Abstract
Macroautophagy is a major intracellular degradation process recognized as playing a central role in cell survival and longevity. This multistep process is extensively regulated at several levels, including post-translationally through the action of conserved longevity factors such as the nutrient sensor TOR. More recently, transcriptional regulation of autophagy genes has emerged as an important mechanism for ensuring the somatic maintenance and homeostasis necessary for a long life span. Autophagy is increased in many long-lived model organisms and contributes significantly to their longevity. In turn, conserved transcription factors, particularly the helix-loop-helix transcription factor TFEB and the forkhead transcription factor FOXO, control the expression of many autophagy-related genes and are important for life-span extension. In this review, we discuss recent progress in understanding the contribution of these transcription factors to macroautophagy regulation in the context of aging. We also review current research on epigenetic changes, such as histone modification by the deacetylase SIRT1, that influence autophagy-related gene expression and additionally affect aging. Understanding the molecular regulation of macroautophagy in relation to aging may offer new avenues for the treatment of age-related diseases.
Abbreviations:
- Atg, autophagy related
- LC3, microtubule-associated protein 1 light chain 3
- acetyl-CoA, acetyl coenzyme A
- AMPK, AMP-activated protein kinase
- BNIP3, BCL2/adenovirus E1B 19kDa interacting protein 3
- CaN, calcineurin; HDAC, histone deacetylase
- HAT, histone acetyltransferase
- MITF, microphthalmia-associated transcription factor
- PDPK1/2, 3-phosphoinositide dependent kinase 1/2
- PtdIns3K, phosphatidylinositol 3-kinase
- PtdIns3P, phosphatidylinositol 3-phosphate
- TFEB, transcription factor EB
- TOR, target of rapamycin
- TSC, tuberous sclerosis complex
- UVRAG, UV radiation resistance associated.
Introduction
The autophagy process
Autophagy is an evolutionarily conserved catabolic process through which damaged organelles and macromolecules are degraded and recycled within the cell. Three forms of autophagy can be distinguished based on the cellular components that are sequestered and degraded: microautophagy, the nonselective sequestration of cytoplasmic components directly into the lysosome; chaperone-mediated autophagy, the selective degradation of specific cargo proteins recognized and delivered to the lysosome by a chaperone complex; and macroautophagy, the degradation of cytoplasmic material via encapsulation in an autophagosome that subsequently fuses with the lysosome. This review focuses on macroautophagy (henceforth referred to as autophagy) since this is the best studied mechanism, including in the context of aging.
The process of autophagy proceeds through at least 5 mechanistically distinct steps: (1) induction, (2) double-membrane nucleation and autophagosome formation, (3) autophagosome elongation and sequestration of cellular debris, (4) autophagosome–lysosome fusion, and (5) degradation of sequestered components in the lysosome () (reviewed in ref. Citation1). Each of these sequential steps involves a number of conserved autophagy-related (ATG) proteins, as briefly described here. Activation of the ULK/Atg1 initiation complex is the first step in the autophagy process, permitting the creation of a phagophore membrane and formation of the autophagosome. The membrane used to form the phagophore can originate from different locations, such as the endoplasmic reticulum, mitochondria, Golgi, endosomes, and plasma membrane (reviewed in refs. Citation2, Citation3). Integration of membrane lipids into the phagophore requires the synthesis of phosphatidylinositol 3-phosphate (PtdIns3P) by the phosphatidylinositol 3-kinase (PtdIns3K) nucleation complex. PtdIns3P is then recognized by PtdIns3P-binding proteins,Citation4,5 which may possibly act as a shuttle by transferring membrane lipids between a membrane donor and the growing phagophore.Citation6 Phagophore elongation is further dependent on 2 ubiquitin-like conjugation reactions. First, the ubiquitin-like protein ATG12 is covalently conjugated to ATG5 by ATG7 and ATG10 (E1- and E2-like enzymes, respectively). The ATG12–ATG5 conjugate, together with its interaction partner ATG16L1 is thought to act in part as a an E3-like ligase to promote conjugation of phosphatidylethanolamine (PE) to soluble LC3-I/Atg8, formed by cleavage of the ubiquitin-like protein LC3/Atg8 by the protease ATG4.Citation7 The resulting PE-conjugated, membrane-bound LC3-II/Atg8–PE is important for phagophore elongation as well as cargo recognition. LC3-II/Atg8–PE can be bound by various cargo receptors; for example, SQSTM1/p62, which recognizes ubiquitinated proteins or organelles targeted for degradation.Citation8
Figure 1. Regulation of autophagy-related gene expression associated with longevity. The process of autophagy and the transcriptional regulation of autophagy genes by the action of specific transcription factors, epigenetic modifications, and miRNAs have emerged as important conserved mechanisms to ensure longevity. (A) The autophagy process is initiated by cytoplasmic nucleation of the phagophore with double-membrane structures. The phagophore then elongates and sequesters cellular components until it matures into an autophagosome, which then docks and fuses with lysosomes to form an autolysosome. Upon fusion, the cargo and inner membrane of the autolysosome are degraded. (B) The nutrient sensor MTORC1 is a major regulator of the autophagy process. MTORC1 inhibits autophagy and positively regulates growth, mRNA translation, and ribosomal and lipid biogenesis. The nutritional status of the cell (e.g., amino acid levels) dictates the recruitment of MTORC1 to the lysosomal membrane and its subsequent activation. Active MTORC1 phosphorylates several targets, including the transcription factors TFEB and FOXO. Phosphorylation of TFEB, and possibly of FOXO, occurs at the lysosomal membrane and leads to their retention in the cytosol. (C) The nuclear translocation of transcription factors TFEB and FOXO, as well as activation of FOXA induces the expression of multiple autophagy-related and lysosomal genes (for FOXA, which is constitutively nuclear; only the C. elegans ortholog PHA-4 has so far been shown to regulate autophagy gene transcription and modulate longevity). TFEB and FOXO are regulators of longevity in several species. (D) Autophagy gene expression can also be regulated epigenetically by histone modifications such as acetylation (Ac) and methylation (Me), which affect the chromatin state thus altering expression of specific genes. For example, the deacetylation of histone mark H4K16 by SIRT1 leads to transcriptional repression of various autophagy-related genes (the acetylation reactions occurs by the acetyltransferase KAT8). High levels of acetyl-CoA can inhibit the expression of autophagy gene ATG7, and conversely, histone hypoacetylation via spermidine treatment can increase gene expression of ATG11, ATG7, and ATG15. The dimethylation of H3K9 through the methyltransferase EHMT2 leads to the repression of autophagy gene transcription. The deacetylase SIRT1, spermidine treatment and levels of nucleocytosolic acetyl-CoA are associated with the regulation of longevity in several species. Mammalian gene names are used exclusively in the figure. See text for reference to specific model systems and for further details.
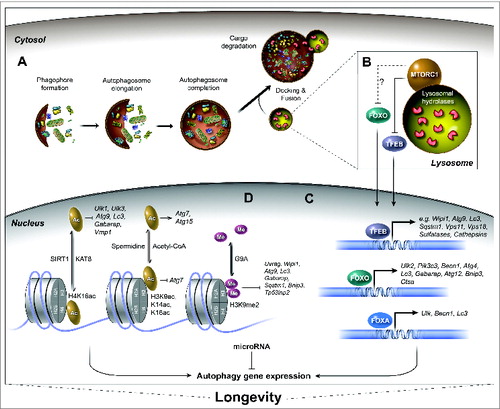
The phagophore undergoes further maturation to fully encapsulate its cargo; the completed autophagosome releases outer membrane-associated autophagy-related proteins, after which it is ready to dock and fuse to the lysosome/vacuole to form an autolysosome. Upon fusion, the inner membrane of the autophagosome and the lumenal cargo are degraded, and the autolysosome reforms as a lysosome that is possibly again available for subsequent vesicular fusion events.Citation9 Incomplete processing of autolysosomes can instead produce a residual body containing indigestible material.Citation10 Notably, several age-related diseases, such as neurodegenerative disorders, are characterized by the accumulation of unprocessed autophagic vacuoles (reviewed in refs. Citation11, Citation12), suggesting that the ability of cells to efficiently coordinate and complete the autophagic process is gradually impaired with age.
Upstream regulators of macroautophagy
Autophagy is critical to cellular homeostasis under both normal and stressed conditions. Basal levels of autophagy ensure intracellular quality control, whereas autophagy induced by starvation and other stressors promotes cellular survival by maintaining adequate amino acid pools and cellular energy levels. Until recently, induction of autophagy was thought to be dependent primarily on post-translational regulation of nutrient-responsive pathways; however, it is now clear that alternative means of regulation exist. The emerging picture is that rapid induction of autophagy, e.g., in response to starvation, is mediated by post-translational protein modifications and protein-protein interactions, whereas transcriptional mechanisms are necessary for a sustained response (reviewed in ref. Citation47).
One key regulator of autophagy is the kinase MTOR/TOR (mammalian target of rapamycin), which is a component of 2 complexes, MTORC1 and MTORC2. As part of the MTORC1 complex, TOR regulates cell growth, proliferation, survival, protein synthesis, and autophagy, whereas the MTORC2 complex primarily regulates different downstream signaling cascades modulating cell shape and metabolism, but can also regulate autophagy indirectly (reviewed in ref. Citation13). The MTORC1 complex (hereafter referred to as MTOR) directly inhibits autophagy through phosphorylation and inactivation of ULK/Atg1 and ATG13/Atg13, which are essential for the induction of autophagy (reviewed in ref. Citation14). TOR activity can be modulated by changes in amino acid abundance via interaction with RAG small GTPases.Citation15 These enzymes are localized at the lysosomal surface through their interaction with the pentameric Ragulator complex.Citation16 Elevated lysosomal amino acid levels, which may reflect the overall cellular abundance of amino acids, promote the guanine-nucleotide exchange factor function of the Ragulator complexCitation17 and subsequent activation of RAG GTPases. These proteins recruit TOR to the lysosomal membrane where it is ultimately activated by the small GTPase RHEB.Citation18
MTOR serves as a hub to integrate additional upstream signals from various sources, including INS/insulin, growth factors, and cellular energy levels (reviewed in refs. Citation13, Citation19). One notable pathway intersecting with MTOR is the INS-IGF1 pathway, which is involved in many functions necessary for metabolism, growth, and longevity.Citation20 Formation of PtdIns(3,4,5)P3 by activated PtdIns3K, a downstream effector of the INS-IGF1 pathway, recruits the protein kinase AKT to the plasma membrane, where it is phosphorylated and activated by the PDPK1/2 (3-phosphoinositide dependent protein kinase 1/2). AKT modulates the function of TSC2, a component of the heterodimeric tuberous sclerosis complex (TSC). The TSC1/2 complex can inhibit MTOR signaling by preventing activation of RHEB.Citation21 Upon growth factor or INS-IGF1 signaling, TSC2 is phosphorylated and inactivated by AKT, thereby allowing RHEB to activate MTOR.Citation22 The TSC-RHEB-MTOR cascade therefore represents a major signaling axis for the control of growth and autophagy.
Autophagy is also regulated by intracellular energy levels via the energy sensor AMPK (AMP-activated protein kinase), which directly activates the ULK/Atg1 initiation complex (reviewed in ref. Citation23). AMPK activity is sensitive to AMP levels and is further regulated by phosphorylation by upstream kinases such as STK11/LKB1 and CAMKK2/CamKKβ (calcium/calmodulin-dependent protein kinase kinase 2, β).Citation24 Although STK11/LKB1 is constitutively active, CAMKK2 has been implicated in AMPK regulation and autophagy induction only in response to changes in intracellular Ca2+ levels.Citation25 AMPK can also influence autophagy by inhibiting MTOR, either by phosphorylating the MTORC1 subunit RPTOR/raptor or by inhibiting phosphorylation of TSC1/2.Citation26 Both AMPK and MTOR can themselves be phosphorylated by ULK/Atg1, providing an additional level of regulatory feedback to modulate and fine-tune autophagy (reviewed in ref. Citation27)
In addition to regulating autophagy, AMPK and MTOR also modulate organismal aging in a conserved fashion. Specifically, inhibition of MTOR extends the life span of organisms ranging from yeast to mice,Citation28 and overexpression of AMPK promotes longevity in worms and flies.Citation24 Collectively, these findings demonstrate that nutrient sensors are major regulators of life span in a variety of species and share autophagy as a downstream effector mechanism for the maintenance of health.
Direct links between autophagy and aging
Accumulating evidence over the past decade supports a direct role for autophagy in the aging process. Multiple genetic experiments have demonstrated a requirement for autophagy-related genes in many longevity paradigms, including inhibition of TOR activity in yeast (ATG1, ATG11, ATG7),Citation29 worms (unc-51/Ulk/ATG1, bec-1/Becn1/VPS30/ATG6, vps-34, atg-18/Wipi1/2)Citation30,31 and flies (Atg5).Citation32 Similarly, life-span extension by dietary restriction is abrogated in autophagy-deficient yeast (ATG7, ATG5, ATG8, ATG15, and v-SNARE genes VAM3, and VAM7)Citation33,34 and in worms (unc-51/Ulk/ATG1, bec-1/Becn1/VPS30/ATG6, vps-34, atg-7).Citation30,31,35 Similar links have been observed in other conserved longevity models (i.e., reduced INS-IGF1 signaling, germline removal; mitochondrial respiration, mRNA translation; as well as resveratrol and spermidine supplementation), which all cause induction of autophagy markers, and in each case, the resulting life-span extension is dependent on at least one autophagy gene (see for a summary of direct links between autophagy-related genes and longevity). In support of such a link, overexpression of specific autophagy genes has been found to promote longevity in several different species. In mice, heterologous overexpression of ATG5 is sufficient to stimulate autophagy, promote a youthful appearance, and extend life span.Citation36 In Drosophila, overexpression of Atg8a in the neurons and muscle of adult flies extends their life span.Citation37,38 Similarly, neuron-specific overexpression of Atg1 in adult Drosophila induces autophagy both cell autonomously and non-cell autonomously, and also results in life-span extension.Citation39 Consistent with the physiological relevance of these observations, many autophagy genes (i.e., Atg1, Atg6, Atg7, Atg5, Atg8) show reduced expression with age in flies,Citation37,40 and LC3 and ATG7 protein levels in muscle also decline with age in mice and humans.Citation41 This culminates in the loss of autophagic capacity generally observed with normal aging (reviewed in ref. Citation42).
Table 1. Effects of modulation of autophagy-related and lysosomal genes on life span including in conserved longevity paradigms.
Notably, genetic and age-related loss of adequate autophagic and lysosomal function has been linked to the development of several metabolic and neurodegenerative diseases (reviewed in ref. Citation11). For example, loss-of-function mutations in several genes with autophagy-related functions (e.g., Becn1/VPS30/ATG6,Citation43 Atg7Citation44, Atg5Citation45) result in decreased autophagy and increased accumulation of disordered and aggregated proteins in neurodegenerative disorders such as Huntington disease (HTT/huntingtin), Alzheimer disease (Aβ and MAPT/tau), and Parkinson disease (SNCA/α-synuclein) (reviewed in ref. Citation46).
Accumulating evidence thus supports a beneficial role for autophagy in aging, although the underlying mechanisms of autophagy regulation in long-lived organisms are not fully understood. In this review, we focus on the role of transcriptional and epigenetic regulation of autophagy in the context of aging by highlighting studies in longevity models, and noting relevance to age-related diseases where applicable.
Transcriptional Regulation of Autophagy Relevant to Aging
Transcriptional mechanisms are emerging to play an important role in the regulation of autophagy. Specifically, several transcription factors are now known to regulate the sustained expression of specific autophagy-related or lysosomal genes (reviewed in refs. Citation47–48, see also refs. 49-50); however, only a few have been carefully examined to determine their conserved roles in inducing autophagy to promote longevity. Nevertheless, the helix-loop-helix transcription factor TFEB and the forkhead transcription factors FOXO and FOXA have been shown to be key transcriptional regulators of autophagy and lysosomal biogenesis. These factors have also been associated with the transcriptional induction of beneficial autophagy found in long-lived organisms. Below, we review recent research on the role of these transcription factors in autophagy regulation and aging.
TFEB and MITF/microphthalmia-associated transcription factors
Beyond its role in phosphorylating autophagy-related proteins, MTOR kinase phosphorylates several transcription factors with roles in autophagy, thereby preventing their translocation to the nucleus.Citation51 The most prominent example of an autophagy-related MTOR-regulated transcription factor is TFEB, a member of the MITF (microphthalmia-associated transcription factor) family.Citation52-57 TFEB was first described as a key transcription factor in lysosomal biogenesis,Citation58 and subsequent studies revealed additional roles in the regulation of genes involved in autophagosome formation (e.g., Atg9, Wipi1/2/Atg18, Lc3/Atg8), cargo recognition (e.g., Sqstm1), vacuolar fusion (e.g., Vps11, Vps18), vacuolar proton pumping (e.g., V-ATPase subunits), and lysosomal degradation (e.g., sulfatases and cathepsins).Citation52 Thus, TFEB regulates autophagic flux by coordinating the expression of genes with functions at all stages of the autophagy process, from vesicle initiation to cargo degradation.
Under nutrient-rich conditions, MTOR phosphorylates TFEB at the lysosomal surface, which results in the binding of YWHA/14–3–3 proteins and the retention of TFEB in the cytosol.Citation56,57 The TFEB sequence contains several predicted MTOR and MAPK1/ERK2 phosphorylation sites, and mutation of serine 142 or serine 211 to nonphosphorylatable residues has been reported to induce nuclear localization of TFEB.Citation55–57 Additionally, TFEB nuclear localization is induced by the TOR inhibitors rapamycin and Torin.Citation55,56 Similarly, TFEB translocates to the nucleus in response to nutrient deprivation, causing increased lysosomal calcium release and activation of the phosphatase calcineurin (CaN) to directly dephosphorylate TFEB.Citation169 Interestingly, another member of the MITF family, TFE3, can bind to similar promoter sequences found on autophagy-related target genes of TFEB (so-called CLEAR sites), displays conserved regulation by TOR, and has overlapping functions in lysosomal biogenesis and autophagy gene regulation under nutrient-scarce conditions. Moreover, overexpression of TFE3 promotes lysosomal biogenesis and autophagy. These findings suggest the potential nuclear coordination of MITF transcription factors in controlling autophagy.Citation59 Nevertheless, TFEB and TFE3 must have distinct functions, as deletion of Tfeb in mice results in embryonic lethality, whereas deletion of Tfe3 has no apparent phenotype.Citation60,61 Recent work has shown that TFEB is recruited to the lysosome for TOR phosphorylation by interaction with FLCN/folliculin,Citation62,63 suggesting a complex regulatory network governs the intracellular localization of TFEB, and thus its activity (). Taken together, recent studies demonstrate an important function for lysosomes, beyond their role in degradation, as sites for the integration of signaling by the nutrient sensor TOR.
The TFEB homolog in C. elegans, HLH-30, plays a role similar to that of TFEB in the induction of orthologous target genes and activation of autophagy.Citation64 Indeed, HLH-30 localization to the nucleus is induced by RNA interference (RNAi)-mediated inhibition of let-363/Tor,Citation64 nutrient deprivation,Citation65 and by 4 additional conserved longevity paradigms (reduced insulin signaling, mRNA translation, and mitochondrial respiration, and removal of the germline) that induce autophagy markers and require autophagy genes for life-span extension.Citation64 The induction of autophagy gene expression by either let-363/Tor inhibition or germline removal requires hlh-30, and hlh-30 is indispensable for the life-span extension observed in all of these 6 autophagy-dependent C. elegans longevity models.Citation64,66 These observations are consistent with HLH-30 transactivation causing an increase in autophagic flux, which is necessary for life-span extension in these longevity paradigms. In support of this, overexpression of HLH-30/TFEB is sufficient to activate autophagy and to moderately extend C. elegans life span.Citation64 HLH-30/TFEB is negatively regulated by another helix-loop-helix transcription factor, MXL-3, which provides precise temporal control of HLH-30/TFEB activity to maintain homeostasis.Citation67 In mammals, the transcription factor ZNF24/ZSCAN3 works in opposition to TFEB by acting as a repressor of TFEB target genes.Citation68 Nuclear TFEB levels are elevated in liver cells of long-lived, dietary-restricted mice,Citation64 indicating that TFEB is a component of a conserved longevity mechanism.
Other targets of HLH-30/TFEB suggested to play a role in life-span extension () include the lysosomal acid lipases lipl-1 and lipl-3,Citation67 supporting previous observations suggesting that increased lipolysisCitation69 via lipophagy by the homologous lysosomal acid lipase lipl-4,Citation70 may represent a central life-span-extending mechanism. Of note, TFEB has been linked to changes in lipid metabolism induced by starvation.Citation65,67 Lipophagy-mediated remodeling and utilization of lipids for energy generation and signaling via lipophagic products may represent possible mechanisms by which cellular homeostasis, organelle biogenesis, and survival are ensured during nutrient deprivation. Indeed, elevation of TFEB activity by overexpression or by starvation leads to an increase in PPARA/PPARα and PPARGC1A/PGC1α expression,Citation65,71 suggesting an enhanced cellular ability to respond to lipid signals. Thus, TFEB-mediated transcriptional induction of autophagy may be central for increasing autophagic flux in order to provide a dynamic pool of metabolites, particularly lipids, for synthetic and signaling pathways conducive to longevity. The conserved regulation of TFEB activity by MTOR supports the notion that nutrient signaling is pivotal for the control of autophagic flux. Thus, TFEB stimulation of autophagy-related and lysosomal gene expression coordinates each step of the process to provide a vital source of metabolites during periods of nutrient deprivation.
Recent work suggests an emerging role for TFEB, beyond aging and metabolism, in the pathology of diseases. For example, overexpression of TFEB has beneficial effects on cellular clearance in models of lysosomal storage diseases, Parkinson disease, and α1-antitrypsin deficiency.Citation52,72,73 In addition, heterologous overexpression of TFEB in mouse brain increases the clearance of aberrant MAPT/tau protein, a key player in Alzheimer disease, in part by improving lysosomal function.Citation74,74 Benefits of TFEB induction in the turnover of disease-related proteins such as HTT/huntingtin also involves interactions with PPARGC1A,Citation75 a transcriptional cofactor involved in mitochondrial biogenesis and recently linked to longevity in Drosophila.Citation76 Conversely, in X-linked spinal and bulbar muscle atrophy, TFEB transactivation is inhibited by interaction with polyglutamine-expanded ARs/androgen receptors, with a subsequent impairment of autophagy.Citation77 This recent finding raises the possibility that TFEB activity may be impaired by the age-related accumulation and aggregation of disordered proteins in a variety of neurodegenerative diseases. Interestingly, TFEB is also required for the response to infection in both worms and mice, suggesting a broad and conserved role for this transcription factor in survival associated with a variety of stressors.Citation78 Taken together, these studies highlight a critical role of TFEB in promoting autophagy under physiological and pathological conditions, including the increasingly prevalent age-related diseases.
Forkhead transcription factors
Another major family of transcriptional regulators of autophagy with a conserved role in aging is the forkhead transcription factors (FOXO), which play well-recognized and central roles in cellular homeostasis through the regulation of genes involved in lipid and glucose metabolism and in mitochondrial function.Citation79 A role for the FOXO family in autophagy was first described in murine models of muscle atrophy, an age-related conditionCitation80,81 to which several degradative pathways contribute, especially autophagy.Citation82 In the muscle, FOXO1 and FOXO3 elevate the autophagic flux by increasing the expression of autophagy genes mainly working as part of the core machinery (Ulk2/ATG1, Pik3c3/VPS34, Becn1/VPS30/ATG6, Atg4B/ATG4, Lc3b/ATG8, GabarapL1/ATG8, Atg12/ATG12, Bnip3)Citation80,81,83,84 and additionally increase protein degradation via the proteasomal pathway.Citation80,85,171 In particular, FOXO3 increases the capacity of the lysosome to degrade incoming cargo, indicating a role for lysosomal function in muscle atrophy. Other FOXOs (FOXO1, FOXO4, and FOXO6) also play roles in proteostasis and autophagy (reviewed in refs. Citation40, Citation86, Citation87). Interestingly, AMPK directly activates FOXO family members in mammalian cells to promote protein breakdown and the production of alternative energy sources, even in the presence of INS/insulin and normal AKT activation.Citation88-90 AMPK also activates FOXO in wormsCitation88 and possibly in flies,Citation39 and overexpression of AMPK in flies is sufficient to induce autophagy gene expression (Ulk/ATG1, Lc3a/ATG8, Lc3b/ATG8).Citation39 Moreover, human FOXO3 and AMPK coordinately enhance autophagy during ultra-endurance running, suggesting that transcriptional upregulation of autophagy genes may also contribute to induced autophagic flux in exercised skeletal muscles.Citation91 Thus, several nutrient-signaling pathways converge to modulate FOXO for activation of autophagy and metabolism.
Reduced INS-IGF1 signaling, which increases the activity of FOXO, extends longevity in C. elegans, Drosophila, and mice, and is observed in human centenarians.Citation20 In C. elegans, life-span extension through reduced INS signaling is mediated by DAF-16/FOXO, which regulates the transcription of numerous genes, including those with relevance to proteostasis.Citation92,93 Similar to HLH-30/TFEB, FOXOs can enhance autophagic flux in C. elegans, at least in part by increasing the expression of autophagy-related genes. For instance, C08H9.1, a lysosomal serine carboxypeptidase similar to CTSA/cathepsin A, is induced by DAF-16/FOXO in long-lived daf-2/Igf1r (insulin-like growth factor 1 receptor) mutants and is linked to thermal stress resistance in these animals.Citation94 Consistent with these observations, animals overexpressing DAF-16/FOXO have an increased abundance of autophagosomes and enhanced resistance to bacterial infections.Citation95 Nevertheless, autophagosome numbers are not affected by the loss of daf-16/FoxO in daf-2/Igf1r mutants,Citation70 suggesting a more complex and/or late-acting role for DAF-16/FOXO in autophagy regulation.Citation70 Another Forkhead transcription factor, PHA-4/FOXA, stimulates autophagic flux in C. elegans by binding to the promoter region of several early-acting genes involved in autophagosome formation, including unc-51/Ulk/ATG1, bec-1/Becn1/VPS30/ATG6, and lgg-1/Lc3/ATG8.Citation96,97 PHA-4/FOXA-mediated induction of these genes may stimulate the longevity-inducing autophagy observed in germline-less, dietary-restricted, and let-363/Tor-inhibited animals, which all require pha-4/FoxA for their survival.Citation31,70,98,99 let-363/Tor is known to regulate DAF-16/FOXO in C. elegansCitation70,100 and may also function as an upstream regulator of PHA-4/FOXA in longevity models.Citation70,99,100 Current studies are aimed at understanding the complexity of signaling between FOXO and MTORCitation101 and its impact on autophagy, metabolism, and aging.
The role of FOXO in intracellular proteostasis is well established in C. elegansCitation20 and is conserved in Drosophila muscle.Citation40 Studies in these organisms have also highlighted the importance of global, tissue-specific, and cell-specific functions of FOXO in modulating aging and age-related diseases.Citation20,102 Long-lived C. elegans display increased proteasomal degradationCitation103 that likely acts in concert with elevated autophagy to facilitate the maintenance of proteostasis.Citation104,105 Interestingly, autophagy and proteasomal activity increase concomitantly in embryonic and hematopoietic stem cells.Citation106,107 This is in line with a report that somatic cells acquire stem cell-like properties in long-lived daf-2/Igf1r mutants.Citation108 These findings suggest that FOXO plays a central role in aging by activating longevity-related gene expression and by maintaining a proteostatic status conducive to cell survival and longevity.
Epigenetic Regulation of Autophagy Relevant to Aging
In addition to transcriptional regulation, sustained expression of autophagy genes can be regulated by several epigenetic mechanisms, such as chromatin modulation, histone modification, and microRNAs (miRNAs). The histone deacetylase SIRT1 influences aging and age-related disorders, at least in part, via effects on autophagy; however, the extent to which other epigenetic mechanisms of autophagy regulation contribute to aging and age-related diseases remains unclear. Here, we review some of the epigenetic factors that have been implicated in the regulation of autophagy gene expression in the context of aging.
Chromatin-modifying enzymes
Chromatin is defined as the complex of condensed DNA molecules wound around histones and associated proteins. Histones are alkaline proteins that act as spools to support the compact packaging of DNA into nucleosomes. Posttranslational modification of histones, including acetylation, methylation, phosphorylation, SUMOylation, ubiquitination, and ADP-ribosylation, occur mainly in the N-terminal tails and have profound effects on chromatin structure and gene expression. Of these modifications, several histone acetylation and methylation marks have been linked to autophagy regulation (reviewed in ref. Citation109) (). Histone-modifying enzymes and ATP-dependent chromatin-remodeling complexes can reorganize chromatin structure, and both types of modifiers can affect longevity. For instance, ATP-dependent remodeling complexes have been implicated in longevity in yeast (Isw2)Citation110 and in worms (SWI/SNF Switch/Sucrose Non-fermentable), which colocalizes with DAF-16/FOXO at the promoters of longevity-promoting genes).Citation111
Among the histone-modifying enzymes, the NAD-dependent deacetylase SIRT1 (sirtuin 1) is a particularly well-known modulator of aging. Specifically, the life spans of yeast, worms, and flies can be extended by overexpression and/or pharmacological activation of SIRT1 (by small molecule activators and resveratrol) (reviewed in refs. Citation112,113) and the life span of mice is extended by ubiquitous overexpression of another sirtuin, SIRT6, or brain-specific overexpression of SIRT1 (reviewed in ref. Citation114). Notably, in C. elegans, the life span-extending effect of the SIRT1 activator resveratrol requires the expression of bec-1/Becn1/VPS30/ATG6,Citation31,115,116 suggesting that autophagy is necessary for this longevity paradigm. Several lines of evidence support this notion. SIRT1 regulates autophagy gene expression through histone deacetylation, with lysine 16 on histone H4 (H4K16) as the primary deacetylation target (reviewed in ref. Citation117). H4K16 deacetylation inhibits the transcription of genes involved in the early and late steps of autophagy in multiple cell types, including yeast and mouse embryonic fibroblasts (Ulk1/ATG1, Ulk3/ATG1, Atg9A/ATG9, Lc3/ATG8, GabarapL2/ATG8, and vacuolar membrane protein Vmp1), resulting in decreased autophagic flux measured by LC3/Atg8 conversion and turnover.Citation48 An antagonist to the histone deacetylation activity of SIRT1 is the histone acetyltransferase (HAT) KAT8/MOF/MYST1/YBF2. Induction of autophagy by starvation or rapamycin treatment is accompanied by deacetylation of H4K16 and downregulation of KAT8. Therefore, the acetylation status of H4K16 represents a molecular switch that can regulate autophagy.Citation118
While the activity of SIRT1 in the nucleus limits autophagy, SIRT1-mediated deacetylation of cytoplasmic proteins is required for autophagy induction, as shown in enucleated cells subjected to starvation or rapamycin treatment.Citation119 This may be due to the direct interaction of SIRT1 with autophagy proteins, since SIRT1 co-immunoprecipitates with ATG5, ATG7, and LC3/Atg8, and directly binds and deacetylates these proteins in vitro.Citation120 Moreover, SIRT1 indirectly regulates autophagy by deacetylation of FOXO3, leading to increased expression of autophagy-related genes, including Bnip3, which are critical for autophagy induction.Citation121,122 Thus, although it is clear that SIRT1 regulates autophagy by both epigenetic and posttranslational mechanisms, the relative contribution of each mechanism to SIRT1-mediated longevity remains to be fully established.
SIRT1 has protective effects in several neurodegenerative disease models, which may in part be due to its role in autophagy induction. For example, in cellular models of Parkinson disease, resveratrol increases the degradation of SNCA/α-synuclein via autophagy induction,Citation123 and SIRT1 overexpression can protect against prion-peptide toxicity in cultured neurons.Citation124 Similarly, SIR-2.1/SIRT1 suppresses the formation of SNCA inclusions in worms.Citation125 In yeast, expression of human SNCA is involved in determining chronological life span, which is accompanied by an increase in autophagy via Sir2/SIRT1.Citation126 Collectively, the evidence to date suggest that SIRT1 is a regulator of life span and autophagy, and plays a role in age-related diseases.
In both yeast and mammals, autophagy can be regulated by acetyl coenzyme A (acetyl-CoA), which serves as the donor for acetylation reactions. In yeast, nucleocytosolic acetyl-CoA is produced through the action of the synthetase Acs2, and inhibition of ACS2 enhances autophagy during chronological aging.Citation127 On the one hand, reduced acetyl-CoA levels have conserved beneficial effects, as illustrated by the extended life span of Drosophila in which AcCoAS (acetyl-CoA synthetase) is specifically reduced in the brain.Citation127 On the other hand, accumulation of nucleocytosolic acetyl-CoA represses autophagy in yeast via hyperacetylation of histone 3 (on K9, K14, and K18) and repression of ATG7 transcription.Citation127 In mice and cultured human cells, inhibition of the enzymes required to maintain cytosolic acetyl-CoA levels leads to induction of autophagy, and conversely, high cytosolic acetyl-CoA levels inhibit the ability of starvation to induce autophagy.Citation128 In human cell lines, the HAT EP300 is required for the repression of autophagy under these conditions. In contrast to yeast, in human cell lines acetyl-CoA influences autophagy through cytosolic effects that are independent of transcriptional changes of autophagy genes.Citation128 Taken together, these findings point to the emergence of nucleocytoplasmic acetyl-CoA as a novel modulator of autophagy and life span; future experiments should clarify its impact on age-related diseases.
Histone acetylation and the activity of HATs and HDACs are also affected by the action of cationic polyamines, such as spermine and spermidine. The levels of specific polyamines decrease with age in several tissues of many organisms and the external addition of spermidine has beneficial affects on life span and aging in model organisms including yeast, worms, flies, mice, and human cells.Citation129,130 These effects are likely to involve, at least in part, changes in autophagy since spermidine extends life span in a manner dependent on autophagy genes in both C. elegans and Drosophila (bec-1/Becn1/VPS30/ATG6 and Atg7, respectively). Consistent with this notion, spermidine treatment stimulates autophagy in S. cerevisiae, C. elegans, Drosophila, and cultured mammalian cells, as detected by LC3 lipidation and increased formation of LC3-positive puncta.Citation129,131 Spermidine treatment leads to hypoacetylation of histone 3 (K9, K14, K18) due to the inhibition of histone acetyltransferase activity. Interestingly, the ATG7 promoter remains hyperacetylated upon spermidine treatment and the expression of ATG11, ATG7, and ATG15 is increased.Citation129 These data support a model in which spermidine treatment leads to the favored transcription of autophagy-related genes, whereas the majority of genes are downregulated by global hypoacetylation.Citation129 Despite the beneficial effects of spermidine treatment on life span, high levels of polyamines are associated with diseases, especially various cancers. In vitro studies as well as studies in yeast suggest that polyamines, including spermidine, increase aggregation of SNCA implicated in Parkinson disease.Citation132–134 However, it remains unclear whether the increase in polyamines is directly implicated in causing disease and whether changes in autophagy-gene expression occur under these circumstances.
In addition to histone acetylation, several histone methylation marks have been implicated in autophagy regulation. The H3K9 methyltransferase EHMT2/G9A is thought to act as a repressor of autophagy under basal conditions. Upon autophagy induction EHMT2 dissociates from histone H3K9, and H3K9 dimethylation decreases. In human cell lines, this is accompanied by an increase in H3K9 acetylation and increased transcription of multiple autophagy genes such as UVRAG, WIPI1/ATG18, ATG9B, ATG9 LC3B/ATG8, GABARAPL1/ATG8, GABARAPL2/ATG8, SQSTM1, BNIP3, and TP53INP2/DOR.Citation135 Interestingly, H3K9me3 repressive marks accumulate in aging flies, although it is unclear whether this age-dependent increase is causally linked to changes in autophagy gene expression.Citation136
Another histone mark, H3K4me3, has also been associated with longevity in C. elegans and Drosophila, as well as with autophagy regulation in yeast and mammalian cells.Citation118,137 In C. elegans, reducing the expression of the H3K4 demethylase RBR-2 leads to a concomitant increase in H3K4me3 and a decrease in life span, and conversely, reducing components of the H3K4-methylation complex (ASH-2/Set1/Ash2, WDR-5.1/Tag-125, and SET-2) decreases H3K4 methylation levels and extends longevity.Citation138 Likewise, downregulation of the Drosophila ortholog of RBR-2, Lid, shortens the life span while increasing H3K4me3 levels.Citation139,140 Although these observations are consistent with the notion that longevity is associated with increased autophagy, it remains to be determined whether long-lived animals with reduced H3K4 methylation levels display increased autophagy flux and require autophagy genes for their longevity. Overall, the emerging concept from these studies is that age-related alterations in histone marks can affect autophagy gene expression with relevance to longevity.Citation141 Future investigations of the relationship between histone marks in autophagy regulation and in life-span determination will be important to assess the impact of epigenetically regulated autophagy in age-related diseases.
MicroRNAs
RNA interference is an epigenetic mechanism of gene regulation that, in contrast to chromatin and histone alterations, occurs posttranscriptionally. miRNAs are small (21–25 nucleotides) single-stranded RNAs that bind to complementary nascent mRNAs and render them susceptible to degradation prior to translation, thereby inhibiting the expression of specific target genes. miRNAs are highly conserved, and to date, thousands have been identified in plants and animals.Citation142 Several miRNAs (e.g., lin-4, lin-14, miR-34, miR-71, miR-238, miR-239, and miR-246) regulate aging in C. elegans, and many miRNAs that affect tissue- and cell-specific aging phenotypes have been identified in mammals (reviewed in ref. Citation142). Similarly to other epigenetic regulators, numerous miRNAs exhibit age-related changes in expression.Citation143-145
One particular example is miR-34, which is linked to autophagy and longevity in several species. C. elegans with miR-34 loss-of-function mutations show an extended life span that is eliminated by RNAi of autophagy genes bec-1/Becn1/VPS30/ATG6, atg-9, and atg-4.1.Citation146 Accordingly miR-34 levels are decreased in long-lived dietary-restricted mice.Citation147 miR-34 is upregulated in C. elegans during aging and inhibits the expression of the autophagy gene Atg9a in vitro.146 In contrast, increased levels of Mir34 extend life span and reduce neurodegeneration induced by polyglutamine expansion proteins in Drosophila;Citation148 however, the contribution of autophagy genes to these miR-34-mediated beneficial effects has yet to be addressed. Several additional studies have linked MIR34 to autophagy regulation in mammalian cells, but the contribution to aging in mammals remains unclear. For example, in human cells, MIR34 targets the cell death-regulating protein BCL2, which directly binds to and inhibits the autophagy protein BECN1/VPS30.Citation149 MIR34 also inhibits the expression of the HDAC SIRT1.Citation150 Thus, the age-related upregulation of MIR34 could contribute to the aging process by directly modulating the expression of autophagy-related proteins.
Several other miRNAs (e.g., MIR376A and MIR376B) prevent starvation-induced autophagy in human cell lines by blocking the expression of BECN1 and ATG4C.Citation151,152 Similarly, overexpression of MIR181A in starved human cells inhibits the induction of ATG5 expression and reduces autophagy, as measured by LC3 lipidation.Citation153 Autophagy can also be regulated by MIR30D, which inhibits BECN1 expression,Citation154,155 and by MIR101, which blocks ATG4D, RAB5A, and STMN1 expression.Citation156,157 In addition to targeting autophagy-related gene expression, miRNAs also target transcription factors and MTOR pathway components with roles in aging. For instance, hypoxia-induced Mir155 blocks Rheb1 expression and stimulates autophagy.Citation158 Interestingly, TFEB is a target of the miRNA Mir128, highlighting a mechanism for fine-tuning of the transcriptional regulation of autophagy.Citation52
The expression of several autophagy-regulating miRNAs is altered during physiological aging as well as in neurodegenerative diseases such as Alzheimer, Huntington, and Parkinson diseases (reviewed in ref. Citation159). However, it is not yet known whether the disease pathology is directly affected by such deregulated miRNAs. Future studies should continue to identify small RNAs with effects on autophagy regulation and determine their corresponding impact on aging.
Conclusions and Future Perspectives
Common denominators in the development of age-related diseases are the inability of cells to maintain adequate metabolism, proteostasis, and organelle function. Emerging evidence strongly supports autophagy as a major mechanism to maintain cellular homeostasis and prolong organismal life span. The capacity of cells to induce and sustain autophagic flux is predominantly dependent on the function of nutrient sensors such as MTOR and AMPK, and on the INS-IGF1-signaling pathway. While MTOR and AMPK regulate metabolism and autophagy via direct phosphorylation of proteins, likely to ensure rapid induction of autophagy, studies in various organisms suggest that these master regulators of homeostasis also govern the transcriptional regulation of autophagy necessary to sustain long-term responses. In turn, several studies have linked the dysfunction of autophagy-relevant transcription factors to diseases (reviewed in ref. Citation160), and it is becoming evident that alterations in the function of such transcription factors can modulate the aging process itself (reviewed in ref. 66 ).
Although numerous transcription factors influence autophagy (reviewed in refs. Citation47, Citation48, see also refs. 49-50), TFEB and FOXO are the most prominent factors currently known to enhance autophagic flux by direct stimulation of autophagy-related and lysosomal gene expression. While FOXO so far has been reported to regulate genes especially in the core machinery, TFEB controls genes with functions in autophagy initiation, phagophore formation and elongation, and lysosomal degradation and lipolysis, suggesting that initiating autophagy or enhancing lysosomal lipid remodeling may suffice to stimulate autophagic flux. Importantly, many of these autophagy-related and lysosomal genes modulate life span and aging in various genetic contexts (summarized in ). An increasing number of transcriptionally regulated autophagy genes are being identified,Citation161 and determining their specific contributions to autophagy flux and aging may help to define novel targets for the pharmacological manipulation of autophagy for the treatment of age-related diseases.
Cellular regulation of FOXO and TFEB relies predominantly on phosphorylation and nuclear exclusion to prevent the activation of autophagy gene expression. MTOR phosphorylation of TFEB occurs at the lysosomal membrane surface, suggesting that this is an important site for autophagic flux regulation. Indeed, lysosomal–nuclear signaling is emerging as a regulatory platform not only for aging but also for metabolism.Citation46 From this standpoint, it is possible that hyperactivation of MTOR makes a major contribution to aging by actively preventing autophagy activation in various metabolic and pathological contexts. As an example, sustained activation of skeletal muscle MTORC1 causes myopathy by inhibiting autophagy.Citation162 Taken together, current evidence suggests that nutrient signaling via MTOR provides a central mechanism by which cells regulate the induction of autophagy-related and lysosomal gene expression, which in turn, modulates stress resistance and life span.
Expression of autophagy-related genes can also be regulated epigenetically. Epigenetic changes are thought to be responsible for acute fine-tuning and long-term transcriptional regulation of the autophagy response. Histone modifications also play a role in preventing prolonged autophagy through negative regulatory effects.Citation109 Epigenetic mechanisms have been implicated in the regulation of life span, but it is not yet clear whether epigenetic regulation of autophagy genes is linked to their involvement in longevity. Spermidine treatment is a prominent and conserved example of a life-span-extending regimen that epigenetically regulates autophagy gene expression. In addition, several histone modifications and chromatin-modifying enzymes have been identified that can alter the expression pattern of multiple early-acting autophagy-related genes, such as Ulk/ATG1, ATG11, Atg9B/ATG9, Atg7/ATG7, Lc3/ATG8 family, and Bnip3. Among these, Atg9B and Lc3 are upregulated in mice that overexpress TFEB,Citation52 but they have yet to be directly implicated in mammalian longevity. It will be interesting to further explore autophagy gene regulation in model organisms in which the homologous genes have known effects on life span; for example, Atg7, which is required for normal life span in mice and for spermidine-induced life span extension in Drosophila.
Aging is accompanied by several prominent changes that influence chromatin structure, including a decline in histone protein levels, an increase in histone acetylation, and alterations in the histone methylation signature.Citation163,164 In age-related pathologies, however, the nature of these chromatin changes, the identity of the enzymes associated with these changes, and the contribution of transcriptional changes of specific autophagy-related genes have yet to be defined. Additionally, it is conceivable that stable changes in the chromatin state could reprogram the profile of transcription factors occupying the promoters of autophagy-related genes, leading to a sustained change in the autophagy response throughout the organism's life span. Future studies should determine whether such chromatin changes can be engineered to sustain and increase autophagic flux in aging cells to improve proteostasis, thereby delaying aging and the onset of age-related diseases.
To summarize, recent evidence has emphasized a major role for autophagy in life-span extension in multiple organisms. The pathways underlying this role, including the transcriptional and epigenetic regulatory mechanisms described here, may provide attractive entry points to explore pharmacological avenues for the therapeutic induction of autophagy; for example, by enhancing the function of longevity-enhancing transcription factors. Alternatively, appropriately increased levels of autophagy induced by exerciseCitation91,165 might be sufficient to promote organismal health, in particular by combating autophagy-related diseases. It seems clear that the successful development of therapeutic autophagy enhancers will require an increased understanding of the regulatory networks surrounding autophagy-promoting transcription factors, their potential interactions with other factors in the nucleus, and the epigenetic modifications necessary to enhance autophagy.
Note
Nomenclature: mammalian genes/proteins are stated first, followed by the yeast name, if different. The nomenclature for other model organisms is used where applicable.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
We would like to thank Jamie Simon (Salk Institute for Biological Studies) for help with the figure.
Funding
LRL was funded by NIH/NIA (K99 AG042494); CK was the recipient of an Ellison/American Association for Aging Research Fellowship; AB was funded by the Italian Telethon Foundation (TGM11CB6), the European Research Council (Advanced Investigator grant no. 250154 CLEAR) and the NIH (R01 NS078072); MS was funded by the Telethon-Italy (TCP04009), ERC (282310-MyoPHAGY), and Foundation Leducq; and MH was funded by NIH/NIA (R01 AG038664 and R01 AG039756) and a Julie Martin Mid-Career Award in Aging Research from the American Federation for Aging Research/The Ellison Medical Foundation.
References
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/10.1038/cr.2013.168.
- Chan SN, Tang BL. Location and membrane sources for autophagosome formation - from ER-mitochondria contact sites to Golgi-endosome-derived carriers. Mol Membr Biol 2013; 30:394-402; PMID:24175710; http://dx.doi.org/10.3109/09687688.2013.850178.
- Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol 2013; 25:455-60; PMID:23578367; http://dx.doi.org/10.1016/j.ceb.2013.03.004.
- Obara K, Ohsumi, Y. Dynamics and function of PtdIns(3)P in autophagy. Autophagy 2008; 4:952-4; PMID:18769109; http://dx.doi.org/10.4161/auto.6790.
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 2009; 186:773-82; PMID:19797076; http://dx.doi.org/10.1083/jcb.200907014.
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6:79-90; PMID:14723849; http://dx.doi.org/10.1016/S1534-5807(03)00402-7.
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823-30; PMID:20811354; http://dx.doi.org/10.1038/ncb0910-823.
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7:279-96; PMID:21189453; http://dx.doi.org/10.4161/auto.7.3.14487.
- Chen Y, Yu L. Autophagic lysosome reformation. Exp Cell Res 2012; 319:142-6; PMID:22999865; http://dx.doi.org/10.1016/j.yexcr.2012.09.004.
- Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 2009; 1793:664-73; PMID:18706940; http://dx.doi.org/10.1016/j.bbamcr.2008.07.014.
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell 2011; 146:682-95; PMID:21884931; http://dx.doi.org/10.1016/j.cell.2011.07.030.
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 2010; 13:805-11; PMID:20581817; http://dx.doi.org/10.1038/nn.2575.
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/10.1016/j.cell.2012.03.017.
- Chan EY, Tooze, SA. Evolution of Atg1 function and regulation. Autophagy 2009; 5:758-65; PMID:19411825; http://dx.doi.org/10.4161/auto.8709.
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496-501; PMID:18497260; http://dx.doi.org/10.1126/science.1157535.
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141:290-303; PMID:20381137; http://dx.doi.org/10.1016/j.cell.2010.02.024.
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678-83; PMID:22053050; http://dx.doi.org/10.1126/science.1207056.
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 2003; 5:566-71; PMID:12766776; http://dx.doi.org/10.1038/ncb996.
- Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013; 14:133-9; PMID:23361334; http://dx.doi.org/10.1038/nrm3522.
- Kenyon CJ. The genetics of ageing. Nature 2010; 464:504-12; PMID:20336132; http://dx.doi.org/10.1038/nature08980.
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 2003; 5:578-81; PMID:12771962; http://dx.doi.org/10.1038/ncb999.
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 2010; 40:310-22; PMID:20965424; http://dx.doi.org/10.1016/j.molcel.2010.09.026.
- Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res 2013; 24:42-57; PMID:24343578; http://dx.doi.org/10.1038/cr.2013.166.
- Burkewitz K, Zhang Y, Mair WB. AMPK at the Nexus of Energetics and Aging. Cell Metab 2014; 20(1):10-25. PMID:24726383.
- Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 2005; 280:29060-6; PMID:15980064; http://dx.doi.org/10.1074/jbc.M503824200.
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004; 6:91-9; PMID:15261145; http://dx.doi.org/10.1016/j.ccr.2004.06.007.
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol 2011; 32:2-11; PMID:22025673; http://dx.doi.org/10.1128/MCB.06159-11.
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab 2010; 11:453-65; PMID:20519118; http://dx.doi.org/10.1016/j.cmet.2010.05.001.
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA Jr, Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy 2009; 5:847-9; PMID:19458476; http://dx.doi.org/10.4161/auto.8824.
- Toth ML, Sigmond T, Borsos E, Barna J, Erdélyi P, Takács-Vellai K, Orosz L, Kovács AL, Csikós G, Sass M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 2008; 4:330-8; PMID:18219227; http://dx.doi.org/10.4161/auto.5618.
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 2008; 4:e24; http://dx.doi.org/10.1371/journal.pgen.0040024.
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 2010; 11:35-46; PMID:20074526; http://dx.doi.org/10.1016/j.cmet.2009.11.010.
- Tang F, Watkins JW, Bermudez M, Gray R, Gaban A, Portie K, Grace S, Kleve M, Craciun G. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy 2008; 4:874-86; PMID:18690010; http://dx.doi.org/10.4161/auto.6556.
- Ruckenstuhl C, Netzberger C, Entfellner I, Carmona-Gutierrez D, Kickenweiz T, Stekovic S, Gleixner C, Schmid C, Klug L, Sorgo AG, et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet 2014; 10:e1004347; PMID:24785424; http://dx.doi.org/10.1371/journal.pgen.1004347.
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 2007; 3:597-9; PMID:17912023; http://dx.doi.org/10.4161/auto.4989.
- Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun 2013; 4:2300; PMID:23939249; http://dx.doi.org/10.1038/ncomms3300.
- Simonsen A, Cumming RC, Brech A,Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 2008; 4:176-84; PMID:18059160; http://dx.doi.org/10.4161/auto.5269.
- Bai H, Kang P, Hernandez AM, Tatar M. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet 2013; 9:e1003941; PMID:24244197; http://dx.doi.org/10.1371/journal.pgen.1003941.
- Ulgherait M, Rana A, Rera M, Graniel J, Walker DW. AMPK Modulates Tissue and Organismal Aging in a Non-Cell-Autonomous Manner. Cell Rep 2014; 8:1767-80; PMID:25199830; http://dx.doi.org/10.1016/j.celrep.2014.08.006.
- Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 2010; 143:813-25; PMID:21111239; http://dx.doi.org/10.1016/j.cell.2010.10.007.
- Carnio S, LoVerso F, Baraibar MA, Longa E, Khan MM, Maffei M, Reischl M, Canepari M, Loefler S, Kern H, et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep 2014; 8:1509-21; PMID:25176656; http://dx.doi.org/10.1016/j.celrep.2014.07.061.
- Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med 2009; 15:217-24; PMID:19380253; http://dx.doi.org/10.1016/j.molmed.2009.03.004.
- Pickford F, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid β accumulation in mice. J Clin Invest 2008; 118:2190-9; PMID:18497889.
- Komatsu M, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880-4; PMID:16625205; http://dx.doi.org/10.1038/nature04723.
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441:885-9; PMID:16625204; http://dx.doi.org/10.1038/nature04724.
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 2013; 14:283-96; PMID:23609508; http://dx.doi.org/10.1038/nrm3565.
- Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol 2013; 23:310-22; PMID:23726895; http://dx.doi.org/10.1016/j.semcancer.2013.05.008.
- Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 2013; 15:65-74; PMID:24326622; http://dx.doi.org/10.1038/nrm3716.
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014; 516(7529):112-5
- Seok S, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 2014; 516(7529):108-11; PMID:25383523.
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 1999; 402:689-92; PMID:10604478; http://dx.doi.org/10.1038/45287.
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/10.1126/science.1204592.
- Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, Wolff NC, Tran TA, Zou L, Xie XJ, Corey DR, Brugarolas J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J 2011; 30:3242-58; PMID:21804531; http://dx.doi.org/10.1038/emboj.2011.257.
- Settembre C, Ballabio A. TFEB regulates autophagy: an integrated coordination of cellular degradation and recycling processes. Autophagy 2011; 7:1379-81; PMID:21785263; http://dx.doi.org/10.4161/auto.7.11.17166.
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/10.1038/emboj.2012.32.
- Roczniak-Ferguson A, Petit CS, FroehlichF, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423.
- Martina JA, Chen Y, Gucek M, Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903-14; PMID:22576015; http://dx.doi.org/10.4161/auto.19653.
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473-7; PMID:19556463.
- Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014; 7:ra9; PMID:24448649.
- Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci U S A 2002; 99:4477-82; PMID:11930005; http://dx.doi.org/10.1073/pnas.072071099.
- Steingrimsson E, Tessarollo L, Reid SW, Jenkins NA, Copeland NG. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 1998; 125:4607-16; PMID:9806910.
- Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 2013; 202:1107-22; PMID:24081491; http://dx.doi.org/10.1083/jcb.201307084.
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 2013; 52:495-505; PMID:24095279; http://dx.doi.org/10.1016/j.molcel.2013.09.016.
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 2013; 4:2267; PMID:23925298.
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013; 15:647-58; PMID:23604321; http://dx.doi.org/10.1038/ncb2718.
- Lapierre LR, Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol Metab 2012; 23:637-44; PMID:22939742; http://dx.doi.org/10.1016/j.tem.2012.07.007.
- O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 2013; 15:668-76; PMID:23604316; http://dx.doi.org/10.1038/ncb2741.
- Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell 2013; 50:16-28; PMID:23434374; http://dx.doi.org/10.1016/j.molcel.2013.01.024.
- Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science 2008; 322:957-60; PMID:18988854; http://dx.doi.org/10.1126/science.1162011.
- Lapierre LR, Gelino SR, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate aging in germline-less C. elegans. Curr Biol 2011; 21:1507-1514; PMID:21906946; http://dx.doi.org/10.1016/j.cub.2011.07.042.
- Settembre C, Ballabio A. Lysosome: regulator of lipid degradation pathways. Trends Cell Biol 2014; 24(12):743-50; PMID:25061009.
- Pastore N, Blomenkamp K, Annunziata F, Piccolo P, Mithbaokar P, Maria Sepe R, Vetrini F, Palmer D, Ng P, Polishchuk E, et al. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in α-1-anti-trypsin deficiency. EMBO Mol Med 2013; 5:397-412; PMID:23381957; http://dx.doi.org/10.1002/emmm.201202046.
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 2011; 21:421-30; PMID:21889421; http://dx.doi.org/10.1016/j.devcel.2011.07.016.
- Polito VA, Li H, Martini-Stoica H, Wang B, Yang L, Xu Y, Swartzlander DB, Palmieri M, di Ronza A, Lee VM, et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med 2014; 6(9):1142-60; PMID:25069841.
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E, La Spada AR. PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med 2012; 4:142ra97; PMID:22786682; http://dx.doi.org/10.1126/scitranslmed.3003799.
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab 2011; 14:623-34; PMID:22055505; http://dx.doi.org/10.1016/j.cmet.2011.09.013.
- Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, Le A, Ivanov N, Sopher BL, Carromeu C, Muotri AR, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci 2014; 17(9):1180-9; PMID:25108912.
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AM, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. Innate Host Defense Requires TFEB-Mediated Transcription of Cytoprotective and Antimicrobial Genes. Immunity 2014; 40(6):896-909; PMID:24882217.
- Sandri M. FOXOphagy path to inducing stress resistance and cell survival. Nat Cell Biol 2012; 14:786-8; PMID:22854812; http://dx.doi.org/10.1038/ncb2550.
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007; 6:472-83; PMID:18054316; http://dx.doi.org/10.1016/j.cmet.2007.11.004.
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007; 6:458-71; PMID:18054315; http://dx.doi.org/10.1016/j.cmet.2007.11.001.
- Sandri M. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol 2013; 45:2121-9; PMID:23665154; http://dx.doi.org/10.1016/j.biocel.2013.04.023.
- Mammucari C, Schiaffino S, Sandri M. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy 2008; 4:524-6; PMID:18367868; http://dx.doi.org/10.4161/auto.5905.
- Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem 2009; 284:28319-31; PMID:19696026; http://dx.doi.org/10.1074/jbc.M109.024406.
- Zhao J, Brault JJ, Schild A, Goldberg, AL. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy 2008; 4:378-80; PMID:18227643; http://dx.doi.org/10.4161/auto.5633.
- Sanchez AM, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci 2013; 71(9):1657-71.
- Calnan DR, Brunet A. The FoxO code. Oncogene 2008; 27:2276-88; PMID:18391970; http://dx.doi.org/10.1038/onc.2008.21.
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 2007; 17:1646-56; PMID:17900900; http://dx.doi.org/10.1016/j.cub.2007.08.047.
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem 2007; 282:30107-19; PMID:17711846; http://dx.doi.org/10.1074/jbc.M705325200.
- Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J 2010; 29:1774-85; PMID:20400940; http://dx.doi.org/10.1038/emboj.2010.60.
- Jamart C, Francaux M, Millet GY, Deldicque L, Frère D, Féasson L. Modulation of autophagy and ubiquitin-proteasome pathways during ultra-endurance running. J Appl Physiol (1985) 2012; 112:1529-37; PMID:22345427; http://dx.doi.org/10.1152/japplphysiol.00952.2011.
- Murphy CT. The search for DAF-16/FOXO transcriptional targets: approaches and discoveries. Exp Gerontol 2006; 41:910-21; PMID:16934425; http://dx.doi.org/10.1016/j.exger.2006.06.040.
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003; 424:277-83; PMID:12845331; http://dx.doi.org/10.1038/nature01789.
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab 2010; 12:260-72; PMID:20816092; http://dx.doi.org/10.1016/j.cmet.2010.08.004.
- Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A 2009; 106:14564-9; PMID:19667176; http://dx.doi.org/10.1073/pnas.0813319106.
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 2010; 6:e1000848; PMID:20174564; http://dx.doi.org/10.1371/journal.pgen.1000848.
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 2010; 21:245-54; PMID:21177963; http://dx.doi.org/10.1101/gr.114587.110.
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 2007; 447:550-5; PMID:17476212; http://dx.doi.org/10.1038/nature05837.
- Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol 2008; 18:1355-64; PMID:18804378; http://dx.doi.org/10.1016/j.cub.2008.07.097.
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR Signaling and Rapamycin Influence Longevity by Regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 2012; 15:713-24; PMID:22560223; http://dx.doi.org/10.1016/j.cmet.2012.04.007.
- van der Vos KE, Gomez-Puerto C, Coffer PJ. Regulation of autophagy by Forkhead box (FOX) O transcription factors. Adv Biol Regul 2012; 52:122-36; PMID:22115564; http://dx.doi.org/10.1016/j.advenzreg.2011.10.002.
- Zhang P, Judy M, Lee SJ, Kenyon C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab 2013; 17:85-100; PMID:23312285; http://dx.doi.org/10.1016/j.cmet.2012.12.013.
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 2012; 489:263-8; PMID:22922647; http://dx.doi.org/10.1038/nature11315.
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol 2010; 8:e1000450; PMID:20711477; http://dx.doi.org/10.1371/journal.pbio.1000450.
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A 2009; 106:14914-9; PMID:19706382; http://dx.doi.org/10.1073/pnas.0902882106.
- Mortensen M, Watson AS, Simon AK. Lack of autophagy in the hematopoietic system leads to loss of hematopoietic stem cell function and dysregulated myeloid proliferation. Autophagy 2011; 7:1069-70; PMID:21552009; http://dx.doi.org/10.4161/auto.7.9.15886.
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 2012; 489:304-8; PMID:22972301; http://dx.doi.org/10.1038/nature11468.
- Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature 2009; 459:1079-84; PMID:19506556; http://dx.doi.org/10.1038/nature08106.
- Fullgrabe J, Heldring N, Hermanson O, Joseph B. Cracking the survival code: Autophagy-related histone modifications. Autophagy 2014; 10:556-561; PMID:24429873; http://dx.doi.org/10.4161/auto.27280.
- Dang W, Sutphin GL, Dorsey JA, Otte GL, Cao K, Perry RM, Wanat JJ, Saviolaki D, Murakami CJ, Tsuchiyama S, et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab 2014; 19:952-66; PMID:24814484; http://dx.doi.org/10.1016/j.cmet.2014.04.004.
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol 2013; 15:491-501; PMID:23604319; http://dx.doi.org/10.1038/ncb2720.
- Hubbard BP, Sinclair DA. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol Sci 2014; 35:146-54; PMID:24439680; http://dx.doi.org/10.1016/j.tips.2013.12.004.
- Park S, Mori R, Shimokawa I. Do sirtuins promote mammalian longevity? A critical review on its relevance to the longevity effect induced by calorie restriction. Mol Cells 2013; 35:474-80; PMID:23661364; http://dx.doi.org/10.1007/s10059-013-0130-x.
- Giblin W, Skinner ME, Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet 2014; 30(7):271-86; PMID:24877878.
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis 2010; 1:e10; PMID:21364612; http://dx.doi.org/10.1038/cddis.2009.8.
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, et al. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy 2010; 6:186-8; PMID:20023410; http://dx.doi.org/10.4161/auto.6.1.10817.
- Salminen A, Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal 2009; 21:1356-60; PMID:19249351; http://dx.doi.org/10.1016/j.cellsig.2009.02.014.
- Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 2013; 500:468-71; PMID:23863932; http://dx.doi.org/10.1038/nature12313.
- Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Bénit P, Rustin P, Criollo A, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol 2011; 192;615-29; PMID:21339330; http://dx.doi.org/10.1083/jcb.201008167.
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 2008; 105:3374-9; PMID:18296641; http://dx.doi.org/10.1073/pnas.0712145105.
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280-93; PMID:20965422; http://dx.doi.org/10.1016/j.molcel.2010.09.023.
- Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest 2010; 120:1043-55; PMID:20335657; http://dx.doi.org/10.1172/JCI41376.
- Wu Y, Li X, Zhu JX, Xie W, Le W, Fan Z, Jankovic J, Pan T. Resveratrol-activated AMPK/SIRT1/autophagy in cellular models of Parkinson disease. Neurosignals 2011; 19:163-74; PMID:21778691; http://dx.doi.org/10.1159/000328516.
- Jeong JK, Moon MH, Lee YJ, Seol JW, Park SY. Autophagy induced by the class III histone deacetylase Sirt1 prevents prion peptide neurotoxicity. Neurobiol Aging 2013; 34:146-56; PMID:22575359; http://dx.doi.org/10.1016/j.neurobiolaging.2012.04.002.
- van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA. C. elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet 2008; 4:e1000027; http://dx.doi.org/10.1371/journal.pgen.1000027.
- Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues M, Tenreiro S, Franssens V, Reichert AS, Outeiro TF, Winderickx J, Ludovico P. SNCA (α-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy 2012; 8:1494-509; PMID:22914317; http://dx.doi.org/10.4161/auto.21275.
- Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Küttner V, Bhukel A, Mariño G, Pietrocola F, Harger A, Zimmermann A, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab 2014; 19:431-44; PMID:24606900; http://dx.doi.org/10.1016/j.cmet.2014.02.010.
- Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 2014; 53:710-25; PMID:24560926; http://dx.doi.org/10.1016/j.molcel.2014.01.016.
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 2009; 11:1305-14; PMID:19801973; http://dx.doi.org/10.1038/ncb1975.
- Minois N, Carmona-Gutierrez D, Madeo F. Polyamines in aging and disease. Aging (Albany NY) 2011; 3:716-32; PMID:21869457.
- Soda K, Dobashi Y, Kano Y, Tsujinaka S, Konishi F. Polyamine-rich food decreases age-associated pathology and mortality in aged mice. Exp Gerontol 2009; 44:727-32; PMID:19735716; http://dx.doi.org/10.1016/j.exger.2009.08.013.
- Antony T, Hoyer W, Cherny D, Heim G, Jovin TM, Subramaniam V. Cellular polyamines promote the aggregation of α-synuclein. J Biol Chem 2003; 278:3235-40; PMID:12435752; http://dx.doi.org/10.1074/jbc.M208249200.
- Grabenauer M, Bernstein SL, Lee JC, Wyttenbach T, Dupuis NF, Gray HB, Winkler JR, Bowers MT. Spermine binding to Parkinson protein α-synuclein and its disease-related A30P and A53T mutants. J Phys Chem B 2008; 112:11147-54; PMID:18693700; http://dx.doi.org/10.1021/jp801175w.
- Lewandowski NM, Ju S, Verbitsky M, Ross B, Geddie ML, Rockenstein E, Adame A, Muhammad A, Vonsattel JP, Ringe D, et al. Polyamine pathway contributes to the pathogenesis of Parkinson disease. Proc Natl Acad Sci U S A 2010; 107:16970-5; PMID:20837543; http://dx.doi.org/10.1073/pnas.1011751107.
- Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol 2013; 33:3983-93; PMID:23918802; http://dx.doi.org/10.1128/MCB.00813-13.
- Wood JG, Hillenmeyer S, Lawrence C, Chang C, Hosier S, Lightfoot W, Mukherjee E, Jiang N, Schorl C, Brodsky AS, et al. Chromatin remodeling in the aging genome of Drosophila. Aging Cell 2010; 9:971-8; PMID:20961390; http://dx.doi.org/10.1111/j.1474-9726.2010.00624.x.
- Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol 2011; 31:1594-609; PMID:21321083; http://dx.doi.org/10.1128/MCB.00524-10.
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 2010; 466:383-7; PMID:20555324; http://dx.doi.org/10.1038/nature09195.
- Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS Genet 2010; 6:e1001221; PMID:21124823; http://dx.doi.org/10.1371/journal.pgen.1001221.
- Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev 2007; 21:537-51; PMID:17311883; http://dx.doi.org/10.1101/gad.1523007.
- Han S, Brunet A. Histone methylation makes its mark on longevity. Trends Cell Biol 2012; 22:42-9; PMID:22177962; http://dx.doi.org/10.1016/j.tcb.2011.11.001.
- Smith-Vikos T, Slack FJ. MicroRNAs and their roles in aging. J Cell Sci 2012; 125:7-17; PMID:22294612; http://dx.doi.org/10.1242/jcs.099200.
- Kato M, Chen X, Inukai S, Zhao H, Slack FJ. Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 2011; 17:1804-20; PMID:21810936; http://dx.doi.org/10.1261/rna.2714411.
- de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 2010; 20:2159-68; PMID:21129974; http://dx.doi.org/10.1016/j.cub.2010.11.015.
- Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 2006; 5:235-46; PMID:16842496; http://dx.doi.org/10.1111/j.1474-9726.2006.00210.x.
- Yang J, Chen D, He Y, Meléndez A, Feng Z, Hong Q, Bai X, Li Q, Cai G, Wang J, et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr) 2011; 35:11-22; PMID:22081425; http://dx.doi.org/10.1007/s11357-011-9324-3.
- Khanna A, Muthusamy S, Liang R, Sarojini H, Wang E. Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging (Albany NY) 2011; 3:223-36; PMID:21415464.
- Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Zhu Y, Wang LS, Bonini NM. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 2012; 482:519-23; PMID:22343898; http://dx.doi.org/10.1038/nature10810.
- Yang F, Li QJ, Gong ZB, Zhou L, You N, Wang S, Li XL, Li JJ, An JZ, Wang DS, et al. MicroRNA-34a targets Bcl−2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat 2014; 13:77-86; PMID:23862748.
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A 2008; 105:13421-6; PMID:18755897; http://dx.doi.org/10.1073/pnas.0801613105.
- Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 2012; 8:165-76; PMID:22248718; http://dx.doi.org/10.4161/auto.8.2.18351.
- Korkmaz G, Tekirdag KA, Ozturk DG, Kosar A, Sezerman OU, Gozuacik D. MIR376A is a regulator of starvation-induced autophagy. PLoS One 2013; 8:e82556; PMID:24358205; http://dx.doi.org/10.1371/journal.pone.0082556.
- Tekirdag KA, Korkmaz G, Ozturk DG, Agami R, Gozuacik D. MIR181A regulates starvation- and rapamycin-induced autophagy through targeting of ATG5. Autophagy 2013; 9:374-85; PMID:23322078; http://dx.doi.org/10.4161/auto.23117.
- Zhang Y, Yang WQ, Zhu H, Qian YY, Zhou L, Ren YJ, Ren XC, Zhang L, Liu XP, Liu CG, et al. Regulation of autophagy by miR-30d impacts sensitivity of anaplastic thyroid carcinoma to cisplatin. Biochem Pharmacol 2014; 87:562-70; PMID:24345332; http://dx.doi.org/10.1016/j.bcp.2013.12.004.
- Yang X, Zhong X, Tanyi JL, Shen J, Xu C, Gao P, Zheng TM, DeMichele A, Zhang L. mir-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochem Biophys Res Commun 2013; 431:617-22; PMID:23274497; http://dx.doi.org/10.1016/j.bbrc.2012.12.083.
- Frankel LB, Wen J, Lees M, Høyer-Hansen M, Farkas T, Krogh A, Jäättelä M, Lund AH. microRNA-101 is a potent inhibitor of autophagy. EMBO J 2011; 30:4628-41; PMID:21915098; http://dx.doi.org/10.1038/emboj.2011.331.
- Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C, Jiang H, Wang X, Li X. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep 2013; 29:2019-24; PMID:23483142.
- Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, Cui K, Liao M, He J, Jiang Y, et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy 2013; 10:70-9; PMID:24262949; http://dx.doi.org/10.4161/auto.26534.
- Tekirdag KA, Ozturk DG, Gozuacik D. Alteration in Autophagic-lysosomal Potential During Aging and Neurological Diseases: The microRNA Perspective. Curr Pathobiol Rep 2013; 1:247-261; http://dx.doi.org/10.1007/s40139-013-0031-x.
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell 2013; 152:1237-51; PMID:23498934; http://dx.doi.org/10.1016/j.cell.2013.02.014.
- Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol 2014; 24:1314-22; PMID:24881874; http://dx.doi.org/10.1016/j.cub.2014.04.048.
- Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, Rüegg MA. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab 2013; 17:731-44; PMID:23602450; http://dx.doi.org/10.1016/j.cmet.2013.03.015.
- Tsurumi A, Li WX. Global heterochromatin loss: a unifying theory of aging? Epigenetics 2012; 7:680-8; PMID:22647267; http://dx.doi.org/10.4161/epi.20540.
- McCauley BS, Dang W. Histone methylation and aging: Lessons learned from model systems. Biochim Biophys Acta 2014; PMID:24859460; 1839(12):1454-62.
- He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 2012; 481:511-5; PMID:22258505; http://dx.doi.org/10.1038/nature10758.
- Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. Autophagy regulates ageing in C. elegans. Autophagy 2007; 3:93-5; PMID:17204841; http://dx.doi.org/10.4161/auto.3636.
- Melendez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003; 301:1387-91; PMID:12958363; http://dx.doi.org/10.1126/science.1087782.
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 2009; 138:314-27; PMID:19632181; http://dx.doi.org/10.1016/j.cell.2009.04.058.
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 2015; 17(3):288-99; PMID:25720963.
- Dwivedi M, Song HO, Ahnn J. Autophagy genes mediate the effect of calcineurin on life span in C. elegans. Autophagy 2009; 5(5):604-7; PMID:19279398.
- Milan G, Romanello V, Pescatore F, Armani A, Paik J-H, Frasson L, Seydel A, Zhao J, Abraham R, Goldberg AL, et al. Regulation of autophagy and the ubiquitin–proteasome system by the FoxO transcriptional network during muscle atrophy. Nature Communications 2015; 6; http://dx.doi.org/10.1038/ncomms7670
