both differed with compact and elongated bunches and pale- and red-colored berries, respectively. The genotypes “Arka Chitra,” “Arka Hans,” “Arka Shweta,” and “Arka Soma” showed seeded berries with greenish yellow color, but varied with their bunch characters. The genotypes “Arka Vathi” and “Arka Neelamani” clustered together showed large and elongated bunches, but contained green- and black-colored berries respectively. Subcluster B consisted of 15 genotypes and clustered into two groups at 23 linkage distances. Group 1 clustered 10 genotypes predominantly characterized by large bunches and dark berry types. The genotype “Angur Kalan” showed golden yellow berries with pinkish tinge, genotype “Sharad Seedless” evidenced a crispy pulp, and genotype E-29/7 had spherical berries. Group II consisted of 5 genotypes clustering at 15 units. All genotypes of the group showed medium to large bunches with dark colored berries. The genotypes E-18/12 and “Arka Trishna” were distinctly characterized by cylindrical-bunch and oval-shaped berries, respectively.
FIGURE 1 Dendrogram showing the clustering patterns of 42 Indian grapevine genotypes based on morphological characters.
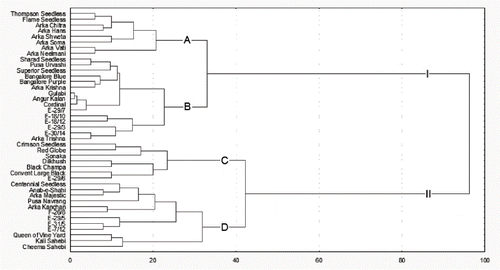
Cluster II consisted of 19 genotypes forming into two subclusters (C and D) at 43 linkage distance in the dendrogram. Subcluster C was comprised of 7 genotypes at 23 distances. All the genotypes under this subcluster showed colored berries and were seeded, except the genotypes “Sonaka” and “Dilkhush,” which had a pale greenish color, and genotypes “Crimson Seedless” and E-29/6 with a seedless nature. The genotypes “Crimson Seedless” and “Red Globe” clustered together, respectively showing light-red and wine-red colored berries. Genotypes “Black Champa” and “Convent Large Black” both had spherical berries. The subcluster D with 12 genotypes was clustered at 32 units. The genotypes were predominantly characterized by greenish golden yellow color and seeded berries with the exception of genotypes “Arka Majestic,” “Pusa Navrang,” and “Arka Kanchan” which evidenced a dark color. The remaining genotypes “Centennial Seedless,” “Arka Kanchan,” and E-29/5 produced seeded berries. The genotypes “Pusa Navrang” and “Kali Sahebi” were characterized by berries with reddish pulp, while genotypes “Queen of Vine Yards,” “Kali Sahebi,” and “Cheema Sahebi” were clustered together and characterized by obovate-shaped berries.
The genetic variability of this germplasm () was evaluated on the basis of the number of alleles (mean: 6.4), gene diversity (GD: 0.71), observed heterozigosity (Ho: 0.849), and probability of coincidence (PC: 0.13). These data indicated the presence of a lower genetic variability in the Indian grapevine germplasm, comparable to the variability found in the Algeria and Mediterranean basin and similar to Spanish grape germplasm (CitationMartin et al., 2003). The most informative locus was VVMD 28 (13 alleles per locus) and the least informative one was VVMD-14 (4) (). The most informative primers VVMD 28 are VVMD 32 were good candidates to be used for paternity testing due to the high direct count heterozygosity, high number of alleles, and even distribution of allelic frequencies. Moreover, in the populations studied, the observed heterozygosity was very similar to the expected heterozygosity at each nuclear SSR locus, suggesting no excess of homozygosity in populations. Since the analysis did not display null alleles hence, the marker should be suitable for a genetic population study among wild relatives and parentage studies (CitationWagner et al., 2006). Such an absence of null alleles is probably due to the choice of nuclear SSR loci which have revealed very low deviation in the observed heterozygosity from the expected heterozygosity (CitationBandelj et al., 2004; CitationKhadari et al., 2003). Hence, a choice criterion should be used to avoid loci displaying null alleles being included.
TABLE 1 Descriptive Statistics and Genetic Diversity for Microsatellite Loci at Each of the Seven Sites Among the Indian Grapevine Genotypes
Pairwise comparisons were made between all genotypes included in this study and the average dissimilarity values were calculated based on microsatellite data. A maximum dissimilarity of 110 units was obtained between the genotypes “Convent Large Black” and “Arka Hans” where the former genotype was characterized by spherical bluish black seeded berries and the later genotype with colorless seeded berries; while a minimum dissimilarity of 37 units were obtained between the genotypes “Anab-e-Shahi” and “Dilkhush,” where both the genotypes were characterized by pale-greenish colored seeded berries.
The dendrogram presented demonstrates clearly the ability of microsatellites to detect a large amount of genetic variation in genetically closely related genotypes of grapevine and to identify groups with different levels of genetic distance. The markers segregated the genotypes into two major clusters (I and II) at 476 linkage distance (). Major cluster I consisted of 30 genotypes grouped into two subclusters (A and B) at 223 linkage distance. Subcluster A with 17 genotypes was grouped into two groups (A1 and A2) with 12 and 5 genotypes, respectively. The genotypes “Anab-e-Shahi” and “Dilkhush” were closely linked in group A1 by 31 linkage distance with both the genotypes showing greenish seeded berries. The genotypes “Pusa Urvasi” and “Cordinal” were closely linked at 39 units and grouped with “Pusa Navrang” at 59 units where “Cordinal” and “Pusa Navrang” were producing pigmented berries. The genotypes “Centenal Seedless” and “Superior Seedless” were linked together at 35 units, both with colorless berries. These genotypes were grouped with “Shrad Seedless” (pigmented berries) at 52 units and linked with cultivars “Sonaka” and “Gulabi” at 73 units. The genotypes “Thompson Seedless” and “Flame Seedless” were grouped together at 45 units and differentiated with colorless and red berries, respectively. The five genotypes of group A2 were linked at 71 units, of which “Black Champa” and “Red Globe” were closely linked at 41 units with both pigmented and seeded berries. The “Convert Large Black” (pigmented and seeded berries) was grouped with “Queen of Vineyards” (golden yellow colored and seeded berries) at 56 units. The genotype “Crimson Seedless” (red colored berries) stood separate and linked to the group at 71 units.
FIGURE 2 Dendrogram showing the clustering patterns of 42 Indian grapevine genotypes based on microsatellite markers.
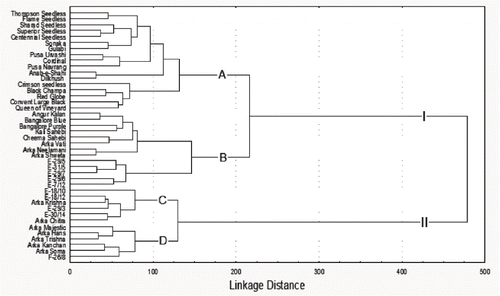
The subcluster B consisted of 13 genotypes segregated into two groups (B1 and B2) at 145 units in the dendrogram. The group B1 with eight genotypes was clustered at 81 units, of which “Arka Neelamani” and “Arka Shweta” were closely linked at 31 units—both were seedless. The large bunches with colored genotypes, namely, “Angur Kalan” (pinkish) and “Bangalore Blue” (purple) were linked at 33 units. “Bangalore Purple” (purple) and “Kali Sahebi” (reddish pulp) were linked at 63 units. The genotypes “Cheema Sahebi” and “Arkavathi” both with pyramidal shaped and tightly packed bunches were clustered together at 45 units. The group B2 consisted of five genotypes linked at 65 units and divided into two minor clusters. The first minor cluster consists of three colorless and seedless genotypes E-29/5, E-31/5, and E-29/7, which are linked at 55 units. The second minor cluster with two genotypes, E-29/6 and E-7/12, is linked at 52 units and both genotypes showed medium-sized bunches and seedless berries. The characteristic feature of cluster I predominantly shows subcluster A with more seeded genotypes and subcluster B with seedless genotypes.
Cluster II consisted of 2 subclusters, C and D, linked at 129 linkage distance with six genotypes in each group. The pigmented genotypes E-18/12 and “Arka Krishna” are closely linked at subcluster C at 40 linkage distance and are linked to E-29/3 at 45 units. The genotypes E-30/14 and “Arka Chitra” are grouped together at 39 units and shared attractive golden-yellow berries. The genotype E-18/10 was clustered separately in the subcluster, characterised by small bunches and pigmented seeded berries. Subcluster D was segregated into two groups (D1 and D2) at 77 linkage distance. Group D1 consisted of three seeded genotypes, “Arka Majestic,” “Arka Hans,” and “Arka Trishna” with seeded fruits and linked at 51 units. The colorless and seeded genotypes “Arka Kanchan” and “Arka Soma” were clustered together in group D2 at 39 units and with E-26/8 at 58 units. The characteristic feature of cluster II predominantly revealed subcluster C with colored and seedless fruits, and subcluster D with colorless and seeded fruits.
In summary, this study, using microsatellite markers on Indian grape vine genotypes, showed considerable genetic diversity existing among the population. This is most likely due to different conditions under which the populations are grown and conserved (CitationSong et al., 2003). The clustering of the genotypes was predominantly based on the pigment and seed characters in fruits. These groupings can be used in selecting diverse parents in breeding improved cultivars and in maintaining variation in the germplasm. Evaluation of genetic diversity among germplasm, particularly of crops, is crucial in utilizing genetic potential to improve traits needed for adaptation to various stress conditions (CitationAmer et al., 2001). To our knowledge, this study is the first attempt in using molecular markers for assessing genetic diversity in Indian grape vine germplasm, and the outcome of this work could be useful for future characterization and exploitation of that germplasm.
Understanding the spatial organization of genetic diversity within the plant populations is of critical importance for the development of strategies designed to preserve genetic variation (CitationBrown and Briggs, 1991; CitationHamrick et al., 1991). It has been shown that species with limited gene flow (i.e., with restricted seed and pollen movement) have considerably more among-population variation for total amount of genetic diversity (CitationSchoen and Brown 1991). Since the ex situ collection cannot exceed a limited number of accessions, it is difficult to preserve the evolutionary potential of the species hence, the future of this fruit depends on the selection of high quality cultivars (CitationTous and Ferguson, 1996). Thus, conservation strategies among grape vine cultivars should be developed with the morphological characters in mind. Taken together, the microsatellites markers can be used for establishing relationships between related species, but their efficiency depends on the amount of variability obtained within the cultivars.
The authors are grateful to the University of Agricultural Sciences, Bangalore and Indian Institute of Horticulture research, Bangalore for providing the facilities.
LITERATURE CITED
- Adam-Blondon , A.F. , Roux , C. , Claux , D. , Butterlin , G. , Merdinoglu , D. and This , P. 2004 . Mapping 245 SSR markers on the . Vitis vinifera genome: A tool for grape genetics. Theor. Appl. Genet. , 109 : 1017 – 1027 .
- Amer , I.M.B. , Borner , A. and Roder , M.S. 2001 . Detection of genetic diversity in Libyan wheat genotypes using wheat microsatellite markers . Genet. Resour. Crop Ev. , 48 : 579 – 585 .
- Anonymous . 1995 . IPGRI, Descriptors for grapevine (Vitis vinifera L.) , Rome : International Plant Genetic Resources Institute .
- Anonymous . 2002 . SPSS for Windows, version 11.5.0 , Chicago : SPSS .
- Anonymous. 2005. Statistical Databases—Agricultural Data http://faostat.fao.org/site/567/default.aspx#ancor
- Bandelj , D. , Jakse , J. and Javornik , B. 2004 . Assessment of genetic variability of olive varieties by microsatellite and AFLP markers . Euphytica , 136 : 93 – 102 .
- Bartish , I.V. , Rumpunen , K. and Nybom , H. 2000 . Combined analyses of RAPDs, cpDNA and morphology demonstrate spontaneous hybridization in the plant genus Chaenomeles . Heredity , 85 : 383 – 392 .
- Bisson , J. 1995 . The principal ecogeographical groups in French grapevines assortment . J. Int. des Sci. de la Vigne et du Vin , 29 : 63 – 68 .
- Bocccacci , P. , Marinoni , D.T. , Gombino , G. , Botta , R. and Schneider , A. 2005 . Genetic characterization of endangered grape cultivars of Reggio Emilia province . Am. J. Enol. Vitic. , 56 : 411 – 416 .
- Bourquín , J.C. , Otten , L. and Walter , B. 1993 . Restriction fragment length polymorphism and molecular taxonomy in Vitis vinifera. Theor . Appl. Genet. , 87 : 157 – 162 .
- Bowers , J.C. and Meredith , C.P. 1997 . The parentage of a classic wine grape, Cabernet Sauvignon . Natl. Genet. , 16 ( 1 ) : 84 – 87 .
- Bowers , J.E. , Bandman , E.B. and Meredith , C.P. 1993 . DNA fingerprint characterization of some California winegrape cultivars . Amer. J. Enol. Viticult. , 44 : 266 – 274 .
- Bowers , J.E. , Boursiquot , J.M. , This , P. , Chu , K. , Johansson , H. and Meredith , C.P. 1999a . Historical genetics: The parentage of Chardonnay, Gamay, and other wine grapes of northeastern France . Science , 285 : 1562 – 1565 .
- Bowers , J.E. , Dangl , G.S. and Meredith , C.P. 1999b . Development and characterization of additional microsatellite DNA markers for grape . Amer. J. Enol. Viticult. , 50 : 243 – 246 .
- Bowers , J.E. , Dangl , G.S. , Vignani , R. and Meredith , C.P. 1996 . Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.) . Genome , 39 : 628 – 633 .
- Brookfield , J.F.Y. 1996 . A simple new method for estimating null allele frequency from heterozygote deficiency . Mol. Ecol. , 5 : 453 – 455 .
- Brown , A.H. and Briggs , J.D. 1991 . Sampling strategies for genetic variation in ex situ collections of endangered plant species , Edited by: Falk , D.A. and Holsinger , K.F. 99 – 119 . New York : Genetics and conservation of rare plants. Oxford University Press .
- Cervera , M.T. , Cabezas , J.A. , Sanchez-Escribano , E. , Cenis , J.L. and Martinez-Zapter , J.M. 2000 . Characterization of genetic variation within table grape varieties based on AFLP markers . Vitis , 39 : 109 – 114 .
- Doligez , A. , Adam-Blondon , A.F. , Cipriani , G. , Di Gaspero , G. , Laucou , V. , Merdinoglu , D. , Meredith , C.P. , Riaz , S. , Roux , C. and This , P. 2006 . An integrated SSR map of grapevine based on five mapping populations . Theor. Appl. Genet. , 113 : 369 – 382 .
- Galet , P. 1990 . Cepages et Vignobles de France. Tome. II. L'Ampelographie Francaise , 2nd , Montpellier, , France : C. Déhan .
- Galet , P. 2000 . Dictionnaire encyclopédique des cépages , Paris : Hachette .
- Grando , M. , De Micheli , L. , Biosetto , L. and Scienza , A. 1995 . RAPD markers in wild and cultivated Vitis vinifera . Vitis , 34 : 37 – 39 .
- Hamrick , J.L. , Godt , M.J. , Murawski , D.A. and Loveless , M.D. 1991 . “ Correlations between species traits and allozyme diversity: Implications for conservation biology. pp. 75–86 ” . In Genetics and conservation of rare plants , Edited by: Falk , D.A. and Holsinger , K.F. New York : Oxford University Press .
- Jean-Jaques , I. , Defontaine , A. and Hallet , J. 1993 . Characterization of Vitis vinifera L. cultivars by random amplified polymorphic DNA markers . Vitis , 32 : 189 – 190 .
- Khadari , B. , Breton , C. , Moutier , N. , Roger , J.P. , Besnard , G. and Berville , A. 2003 . The use of molecular markers for germplasm management in a French olive collection . Theor. Appl. Genet. , 106 : 521 – 529 .
- Labra , M. , Failla , O. , Fossati , T. , Castiglione , S. , Scienza , A. and Sala , F. 1999 . Phylogenetic analysis of grapevine cv. Ansonica growing on the island of Gighio, Italy by AFLP and SSR markers . Vitis , 38 : 161 – 166 .
- Lamboy , W.F. and Alpha , C.G. 1998 . Using simple sequence repeats (SSRs) for DNA fingerprinting germplasm accessions of grape (Vitis L.) species . J. Am. Soc. Hortic. Sci. , 123 ( 2 ) : 182 – 188 .
- Martin , J.P. , Borrego , J. , Cabello , F. and Ortiz , J.M. 2003 . Characterization of Spanish grapevine cultivar diversity using sequence-tagged microsatellite site markers . Genome , 46 : 10 – 18 .
- Martinez , L. , Carvagnaco , P. , Masuelli , R. and Rodriguez , J. 2003 . Evaluation of diversity among Argentine grapevine (Vitis vinifera L.) varieties using morphological data and AFLP markers . Electron. J. Biotechnol. , 6 ( 3 ) : 241 – 250 .
- Maul, E. and R. Eibach. 2003. Vitis international variety catalogue http://www. genres.de/eccdb/vitis/
- Merdinoglu , D. , Butterlin , G. , Baur , C. and Balthazard , J. 2000 . Comparison of RAPD, AFLP and SSR (microsatellite) markers for genetic diversity analysis in Vitis vinifera L . Acta Hortic. (ISHS) , 528 : 195 – 200 .
- Nayak , S. , Rout , G.R. and Das , P. 2003 . Evaluation of the genetic variability in bamboo using RAPD markers . Plant Soil Environ. , 49 : 24 – 28 .
- Nei , M. 1973 . Analysis of gene diversity in subdivided populations . Am. Nat. , 106 : 238 – 292 .
- Olmo , H. P. 1976 . “ Grapes. pp. 294–298 ” . In Evolution of crop plants , Edited by: Simmonds , N.W. London : Longman .
- Paetkau , D. , Calvert , W. , Stirling , I. and Strobeck , C. 1995 . Microsatellite analysis of population structure in Canadian polar bears . Mol. Ecol. , 4 : 347 – 354 .
- Pellerone , F.I. , Edwards , K.J. and Thomas , M.R. 2001 . Grapevine microsatellite repeat: Isolation, characterization and use of genotyping grape germplasm from Southern Italy . Vitis , 40 : 79 – 186 .
- Riaz , S. , Dangl , G.S. , Edwards , K.J. and Meredith , C.P. 2004 . A microsatellite marker based framework linkage map of Vitis vinifera L . Theor. Appl. Genet. , 108 : 864 – 872 .
- Schoen , D.J. and Brown , A.H.D. 1991 . Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants . P. Natl. Acad. Sci. USA , 88 : 4494 – 4497 .
- Scott , K.D. , Eggler , P. , Seaton , G. , Rossetto , M. , Ablett , E.M. , Lee , L.S. and Henry , R.J. 2000 . Analysis of SSRs derived from grape ESTs . Theor. Appl. Genet. , 100 : 723 – 726 .
- Sefc , K.M. , Steinkellner , H. , Wagner , H.W. , Glossl , J. and Regner , F. 1997 . Application of micro satellite markers to parentage studies in grapevine . Vitis. , 36 : 179 – 183 .
- Simon , L. , Shyamalamma , S. and Narayanaswamy , P. 2007 . Morphological and molecular analysis of genetic diversity in jackfruit . J. Hortic. Sci. Biotech. , 82 ( 5 ) : 764 – 768 .
- Sokal , R. R. and Sneath , P. H. A. 1973 . Principles of Numerical Taxonomy , San Francisco : W.H. Freeman .
- Song , Z.P. , Xu , X. , Wang , B. , Chen , J.K. and Lu , B.R. 2003 . Genetic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theor . Appl. Genet. , 107 : 1492 – 1499 .
- Tamhankar , S.A. , Patil , S.G. and Rao , V.S. 2001 . Assessment of the genetic diversity of some important grape genotypes in India using RAPD markers . Vitis , 40 : 157 – 161 .
- Thomas , M.R. and Scott , N.S. 1993 . Microsatellite repeats in grapevine reveal DNA polymorphisms when analyzed as sequence-tagged sites (STSs) . Theor. Appl. Genet. , 86 : 985 – 990 .
- Thomas , M.R. , Cain , P. and Scott , N.S. 1994 . DNA typing of grapevines: A universal methodology and database for describing cultivars and evaluating genetic relatedness . Plant Mol. Biol. , 25 ( 6 ) : 939 – 949 .
- Tous , J. and Ferguson , L. 1996 . “ Mediterranean fruits. pp. 416–430 ” . In Progress in New Crops , Edited by: Janick , J. Arlington, VA : ASHS Press .
- Vidal , J.R. , Moreno , S , Gogorcena , Y. , Masa , A. and Ortiz , J.M. 1999 . On the genetic relationships and origins of six grape cultivars of Galicia (Spain) using RAPD markers . J. Am. Soc. Hortic. Sci. , 50 : 69 – 75 .
- Wagner , A. P. , Creel , S. and Kalinowski , S.T. 2006 . Estimating relatedness and relationships using microsatellite loci with null alleles . Heredity , 97 : 336 – 345 .
- Wagner , H.W. and Sefc , K.M. 1999 . IDENTITY 1.0. Centre for applied genetics , Vienna : University of Agricultural Science .
- Ward , J.H. 1963 . Hierarchic grouping to optimize an objective function . J. Am. Stat. Assn. , 58 : 236 – 239 .
- Ye , G.N. , Soylemezogln , G. , Weeden , N.F. , Lamboy , W.F. , Pool , R.M. and Reisch , B.I. 1998 . Analysis of the relationship between grapevine cultivars, species and clones via DNA fingerprinting . Vitis , 37 : 33 – 38 .