Abstract
Proximally growing individuals of wild, lowbush blueberry (Vaccinium angustifolium Ait.) vary widely in yield despite being grown under conditions in which environmental heterogeneity is minimized by cultivation practices. We recently established that the relative self-fertility of the bearing plant is a significant predictor of its outcross yield. Further, although the species has historically been characterized as largely self-infertile, we, and others, have documented large variation in this trait among individuals within fields, and, thus, relative self-fertility stands as a partial explanation of yield variation in addition to other genetic factors, such as significant general and specific combining abilities. Here, we extend our scope by experimentally addressing whether pollen neighborhood affects yield. Lowbush blueberry is not sown, rather individuals have colonized fields by natural processes. Commercial fields of lowbush are generally pollinated by rented honey bees, which tend to pollinate nearby flowers. Our hypothesis is that clones may have become situated in differentially suitable pollen environments, at least partly explaining yield differences. We identified two high and two low natural yielders from each of two managed fields and collected pollen mixes from five donors surrounding each (n = 8). Under field conditions, each recipient received four pollination treatments including its' own neighborhood and the other three donor neighborhoods within the same field. Results showed that one of the low producers in each field had significantly higher yields when pollinated by any but their own neighborhood, thus substantiating our hypothesis that pollen neighborhood has an effect on yield.
INTRODUCTION
Lowbush blueberry fields, in distinction to the cultivated fields of highbush and rabbiteye blueberries, are wild. Individual clones (genotypes) arose from natural distributional processes (animal dispersal) over long periods of time (CitationBell et al., 2009; CitationHall et al., 1979). Although a decade of breeding by traditional methods in the 1970s provided concrete evidence that genetic improvement and production of high yielding clones is possible (CitationAalders et al., 1975; CitationHall, 1983), to date, no extensive deployment or sowing of these superior cultivars has been undertaken within the cultivated regions of northeastern America. The two most probable reasons are: (1) the desire to preserve the marketing value of the ‘wild’ image, and (2) the time and difficulty in establishing new plants (CitationYarborough, 2010). We also have a lack of knowledge regarding the genetics of yield in this complex autotetraploid (2n = 4x = 48) species (CitationHokanson and Hancock, 1993). Only during the last decade, with the development of highly reproducible molecular markers in blueberry, have genomic approaches, such as genetic fingerprinting, relationship studies, marker assisted selection, and mapping in highbush blueberry (CitationRowland et al., 2003a, Citation2003b), and characterization of spatial genetic structure in wild fields of lowbush blueberry (CitationBell et al., 2008; CitationBell, 2009; CitationBell et al., 2009; CitationBurgher et al., 2002) become real possibilities. The awareness of field genetic structure in lowbush blueberry is highly germane since the species is susceptible to inbreeding depression and carries a high genetic load (CitationBell et al., 2010; CitationHokanson and Hancock, 2000). The crop is pollinated by rented honey bees in a likely localized pattern, as inferred from bee foraging patterns (CitationAras et al., 1996; CitationDrummond 2002), and thus inbreeding depression models have been invoked to explain yield differences among clones (CitationMyra et al., 2004).
The astounding 4-fold increase in productivity of lowbush blueberry, achieved during the last two decades by the incorporation of intensive cultivation practices, such as selective herbicides, ICM (Integrated Crop Management), and the addition of pollinators, has likely reached its upper limit (CitationYarborough, 2009, Citation2004). The research described here is a continuation of a cooperative project between the University of Maine in Orono, and the Genetic Improvement of Fruits and Vegetables Lab at the USDA-ARS in Beltsville, Maryland, which began about 4 years ago. Our goal has been to develop and leverage new genomic tools in order to better understand the genetic basis of yield variation in this complex, highly polymorphic species. Yield can vary among nearly adjacent clones by as much as 15-fold (CitationBell, 2009). Understanding these yield differences would provide growers with additional resources from which to make the challenging management decisions they are facing today. Molecular approaches could be utilized to help select high yielders and develop seed families from these selections (CitationJamieson, 2008a, Citation2008b) for deployment into fields, while maintaining the high degree of genetic diversity now present and likely integral in this system.
During the last 2 years, we have gained important insights into the causes of yield variation by using a combination of hand crosses in several designs (pairwise touching neighbor design and a diallel) with molecularly genotyped experimental clones. Using the pairwise touching neighbor design, we found no evidence that crossing genetically similar individuals (half to full sib levels) necessarily affects yield, as had been hypothesized under the supposition of an inbreeding depression model (CitationBell et al., 2010; CitationMyra et al., 2004). Additionally, we found positive spatial genetic structure among fields, but not within fields. We also showed a significantly higher incidence of self-fertility than had been reported by CitationAalders and Hall (1961), with rates more in agreement with CitationWood (1968) Furthermore, we found that self-fertility was a significant predictor of general outcross yield using a least-squares regression model. Finally, using a diallel factorial design, we found that GCA (general combining ability), SCA (specific combining ability), and ‘narrow-sense’ heritability were significant at the whole model level for the measured post-pollination traits, fruit set, mean berry weight per pollination, and mean large, fertile seed per pollination. Also, various combinations of clones showed both positive and negative specific combining abilities indicating that dominance interactions (SCA), in addition to additive genetic actions (GCA), play roles in determining yield.
Our current research is focused on expanding the single source pairwise crosses described above to address the question of whether pollen neighborhoods (defined as the average genetic similarity of five surrounding likely pollen donors to the focal pollen recipient) and/or other possible spatial genetic structure (SGS) patterns affect yield. To accomplish this goal, the experiment was divided into two components: (1) field based hand pollinations utilizing a 4 × 4 reciprocal (mixed pollen) design to determine if yield changes in response to changing mixed pollen sources from different neighborhoods, and (2) genotyping all individuals involved, determining the average number of marker differences between the five surrounding male donors and the focal recipients, and testing whether these average genetic similarities correlate with yield changes. This study is concurrently serving as an extension for detecting SGS at a more intensive sampling scale than previously done (CitationBell et al., 2009) using population genetic tools, such as AMOVA (analysis of molecular variance) and spatial correlograms.
MATERIALS AND METHODS
Selection of Clones, Pollen Collection, and Experimental Design
From previous work spanning two growing seasons, two high and two low producing clones (as measured by percent fruit set) were selected from two managed fields. One was the Blueberry Hill Farm in Jonesboro, Maine, which is an extension research facility of the University of Maine, and the other was Columbia (Jasper Wyman & Son), which is a commercially managed field near Cherryfield, Maine. These focal clones were relocated in 2009 using previously collected (2005) GPS coordinates. Also, the five touching clones surrounding each focal clone (likely pollen donors) were identified and marked. Thus, within each field, we established four experimental neighborhoods of six plants each (one central focal pollen recipient, and five surrounding pollen donors) for a total of eight neighborhoods (n = 48 individual clones). Pollen was collected from stems cut at midday (a time of intense pollinator activity) from each of the five surrounding donors in each of the eight neighborhoods. These stems were brought to the lab and left to bench dry for one day. Freshly opened flowers were selected with forceps. Pollen was then gathered by sonication with a tuning fork onto a single glass petri dish for each neighborhood. We attempted to gather equal amounts of pollen from each of the five donor clones by collecting from enough flowers to make five equal sized piles of pollen in a small circle on the dish. The five samples were brought together and mixed by vibrating the dish so that all pollen was combined in the center of the dish. Pollen was applied to stigmas using a small brush.
Within each neighborhood, four treatments were established by marking five stems for each of the four pollen treatment mixes. Each focal recipient received pollen mixes from its natural neighborhood and also from each of the other three neighborhoods within the same field. Pollen was not moved between fields since it was judged unlikely that cross field pollinations occur at these distances (∼12.5 km); thus, each field comprised a 4 × 4 factorial treatment design. Terminal clusters were used, and approximately five flowers per stem were hand pollinated within each treatment group during bloom. In August, berries from pollinated stems were collected, counted, and weighed. For this experiment, we chose to measure yield from a single, derived index, proportion fruit set times the mean weight of berries collected for that cross, which simply is the mean berry weight per pollination. As a single index of yield, this measurement is highly informative to growers since it takes into account not only fruit set (numbers of berries harvested per pollination), but also berry size (CitationBell et al., 2010).
DNA Extractions, PCR, and Analysis of EST-PCR Markers
Leaf tissue was collected from each clone and frozen. DNA extractions, as well as Polymerase Chain Reaction (PCR) conditions using primers designed from ESTs of highbush blueberry, followed previously established protocols (CitationBell et al., 2008, Citation2009; CitationRowland et al., 2003a, Citation2003b). In distinction to previous years, however, all Expressed Sequence Tag-Polymerase Chain Reaction (EST-PCR) products were separated and analyzed using the high throughput Qiagen QIAxcel automated capillary gel electrophoresis system (Valencia, CA, USA) with the QIAxcel DNA screening cartridge. A total of 8 EST primer pairs, previously used on other lowbush blueberry genotypes, were chosen for their robust amplification characteristics and high polymorphic information content. From these primers, 37 total bands were scored yielding an average of 4.6 bands per primer. Bands were scored as dominant markers (present or absent) and analyzed both with NTSYS v. 2.20j (Setauket, NY, USA; CitationRohlf, 1998) for clustering and ‘goodness of fit’ estimates, and with GenAlEx v. 6.3 (Peakall, Canberra, ACT, Australia; CitationPeakall and Smouse, 2006) for calculating the average number of band differences between the five donors and the focal recipient in each of the eight neighborhoods.
Statistical Analysis
Two principal statistical tests were conducted both using JMP, Version 7 (CitationSAS, 1989–2007). Least squares regression analysis was performed that tested for significant differences in mean berry weight per pollination between each of the four pollen neighborhoods within each field separately. Specifically, yield from each focal recipient's natural environment was compared to the other three in that same field. Since both yield and mean band differences were normally distributed and observations were independent, a second linear regression was calculated testing if a significant relationship existed between the mean berry weight per pollination and the mean band differences of the composite five donors comprising each pollen neighborhood (a measure of genetic dissimilarity) from their focal pollen recipients (bearing plant).
RESULTS AND DISCUSSION
Physical Map of the Four Neighborhoods within Each Field
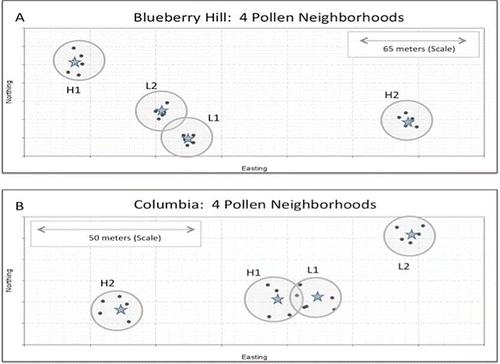
A physical map in UTM (Universal Transmercator) units depicting the relative locations of the four neighborhoods in each field is shown in . Four focal pollen recipients, two that had low and two that had high relative open pollination yield potential (fruit set) averaged over both 2005 and 2007 were chosen from 20 monitored clones in each field. Five touching clones surrounding each focal recipient comprised the pollen neighborhoods.
FIGURE 1 Physical map of Blueberry Hill (top) and Columbia (bottom), showing the locations of the eight pollen neighborhoods and the precise locations of the five clones comprising each neighborhood and the central focal recipients. Neighborhood areas, as measured by the area internal to a linear perimeter connecting the five donors, ranged from 30.2–110.7 m2 with a mean area of 69.7 m2. Scale insets are also shown. The designations H1, H2, L1, and L2 indicate relative natural yield potential under open pollination conditions (fruit set)—H for high, L for low. Blueberry Hill and Columbia are approximately 12.5 km apart (color figure available online).
Neighborhood Yield Results from the 4 × 4 Reciprocal Crossing Experiment as Compared to Mean Band Differences of Donors to Pollen Recipient
shows the results of the 4 × 4 reciprocal crossing experiment along with the mean band differences of the donors to the focal pollen recipient. Diagonals (grayed cells) represent ‘self’ crosses, i.e., the recipient received a pollen mix from its own natural environment. Linear regressions, testing for significant within row differences for each environment, showed that one of the low producers in each field had significantly higher yield when pollinated by any of the other three neighborhoods. In other words, its lowest yield was that produced from its own natural pollen neighborhood. For example, at Blueberry Hill, yield from ♀L2 increased from 0.03 g for mean berry weight to 0.23, 0.17, and 0.26 g, respectively, when pollinated by pollen mixes from each of the other three neighborhoods.
TABLE 1 4 × 4 Grids of Pollen Recipient (Rows) and Mixed Sire (Columns) Neighborhoods Showing Yield Results as Mean Berry Weight per Pollination (Top), and Mean Band Differences between the Five Surrounding Donors and the Focal Pollen Recipient (Bottom) for Both Blueberry Hill and Columbia [H1, H2, L1, and L2 Designate the Two High and Two Low Producing Clones (Recipients; n = 8) as Measured by Open Pollination Fruit Set for 2005 and 2007]
Columbia revealed a similar pattern although at a higher statistical significance (P > 0.05). Yield from L2 increased from 0.14 g for mean berry weight to 0.45, 0.32, and 0.45, respectively, when pollinated by the other three neighborhoods. On the other hand, it is interesting to note the relatively consistent level of the yield values across columns for the other three focal recipients in both fields, remained fairly static under different pollen neighborhood regimes. CitationBell et al. (2010) recently demonstrated the proportional relationship of self-fertility in predicting general outcross yield in lowbush blueberry. This phenomenon has been also observed in V. corymbosum and V. myrtilloides (CitationHokanson and Hancock, 2000). Here, we have shown that mixed sire sourcing (neighborhood) matters as well as self-fertility and have, thus, significantly implicated a composite donor effect. This evidence is supportive of the hypothesis that neighborhood or happenstance of colonization context can affect yield, too.
The field crosses by themselves, however, do not shed any light on the mechanism responsible for our findings. Mean band differences (or genetic dissimilarity) did not correlate with differences in yield as in our previous study, although we understand that analysis of more markers (only 37 markers were used here) is probably needed to render any strong conclusions on this point. Here we considered the ‘average’ genetic dissimilarity of the surrounding donors to the focal recipient, in distinction to our earlier work where we had only considered single source touching donors. Here also, we chose to measure genetic distance differently, based on the ideas of CitationPeakal and Smouse (2006), which basically tallies pairwise band differences between genotypes as a measure of genetic dissimilarity. These data are shown in the second 4 × 4 grid in . Regression analyses found no significant relationship between yield and mean band differences of males to the focal recipient within neighborhoods at both Blueberry Hill (F (1,14) = 0.014; P = 0.91) and Columbia (F (1,14) = 0.569; P = 0.46), respectively. Thus, as reported by CitationBell et al. (2010), even with an expansion of the experimental design to include neighborhoods of donors, no significant relationship between yield and genetic similarities was found. By way of a caveat, however, although we believe we applied equitable amounts of each of the five donor pollen sources in homogeneous mixes, we could not guarantee that all pollen genotypes performed equally within the style and, thus, equally obtained access to ovules. Differences in pollen viability could also be a factor. However, we previously estimated pollen viability percentages from 56 clones and found a range from 61–99% viability. Furthermore, the distribution was heavily skewed with 75% of the clones falling above 78% viability. Thus, there were probably no major differences in pollen viability or amounts of pollen from the donor plants. Variable gametophytic competition in the style could be present, but testing for such kinds of competition was not undertaken in this field experiment.
Interestingly, we did not observe a high yielding focal recipient to be negatively affected by the pollen neighborhood. Although our small sample size may have precluded us from documenting a yield reduction of this kind, we alternatively could speculate from these results and our previous work (CitationBell et al., 2010) that there is something in the genetic nature of higher yielders, such as lower individual genetic load and concomitant superior female fertility that inhibits such downturns in yield regardless of pollen source or neighborhood effects as here defined. The higher yielders do have a tendency to be better selfers than the low yielders.
CONCLUSIONS
The molecular data presented here is preliminary, based on only 37 bands generated from eight EST-PCR primer pairs. Although the cophenetic ‘goodness’ of fit as generated in NTSYS v. 2.20j was quite good (r = 0.82), we plan to score additional markers. We also are planning to attempt to estimate heterozygosity using SSR markers that have been mapped in highbush blueberry. Further, we plan to expand the ‘average’ relatedness concept to include the possibility that SGS, as derived from AMOVA analyses from these neighborhoods, may also be involved. In addition, we are performing an analysis of a set of 95 different contiguous genotypes (sampling every single genotype within a given area) collected within a 0.35 hectare area, to evaluate SGS on a more local scale. We have also germinated open pollinated seedlings from the focal recipients in this study to construct a probabilistic model of gene flow (paternity analysis). Overall, we believe that our synthesis of experimental approaches involving field hand crosses, quantitative genetics, and newly developed genomic approaches have allowed us, and will continue to allow us, to develop a more complete knowledge of the biology and genetics of this wild but cultivated species.
ACKNOWLEDGMENTS
We wish to acknowledge all the student workers, high school interns, and employees for all of their help in conducting both field and laboratory experiments, together with the lowbush blueberry industry (Wild Blueberry Commission of Maine) for providing matching funds in support of this project.
LITERATURE CITED
- Aalders , L.E. and Hall , I.V. 1961 . Pollen incompatibility and fruit set in lowbush blueberries . Can. J. Genet. Cytol. , 3 : 300 – 307 .
- Aalders , L.E. , Ismail , A.A. , Hall , I.V. and Hepler , P.R. 1975 . Augusta lowbush blueberry . Can. J. Plant. Sci. , 55 : 1079
- Aras , P. , de Oliveira , D. and Savoie , L. 1996 . Effect of a honeybee (Hymenoptera: Apidae) gradient on the pollination and yield of lowbush blueberry . J. Econ. Entomol. , 89 : 1080 – 1083 .
- Bell , D.J. 2009 . Spatial and genetic factors influencing yield in lowbush blueberry (Vaccinium angustifolium Ait.) in Maine , Orono , Maine : Ph.D. Dissertation, School of Biological Sciences, University of Maine .
- Bell , D.J. , Rowland , L.J. , Polashock , J.J. and Drummond , F.A. 2008 . Suitability of EST-PCR markers developed in highbush blueberry for genetic fingerprinting and relationship studies in lowbush blueberry and related species . J. Amer. Soc. Hort. Sci. , 133 : 701 – 707 .
- Bell , D.J. , Rowland , L.J. , Zhang , D. and Drummond , F.A. 2009 . The spatial genetic structure of lowbush blueberry, Vaccinium angustifolium, in four fields in Maine . Botany , 87 : 932 – 946 .
- Bell , D.J. , Rowland , L.J. , Stommel , J. and Drummond , F.A. 2010 . Yield variation among clones of lowbush blueberry as a function of genetic similarity and self-compatibility . J. Amer. Soc. Hort. Sci. , 135 : 259 – 270 .
- Burgher , K.L. , Jamieson , A.R. and Lu , X. 2002 . Genetic relationships among lowbush blueberry genotypes as determined by randomly amplified polymorphic DNA analysis . J. Amer. Soc. Hort. Sci. , 127 : 98 – 103 .
- Drummond , F.A. 2002 . Honeybees and blueberry pollination , The University of Maine Cooperative Extension Wild Blueberry Bull. No. 629 .
- Hall , I.V. 1983 . Genetic improvement of the lowbush blueberry, Vaccinium angustifolium . Can. J. Plant. Sci. , 63 : 1091 – 1092 .
- Hall , I.V. , Aalders , L.E. , Nickerson , N.L. and VanderKloet , S.P. 1979 . The biological flora of Canada. 1. Vaccinium angustifolium, sweet lowbush blueberry . Can. Field-Naturalist , 9 : 415 – 430 .
- Hokanson , K. and Hancock , J. 1993 . The common lowbush blueberry, Vaccinium angustifolium Ait., may be an autopolyploid . Can J. Plant Sci. , 73 : 889 – 891 .
- Hokanson , K. and Hancock , J. 2000 . Early-acting inbreeding depression in three species Vaccinium (Ericaceae) . Sex. Plant Reprod. , 13 : 145 – 150 .
- Jamieson , A.R. 2008a . Developing seed-propagated lowbush blueberry families . HortScience , 43 : 1686 – 1689 .
- Jamieson , A.R. 2008b . ‘Novablue’, a seed-propagated lowbush blueberry family . HortScience , 43 : 1902 – 1903 .
- Myra , M. , MacKenzie , K. and Vander Kloet , S.P. 2004 . Investigation of a possible sexual function specialization in the lowbush blueberry (Vaccinium angustifolium Aiton. Ericaceae) . Small Fruits Rev. , 3 : 313 – 324 .
- Peakall , R. and Smouse , P.E. 2006 . GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research . Mol. Ecol. Notes , 6 : 288 – 295 .
- Rohlf , F.J. 1998 . NTSYS-PC: Numerical taxonomy and multivariate analysis system , Setauket , N.Y : Exeter Software . Release 2.20j
- Rowland , L.J. , Dhanaraj , A.L. , Polashock , J.J. and Arora , R. 2003a . Utility of blueberry-derived EST-PCR primers in related Ericaceae species . HortScience , 38 : 1428 – 1432 .
- Rowland , L.J. , Mehra , S. , Dhanaraj , A. , Ogden , E.L. , Slovin , J.P. and Ehlenfeldt , M.K. 2003b . Development of EST-PCR markers for DNA fingerprinting and genetic relationship studies in blueberry (Vaccinium, section Cyanococcus) . J. Amer. Soc. Hort. Sci. , 128 : 682 – 690 .
- SAS . 1989–2007 . JMP , Cary , NC : SAS Institute Inc . Version 7
- Wood , G. W. 1968 . Self fertility in the lowbush blueberry . Can. J. Plant Sci. , 48 : 431 – 434 .
- Yarborough , D. 2004 . Factors contributing to the increase in productivity in the wild blueberry industry . Small Fruits Rev. , 3 : 33 – 43 .
- Yarborough , D. 2009 . Wild blueberry fact sheet: Wild blueberry culture in Maine , Univ. Maine Coop. Ext. Fact Sheet No. 220 .
- Yarborough , D. 2012 . Establishment and management of the cultivated lowbush blueberry (Vaccinium angustifolium) . Int. J. Fruit Sci. , 12 : 14 – 22 .