Abstract
TGFB2-OT1 (TGFB2 overlapping transcript 1) is a newly discovered long noncoding RNA (lncRNA) derived from the 3′UTR of TGFB2. It can regulate autophagy in vascular endothelial cells (VECs). However, the mechanisms of TGFB2-OT1 action are unclear, and whether it is involved in VECs dysfunction needs investigation. Here, the level of TGFB2-OT1 was markedly increased by lipopolysaccharide and oxidized low-density lipoprotein, 2 VECs inflammation triggers. A chemical small molecule, 3-benzyl-5-((2-nitrophenoxy) methyl)-dihydrofuran-2(3H)-one (3BDO) significantly decreased TGFB2-OT1 levels and inhibited the effect of LPS and oxLDL. The NUPR1 level was upregulated by the 2 inflammation inducers and modulated the TGFB2-OT1 level by promoting the expression of TIA1, responsible for TGFB2-OT1 processing. We focused on how TGFB2-OT1 regulated autophagy and inflammation. Use of miRNA chip assay, TGFB2-OT1 overexpression technology and 3BDO revealed that TGFB2-OT1 regulated the levels of 3 microRNAs, MIR3960, MIR4488 and MIR4459. Further studies confirmed that TGFB2-OT1 acted as a competing endogenous RNA, bound to MIR3960, MIR4488 and MIR4459, then regulated the expression of the miRNA targets CERS1 (ceramide synthase 1), NAT8L (N-acetyltransferase 8-like [GCN5-related, putative]), and LARP1 (La ribonucleoprotein domain family, member 1). CERS1 and NAT8L participate in autophagy by affecting mitochondrial function. TGFB2-OT1 increased the LARP1 level, which promoted SQSTM1 (sequestosome 1) expression, NFKB RELA and CASP1 activation, and then production of IL6, IL8 and IL1B in VECs. Thus, NUPR1 and TIA1 may control the level of TGFB2-OT1, and TGFB2-OT1 bound to MIR3960, MIR4488 and MIR4459, which targeted CERS1, NAT8L, and LARP1, respectively, the key proteins involved in autophagy and inflammation.
Abbreviations
| 3BDO | = | 3-benzyl-5-((2-nitrophenoxy) methyl)-dihydrofuran-2(3H)-one |
| 3′UTR | = | 3′ untranslated region |
| ATG13 | = | autophagy-related 13 |
| CeRNA | = | competing endogenous RNA |
| CERS1 | = | ceramide synthase 1 |
| HUVECs | = | human umbilical vein endothelial cells |
| IL6 | = | interleukin 6 |
| IL8 | = | interleukin 8 |
| IL1B | = | interleukin 1, β |
| LARP1 | = | La ribonucleoprotein domain family, member 1 |
| lncRNA | = | long noncoding RNAs |
| LPS | = | lipopolysaccharide |
| MIR3960 | = | microRNA 3960 |
| MIR4459 | = | microRNA 4459 |
| MIR4488 | = | microRNA 4488 |
| miRNA | = | microRNA |
| NAT8L | = | N-acetyltransferase 8-like (GCN5-related, putative) |
| NUPR1 | = | nuclear protein, transcriptional regulator, 1 |
| oxLDL | = | oxidized low-density lipoprotein |
| qPCR | = | quantitative real-time PCR |
| SQSTM1 | = | sequestosome 1 |
| TGFB2 | = | transforming growth factor, β 2 |
| TGFB2-OT1 | = | TGFB2 overlapping transcript 1 |
| TIA1 | = | TIA1 cytotoxic granule-associated RNA binding protein |
| VECs | = | vascular endothelial cells. |
Introduction
In recent years, numerous studies have documented that only 2% of the mammalian genome is composed of genes that encode proteins. This finding is particularly surprising because most human genes are transcribed as noncoding RNA.Citation1 Long noncoding RNAs (lncRNAs) are defined as nonprotein-coding transcripts longer than 200 nucleotides that represent a large portion of the mammalian transcriptome.Citation2 Whether these lncRNAs participate in important cellular functions or merely represent transcriptional noise remains a matter of debate in the initial phases of study.Citation3,4
The roles of lncRNAs in regulating gene transcription have been extensively studied. Some lncRNAs interact with chromatin-modifying enzymes and regulate the transcriptional activation or silencing of some genes. For example, the lncRNA XIST participates in the inactivation of one X chromosome.Citation5-8 Some lncRNAs are involved in post-transcriptional gene regulation. LncRNA MALAT1 (metastasis-associated lung adenocarcinoma associated transcript 1) can sequester serine/arginine proteins to modulate pre-mRNA alternative splicing.Citation9
MicroRNAs (miRNAs) are small, 19- to 22-nucleotide sequences of noncoding RNA that mostly work as gene expression regulators.Citation10 The crossregulation between lncRNAs and miRNAs has attracted increasing interest. This crossregulation can be artificially divided into 4 forms.Citation11 First, miRNAs target lncRNAs and reduce lncRNA stability. The miRNA MIRLET7B contributes to lowering TP53COR1 stability in human cervical carcinoma cells.Citation12 Additionally, the recruitment of MIRLET7 decreases the stability of another lncRNA, HOTAIR.Citation13 Second, lncRNAs compete with miRNAs to bind with the common target mRNAs. BACE1-AS is an antisense lncRNA of BACE1 (β-site APP-cleaving enzyme 1). In HEK293 cells, MIR485 overexpression reduces BACE1 mRNA level, which is rescued by overexpressing BACE1-AS, complementary to BACE1 mRNA in a region that contains the MIR485 binding site.Citation14 Third, lncRNAs generate miRNAs to induce target mRNA silencing. LINCMD1 can generate MIR206 and MIR133B from an intron and an exon.Citation15 LncRNA H19 can also generate MIR675.Citation16 Finally, lncRNAs act as molecular decoys or “sponges” of miRNAs. These lncRNAs are known as competing endogenous RNAs (ceRNAs).Citation17 CeRNAs share the same miRNA recognition sequences with mRNAs and compete for miRNA binding and then affect the regulation and functions of target mRNAs. CeRNA was first described by Poliseno and colleagues,Citation18 who report that the expression of the tumor suppressor PTEN (phosphatase and tensin homolog) depends on the expression of the pseudogene PTENP1 transcript. PTENP1 can act as a decoy for miRNAs targeting PTEN mRNA and affect PTEN expression. Despite the extensive existence and abundant expression of lncRNAs, their functions have been rarely revealed. Research into mammalian lncRNAs that sponge miRNAs from target binding mRNAs has focused on muscle differentiation and stem-cell self-renewal.
In a previous study, we have used the small chemical molecule 3-benzyl-5-((2- nitrophenoxy) methyl)-dihydrofuran-2(3H)-one (3BDO) and have identified a novel long noncoding RNA, TGFB2-OT1 (TGFB2 overlapping transcript 1), located in the 3′UTR (3′ untranslated region) of TGFB2 (transforming growth factor, β 2). TGFB2-OT1 can sequester MIR4459 (microRNA 4459), regulate the level of the MIR4459 target ATG13 (autophagy related 13) and then promote autophagy.Citation19 Our subsequent study shows that 3BDO can inhibit the production of inflammatory cytokines both in vitro and in vivo.Citation20 Autophagy and inflammation are very closely related processes. Autophagy can be induced by the inflammatory response and modulate it.Citation21,22
SQSTM1 (sequestosome 1) is a multifunctional scaffold protein that participates in various processes, including signal transduction, cell proliferation, cell survival and death, inflammation, tumorigenesis, and oxidative stress response. SQSTM1 is an autophagy substrate and widely used marker of autophagic degradation but also acts as a scaffold of numerous interacting proteins that promote the interaction of effector proteins with their substrates, and then transmits the signal downstream to activate NFKB signal pathway.Citation23 Previous studies show that SQSTM1 also activates CASP1 (caspase 1, apoptosis-related cysteine peptidase) and then increases IL1B (interleukin 1, β) levels.Citation24
In this study we first observed that the inflammation inducers lipopolysaccharide (LPS) and oxidized low-density lipoprotein (oxLDL) elevated the level of TGFB2-OT1, and 3BDO inhibited this effect, then we investigated the underlying mechanisms of autophagy and inflammation induced by TGFB2-OT1, possibly via SQSTM1.
Results
LPS and oxLDL increased TGFB2-OT1 levels and 3BDO inhibited this process
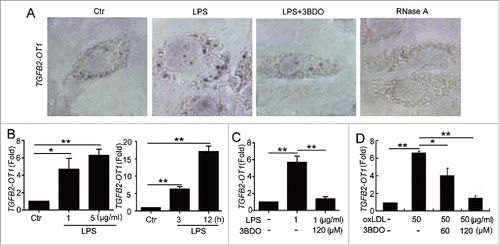
To understand the relationship between TGFB2-OT1 and vascular endothelial cells (VECs) inflammation, we first investigated the effects of LPS and oxLDL, inducers of human umbilical VECs (HUVECs) inflammation, on the level of TGFB2-OT1. LPS stimulation increased TGFB2-OT1 RNA level in HUVECs, which was significantly inhibited by 3BDO (). LPS increased TGFB2-OT1 mRNA level dose- and time-dependently, which was also reversed by 3BDO (). In addition, oxLDL increased TGFB2-OT1 level, which was reversed by 3BDO.()
Figure 1. The increased TGFB2-OT1 level induced by LPS and oxLDL was inhibited by 3BDO. (A) LPS increased TGFB2-OT1 expression and 3BDO (120 μM) inhibited the increase of TGFB2-OT1 induced by 1 μg/ml LPS for 12 h in HUVECs with in situ hybridization. (B) Quantified real-time PCR analysis of RNA levels of TGFB2-OT1 after HUVECs were exposed to 1 μg/ml or 5 μg/ml LPS for 3 h, and treated with 1 μg/ml LPS for 3 h or 12 h. (C) Quantified real-time PCR analysis of TGFB2-OT1 expression in HUVECs treated with 1 μg/ml LPS in presence or absence of 120 μM 3BDO for 3 h. (D) HUVECs were exposed to 50 μg/ml nLDL and oxLDL in presence or absence of 3BDO (60 μM and 120 μM) for 24 h, then TGFB2-OT1 levels were determined by qPCR. *, P < 0.05; **, P < 0.01; n = 3.
LPS and oxLDL elevated NUPR1 (nuclear protein, transcriptional regulator, 1) and TIA1 (TIA1 cytotoxic granule-associated RNA binding protein) levels
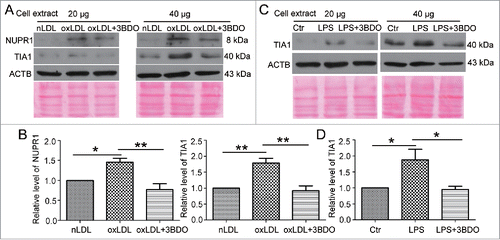
Our previous study shows that LPS induces the expression of the NUPR1,Citation25 and TIA1 is responsible for processing TGFB2-OT1.Citation19 However, the effect of oxLDL on NUPR1 and TIA1 levels and whether LPS affects TIA1 expression are unclear. Here, we showed that oxLDL increased NUPR1 levels, and 3BDO inhibited this effect (). Similarly both oxLDL and LPS increased TIA1 protein levels, and 3BDO inhibited these effects.()
Figure 2. The increased NUPR1 and TIA1 levels induced by LPS and oxLDL were inhibited by 3BDO. (A) HUVECs were exposed to 50 μg/ml nLDL or oxLDL with or without 3BDO (60 μM) for 24 h, then 20 and 40 μg cell extracts was loaded on the SDS-PAGE respectively. Western blot analysis of NUPR1 and TIA1 protein levels, with ACTB and Ponceau staining as the loading control, and (B) quantification. (C) HUVECs were treated with LPS (1 μg/ml, 6 h) with or without 3BDO (60 μM), then 20 and 40 μg cell extracts was loaded on SDS-PAGE respectively. Western blot analysis of TIA1 protein level, with ACTB and Ponceau staining as the loading control, and (D) quantification. *, P < 0.05; **P < 0.01; n = 3.
NUPR1 and TIA1 regulated TGFB2-OT1 level
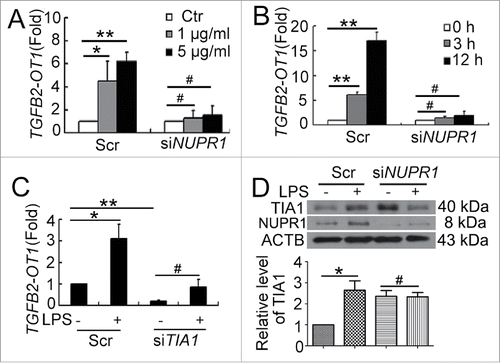
Next, we determined the role of NUPR1 in regulating TGFB2-OT1 level. When NUPR1 was knocked down, LPS did not increase the level of TGFB2-OT1 in HUVECs (). In our previous study, we demonstrated that TIA1 is responsible for TGFB2-OT1 processing, and knockdown of TIA1 decreases the TGFB2-OT1 level.Citation19 Here we further indicated that when TIA1 was knocked down, LPS did not increase TGFB2-OT1 levels (). Furthermore, when NUPR1 was knocked down, LPS did not increase the TIA1 level (). NUPR1 may modulate the expression of TIA1 responsible for processing TGFB2-OT1.
Figure 3. NUPR1 and TIA1 were involved in regulation of TGFB2-OT1 level. qPCR analysis of TGFB2-OT1 mRNA levels in HUVECs subjected to scrambled siRNA (Scr) or NUPR1 siRNA (siNUPR1, 40 nM) for 24 h, then exposed to LPS at various concentrations for 3 h (A) or treated with 1 μg/ml LPS for 3 or 12 h (B). (C) qPCR analysis of TGFB2-OT1 mRNA levels in HUVECs subjected to scrambled siRNA (Scr) or TIA1 siRNA (siTIA1, 40 nM) for 24 h, then treated with 1 μg/ml LPS for 6 h. (D) Western blot analysis of the TIA1 protein level in HUVECs subjected to scrambled siRNA (Scr) or NUPR1 siRNA (siNUPR1, 40 nM) for 24 h, then exposed to 1 μg/ml LPS for 6 h. *, P < 0.05; **, P < 0.01; #, P > 0.05; n = 3.
TGFB2-OT1 regulated the levels of MIR3960 and MIR4488
In a previous study, we showed that TGFB2-OT1 binds to MIR4459, modulates the expression of ATG13, an MIR4459 target, and then participates in autophagy.Citation19 Moreover, our bioinformatics analysis revealed that 57 presumed miRNAs could recognize and bind to TGFB2-OT1 (http://bioinfo.uni-plovdiv.bg/microinspector/). So we further detected new miRNAs regulated by TGFB2-OT1 in this study.
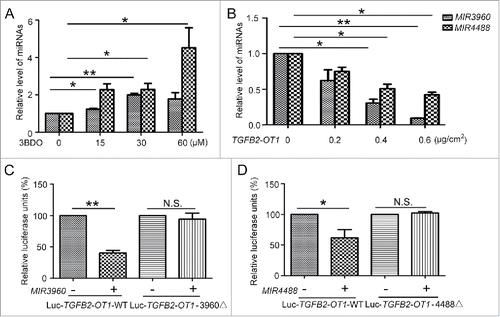
We first performed an miRNA microarray assay to identify miRNAs with changed expression when TGFB2-OT1 was up- or downregulated (Fig. S1). We considered both bioinformatics analysis and microarray assay results and identified the top 5 miRNAs, including MIR3960, MIR4459, MIR149-3p, MIR4688 and MIR4488, with significantly changed expression (Table S2). We excluded MIR4688 because of low expression in HUVECs and MIR149-3p because we did not find an appropriate MIR149-3p target involved in autophagy or inflammation. We further chose MIR3960 and MIR4488, which could bind to TGFB2-OT1 and showed significantly changed expression on miRNA array assay. We verified the miRNA microarray results by qPCR. TGFB2-OT1 downregulation by 3BDO increased and TGFB2-OT1 overexpression decreased the levels of MIR3960 and MIR4488.()
Figure 4. MIR3960 and MIR4488 directly bind to TGFB2-OT1. (A) qPCR analysis of MIR3960 and MIR4488 level in HUVECs treated with or without 3BDO (0, 15, 30 and 60 μM) for 24 h and (B) transfected with the empty vector pCMV6 (0.8 μg/cm2, indicated by 0 in the figure) or pCMV6-TGFB2-OT1 plasmid (0.2, 0.4 and 0.8 μg/cm2) for 24 h. (C) Luciferase activity assay with Luc-TGFB2-OT1-WT and Luc-TGFB2-OT1-3960Δ plasmids cotransfected into HEK293 cells with 80 nM MIR3960 mimics or negative control for 24 h. (D) Luciferase activity assay with Luc-TGFB2-OT1-WT and Luc-TGFB2-OT1-4488Δ plasmids cotransfected into HEK293 cells with 80 nM MIR4488 mimics or negative control for 24 h. *, P < 0.05; **, P < 0.01; n = 3.
MIR3960 and MIR4488 directly bind to TGFB2-OT1
According to the prediction results, TGFB2-OT1 has one putative MIR3960 binding site at positions 623 to 642 and one putative MIR4488 binding site at positions 619 to 636. So we recombined the TGFB2-OT1 cDNA (Luc-TGFB2-OT1-WT) and mutational cDNA with the presumed MIR3960 and MIR4488 deleted recognition sequences (Luc-TGFB2-OT1-3960Δ and Luc-TGFB2-OT1-4488Δ) downstream the luciferase reporter gene (Fig. S2A and B). The vectors were transfected in HEK293 cells along with the corresponding miRNA mimics. With MIR3960 and MIR4488 mimics transfection, luciferase activity was reduced by about 60% and 40% respectively, as compared with control miRNA. When adding mutant substrates for MIR3960 and MIR4488, the decreased luciferase activity was abolished (). Therefore, MIR3960 and MIR4488 directly bind TGFB2-OT1.
MIR3960, MIR4488 and MIR4459 targeted the 3′UTRs of CERS1 (ceramide synthase 1), NAT8L (N-acetyltransferase 8-like [GCN5-related, putative]) and LARP1 (La ribonucleoprotein domain family, member 1), respectively.
Next, we predicted the downstream targets of MIR3960 and MIR4488 by bioinformatics analysis. We selected CERS1 as an MIR3960 target protein and NAT8L as an MIR4488 target protein, both involved in regulating autophagy by affecting mitochondria.Citation26,27 We first examined the modulation of miRNA on the targets. HUVECs were transfected with MIR3960 and MIR4488 mimics or inhibitor. The efficiency of miRNA mimics and inhibitor was determined by qPCR. miRNA mimics and the inhibitor at 25 and 50 nM effectively increased or decreased the corresponding miRNA level, respectively (Fig. S3A). We further determined the mRNA level of miRNA targets by transfecting HUVECs with miRNA mimics or inhibitor. miRNA mimics decreased the corresponding target mRNA levels, and miRNA inhibitor increased the corresponding target mRNA levels (Fig. S3B).
Furthermore, we determined the effect of miRNAs on the protein levels of miRNA targets. MIR3960 and MIR4488 mimics at 50 nM significantly decreased the protein levels of CERS1 and NAT8L. Moreover, the MIR3960 and MIR4488 inhibitor increased the protein levels (). Therefore, MIR3960 and MIR4488 modulate the levels of CERS1 and NAT8L.
Figure 5. MIR3960, MIR4488 and MIR4459 targeted CERS1, NAT8L and LARP1. Western blot analysis of protein levels of CERS1 (A) and NAT8L (B) in HUVECs transfected with 25 and 50 nM MIR3960 (A), MIR4488 (B) mimics, inhibitor or negative control for 24 h and quantification. (C) Western blot analysis of protein levels of LARP1 in HEK293 cells (left) and HUVECs (right) transfected with 25 and 50 nM MIR4459 mimics or negative control for 24 h and quantification. (D) Luciferase activity assay with Lu-CERS1-3′UTR, Lu-NAT8L-3′UTR, and Lu-LARP1-3′UTR plasmids cotransfected into HEK293 cells with 50 nM MIR3960, MIR4488 and MIR4459 mimics for 24 h, a negative miRNA as a control. *, P < 0.05; **, P < 0.01; n = 3.
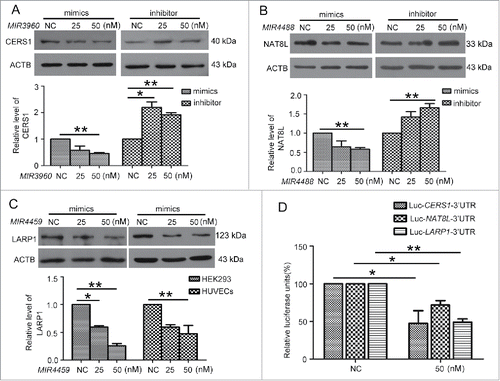
A previous study shows that ATG13 is a target of MIR4459; however, an miRNA may have several targets,Citation28 so we studied other predicted targets of MIR4459. Among the predicted targets, we chose the one with the highest context score, LARP1. We transfected MIR4459 mimics into both HEK293 cells and HUVECs, and found that LARP1 protein level was inhibited.()
Next, we cloned the 3′UTRs of CERS1, NAT8L and LARP1 within the predicted binding sites of miRNAs into the luciferase reporter vector (Fig. S2C) and transfected the vectors into HEK293 cells with the corresponding miRNA mimics or the negative control. The luciferase activity was significantly decreased by the miRNAs as compared with the control (). So CERS1, NAT8L and LARP1 were directly targeted by MIR3960, MIR4488 and MIR4459, respectively.
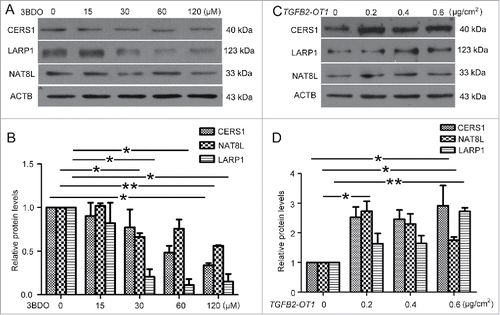
3BDO and TGFB2-OT1 overexpression regulated the protein levels of CERS1, NAT8L and LARP1
We further detected the role of TGFB2-OT1 in the miRNA-targeted proteins. 3BDO treatment in HUVECs decreased the mRNA levels of CERS1, LARP1 and NAT8L (Fig. S4), which was verified by western blot analysis of protein levels. Similarly, TGFB2-OT1 downregulation by 3BDO dose-dependently decreased the protein levels of CERS1, NAT8L and LARP1 (). Next, HEK293 cells were transfected with different amounts of pCMV6-TGFB2-OT1 plasmid or pCMV6 plasmid as a control. TGFB2-OT1 overexpression increased the protein levels of CERS1, NAT8L and LARP1.()
Figure 6. 3BDO and TGFB2-OT1 overexpression regulated the protein levels of CERS1, LARP1 and NAT8L. (A) Western blot analysis of protein levels of CERS1, LARP1 and NAT8L in HUVECs treated with 3BDO (0, 15, 30, 60 and 120 μM) for 24 h and (B) quantification. (C) Western blot analysis of protein levels of CERS1, LARP1 and NAT8L in HEK293 cells transfected with the empty vector pCMV6 (0.8 μg/cm2, indicated by 0 in the figure) or pCMV6-TGFB2-OT1 plasmid (0.2, 0.4 and 0.8 μg/cm2) for 48 h and (D) quantification. *, P < 0.05; **, P < 0.01; n = 3.
Overexpression of TGFB2-OT1 promoted translation of autophagy-related proteins
LARP1 is an RNA binding protein related to transcript stability and translation of mRNAs.Citation29 siRNA knocks down LARP1 expression and inhibits global protein synthesis rates.Citation30 So we further investigated whether protein synthesis was affected by TGFB2-OT1 and MIR4459. We first examined several proteins related to autophagy.
TGFB2-OT1 overexpression increased the protein levels of ATG3 and ATG7 (Fig. S5). SQSTM1 is the substrate of autophagic degradation.Citation31 However, SQSTM1 protein levels were greatly increased after HUVECs transfection with 0.01, 0.05, 0.1, 0.2 μg/ml pCMV6-TGFB2-OT1 vector (), and SQSTM1 puncta were increased in number in HUVECs treated with pCMV6-TGFB2-OT1 plasmid (). Furthermore, the mRNA level of SQSTM1 was not increased in cells transfected with 0.1, 0.2 and 0.4 μg/ml pCMV6-TGFB2-OT1 vector as compared with control transfection (Fig. S6A). In addition, 3BDO inhibited the increased SQSTM1 protein level induced by TGFB2-OT1 (Fig. S6B) but did not affect the SQSTM1 mRNA level (Fig. S6C).
Figure 7. TGFB2-OT1 promoted SQSTM1 translation, RELA nuclear translocation and IL6 and IL8 production in HUVECs. (A) Western blot analysis of SQSTM1 protein level in HUVECs transfected with 0.01, 0.05, 0.1, 0.2 μg/ml of pCMV6 or pCMV6-TGFB2-OT1 for 48 h. This result is representative of 3 independent experiments. (B) Immunostaining of SQSTM1 in HUVECs transfected with pCMV6 or pCMV6-TGFB2-OT1 (0.2 μg/ml) for 48 h, and the proportion of cells containing SQSTM1 puncta (> 5). The arrows indicate SQSTM1 puncta in cells. Bar: 16 μm. (C) Western blot analysis of SQSTM1 protein level in HUVECs transfected with pCMV6 or pCMV6-TGFB2-OT1 (0.2 μg/ml) for 12 h, then treated with or without cycloheximide (CHX) for 12 h. (D) Western blot analysis of SQSTM1 protein level in HUVECs transfected with 25 and 50 nM MIR4459 mimics or negative control for 24 h. (E) Representative photomicrographs of immunofluorescence staining showing RELA nuclear translocation in HUVECs transfected with pCMV6 or pCMV6-TGFB2-OT1 (0.4 μg/ml) for 48 h. Bar: 16 μm. (F) Immunofluorescent graphs of RELA nuclear translocation in HUVECs transfected with pCMV6, pCMV6-TGFB2-OT1, or both siRNA against SQSTM1 and pCMV6-TGFB2-OT1 for 48 h. n indicates the nuclear. Bar: 16 μm. (G) ELISA of IL6 and IL8 production in HUVECs transfected with 0.4 μg/ml pCMV6 or pCMV6-TGFB2-OT1 for 48 h, then exposed to 3BDO (60 μM) for 6 h. (H) Western blot analysis of cleaved-CASP1 protein level in HUVECs transfected with 0.4 μg/ml pCMV6 or pCMV6-TGFB2-OT1 for 48 h and quantification. (I) qPCR analysis of mRNA levels of TGFB2-OT1 and IL1B in HUVECs transfected with 0.4 μg/ml pCMV6 or pCMV6-TGFB2-OT1 for 48 h, then exposed to 3BDO (60 μM) for 12 h. *, P < 0.05; **, P < 0.01; n = 3.
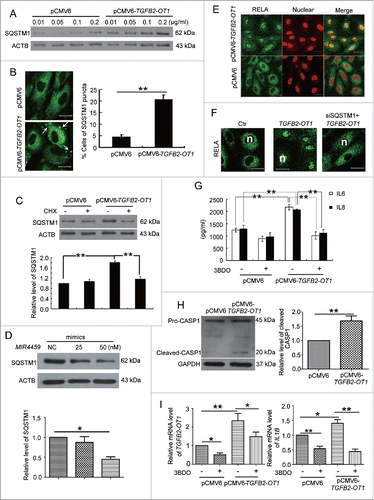
We next investigated whether TGFB2-OT1 regulated SQSTM1 expression via protein synthesis. TGFB2-OT1 overexpression did not increase the level of SQSTM1 in the presence of cycloheximide, a well-known protein synthesis inhibitor (). In addition, transfection with MIR4459 mimics decreased SQSTM1 protein level, indicating that LARP1 was involved in SQSTM1 protein synthesis.()
TGFB2-OT1 was involved in regulating NFKB RELA nuclear translocation and CASP1 activation in HUVECs
SQSTM1 is a receptor protein involved in both autophagy and inflammation. Therefore we deduced that TGFB2-OT1 and MIR4459 might participate in inflammation via SQSTM1. We first detected the location of the NFKB RELA by immunofluorescence assay. Overexpression of TGFB2-OT1 significantly promoted NFKB RELA nuclear translocation (, Fig. S7A). Furthermore, with SQSTM1 knockdown, TGFB2-OT1 overexpression did not induce RELA nuclear translocation.(, S7B and S7C)
RELA plays a crucial role in inflammatory and immune responses. We found that overexpression of TGFB2-OT1 in HUVECs promoted the production of IL6 and IL8 (). Furthermore, TGFB2-OT1 overexpression activated CASP1 and increased the IL1B mRNA level, which was inhibited by 3BDO (). However, overexpression of TGFB2-OT1 did not affect cell viability (Fig. S7D) or reactive oxygen species levels in HUVECs (Fig. S7E).
LOC100129973 (Genbank accession no. NR_102751.1, also known as AF007131) is annotated as an lncRNA in the NCBI database, but its functions have not been identified. We chose the lncRNA LOC100129973 as the nonrelated lncRNA expression plasmid control to avoid a nonspecific, stress-related effect on the expression of the lncRNA. Transfection with pcDNA-LOC100129973 plasmid did not affect SQSTM1 protein level or NFKB RELA nuclear translocation.(Fig. S8)
Discussion
Recent research has demonstrated that lncRNAs play an essential role in a number of cellular, developmental and pathological processes, such as cell apoptosis and differentiation,Citation32,33 tumorigenesis.Citation34 and X-inactivation.Citation35 Numerous reports suggest the existence of a widespread interaction network involving ceRNAs, with lncRNAs regulating miRNA by binding and titrating them off their binding sites on protein coding messengers. A recent study indicates that the lncRNA MIAT functions as a ceRNA and forms a feedback loop with vascular endothelial growth factor and MIR150-5p to regulate endothelial cell function.Citation36 However, the involvement of lncRNAs in endothelial cells biology is just beginning to be studied, and research of lncRNAs in controlling NFKB signaling and inflammation has mainly concentrated on other cell types such as macrophages.Citation37 Here, we verified that an lncRNA, TGFB2-OT1 regulated autophagy and inflammation in VECs.
TGFB2-OT1 is a newly discovered lncRNA located in the 3′UTR of TGFB2 and involved in VECs autophagy. In this study, we found that 2 inflammation inducers (LPS and oxLDL) promoted TGFB2-OT1 expression, and the small chemical molecule 3BDO inhibited the increased TGFB2-OT1 level in HUVECs (). How LPS and oxLDL affect TGFB2-OT1 production was not clear. Our previous study shows that LPS can increase NUPR1 protein level and autophagy in VECs, and 3BDO inhibits this effect.Citation25 We also discovered that TIA1 is responsible for TGFB2-OT1 processing, and TIA1 phosphorylation inhibits TGFB2-OT1 production.Citation19 In this study, we find that NUPR1 regulates TGFB2-OT1 expression and acts upstream TIA1 (). LPS and oxLDL modulated TGFB2-OT1 via NUPR1 and TIA1. Moreover, the TIA1 protein level was increased with siRNA-mediated knockdown of NUPR1 (, lane 3). NUPR1 is a stress-inducible nuclear protein involved in cellular stress responses as a transcription regulator. NUPR1 deficiency induces apoptosis and intracellular reactive oxygen species level.Citation38-40 TIA1 is a proapoptosis protein involved in stress granule formation during cellular stress.Citation41-43 So the increase in TIA1 level might be due to the apoptosis and stress induced by NUPR1 knockdown.
TGFB2-OT1 acts as a ceRNA, competes to bind with MIR4459 and regulates the level of ATG13.Citation19 We have further determined the mechanisms by which TGFB2-OT1 induced autophagy and inflammation. Here, we further identify 2 other miRNAs (MIR3960 and MIR4488) that bind TGFB2-OT1.()
A previous study indicates that mouse Mir3960 promotes osteoblast differentiation by targeting HOXA2 (homeobox A2).Citation44 There are no reports about human MIR3960 functions. Among the various predicted targets of MIR3960, we first selected CERS1. Luciferase activity assay confirmed the binding of MIR3960 to the 3′UTR of CERS1 (). Ceramide synthases regulate the de novo generation of ceramides with specific fatty acid chain lengths.Citation45 CERS1 preferentially generates C18-ceramide. CERS1-driven production of C18-ceramide is defined as a tumor suppressor in preclinical and clinical research.Citation46 C18-ceramide induces autophagy via selective targeting of mitochondria by MAP1LC3B-containing phagophores via direct interaction between ceramide and MAP1LC3B on mitochondrial membranes.Citation47 Furthermore, excessive mitophagy induced by CERS1 is lethal and leads to tumor suppression.Citation26,47 TGFB2-OT1 downregulation by 3BDO increased MIR3960 levels and reduced those of CERS1, whereas TGFB2-OT1 overexpression downregulated MIR3960 levels and increased the CERS1 protein level (). CERS1 indirectly regulated by TGFB2-OT1 contributes to explaining the autophagy alterations observed with changes in TGFB2-OT1 level. Excessive promotion of CERS1 might lead to injury of VECs, which is consistent with overexpression of TGFB2-OT1 inducing VECs inflammation.
So far, there are no studies of MIR4488 functions. By using bioinformatics prediction analysis and luciferase activity assay (), we found that NAT8L was an important target of MIR4488. NAT8L catalyzes the formation of N-acetylaspartate (NAA) from acetyl-CoA and aspartate. In the brain, NAA delivers the acetate moiety for synthesis of acetyl-CoA, which is further used for fatty acid generation.Citation48,49 Recent studies discovered new functions for NAT8L in adipose tissues. NAT8L is highly expressed in adipose tissues, murine and human adipogenic cell lines and localized in mitochondria of brown adipocytes. Upon NAT8L overexpression, mitochondrial mass and number as well as oxygen consumption are elevated.Citation27 So NAT8L may also be involved in autophagy by affecting mitochondria. TGFB2-OT1 modulates NAT8L and then autophagy by sponging MIR4488.
LARP1 is an RNA binding protein and related to transcript stability and translation of mRNAs.Citation29,50 siRNA-depleted LARP1 level inhibits global protein synthesis rate.Citation30 As a ceRNA, TGFB2-OT1 competed with MIR4459 to modulate LARP1 protein level (). Previous studies show that TGFB2-OT1 acts downstream MTOR,Citation19 a master controller of protein synthesis, both in the initiation and elongation steps of protein synthesis.Citation14
Here, we show that TGFB2-OT1 overexpression increases ATG3, ATG7, and SQSTM1 protein levels (, Fig. S4), which suggests that TGFB2-OT1 is a key regulator in modulating protein synthesis. Furthermore, the decreased LARP1 protein level induced by MIR4459 mimics transfection also decreased SQSTM1 expression (), so TGFB2-OT1 modulated SQSTM1 protein level by LARP1.
SQSTM1 acts as a signaling hub by recruiting and oligomerizing important signaling molecules in cytosolic speckles to determine cell survival or death by triggering the TRAF6-NFKB pathway or activating the aggregation of caspase 8 and its downstream effector caspases.Citation23,31 SQSTM1 similarly plays a role in the activation of CASP1, thereby inducing IL1B production, as an important component in the inflammasome complex.Citation24 The data showing that pCMV6-TGFB2-OT1 elevated SQSTM1 but did not affect cell viability (Fig. S6C) indicate that caspase 8 and its downstream effector caspases were not activated. So we further examined the inflammation-related signal pathways.
The evidence that overexpression of TGFB2-OT1 induced NFKB RELA nuclear translocation and IL6 and IL8 production in VECs suggested that the NFKB pathway was activated (). Furthermore, with SQSTM1 knockdown, TGFB2-OT1 overexpression did not induce RELA nuclear translocation (). Activation of the NFKB pathway leads to the expression of various inflammation-associated genes, including cytokines, chemokines, and adhesion molecules.Citation51 Furthermore, TGFB2-OT1 overexpression induced CASP1 activation and then IL1B expression (), so TGFB2-OT1 might be involved in activation of inflammasomes. Therefore, we demonstrated that TGFB2-OT1 promoted inflammation responses via elevating SQSTM1 level. In contrast, a recent study shows that SQSTM1 deficiency in the tumor stroma results in the creation of a protumorigenic inflammatory environment driven by IL6.Citation52 SQSTM1 may present different outcomes depending on the cell type.
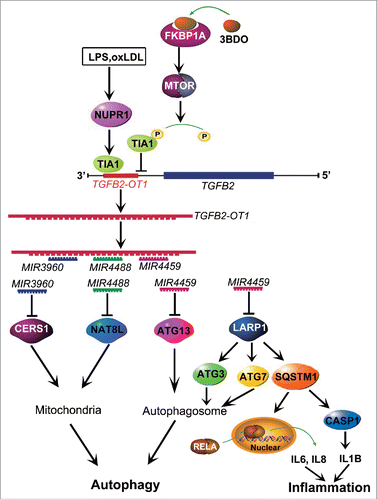
In summary, as shown in , lncRNA TGFB2-OT1, with decoy activity for MIR3960, MIR4488 and MIR4459 and by sequestering these miRNAs, increases the levels of CERS1, NAT8L, ATG13 and LARP1. LARP1 further elevates the levels of ATG3, ATG7 and SQSTM1, and then TGFB2-OT1 promotes autophagy via CERS1, NAT8L, ATG13, ATG3 and ATG7 and participates in inflammation by promoting SQSTM1 protein synthesis and activating RELA and CASP1. LPS and oxLDL promotes TGFB2-OT1 processing via NUPR1 and TIA1, so TGFB2-OT1 is involved in autophagy and inflammation. 3BDO induces TIA1 phosphorylation via FKBP1A and MTORC1, which inhibits TGFB2-OT1 processing,Citation19 then autophagy and inflammation as induced by TGFB2-OT1. Therefore, TGFB2-OT1 is an attractive target against inflammation in VECs, and 3BDO might be a potential therapeutic compound for the development of new drugs for vascular diseases.
Figure 8. Conceptual schematic of the miRNA signal pathway regulated by TGFB2-OT1 in autophagy and inflammation. LPS and oxLDL promote TIA1 expression via NUPR1. TIA1 is responsible for TGFB2-OT1 processing from 3′UTR of TGFB2. TGFB2-OT1, as a miRNA sponge, decoys MIR3960, MIR4488, and MIR4459, then elevates CERS1, NAT8L, ATG13, and LARP1 protein levels. LARP1 further increases the levels of ATG3, ATG7 and SQSTM1. CERS1 and NAT8L regulate autophagy by affecting mitochondria. ATG13, ATG3 and ATG7 modulate autophagy. The elevated synthesis of SQSTM1 protein activates RELA and CASP1, increase levels of inflammatory cytokines IL6, IL8 and IL1B, and thereby induce inflammation. 3BDO induces TIA1 phosphorylation via FKBP1A and MTORC1, which may result in inhibition of TGFB2-OT1 processing, autophagy and inflammation induced by TGFB2-OT1.
Materials and Methods
Cell culture and treatment
All cells were cultured at 37°C with 5% CO2 in a humidified incubator. HUVECs were obtained as previously described,Citation53 and cultured in M199 medium (Gibco, 31100-035) with 10% fetal bovine serum (Hyclone, SV30087.02) and FGF2 (2 ng/ml). The cells used were not greater than passage 10. Human embryonic kidney 293 (HEK293) cells were cultured in DMEM medium (Gibco, 12800-017) with 10% fetal bovine serum. Cells at 80% confluence were activated by LPS (Sigma–Aldrich, L2880) and oxLDL (Biomedical Technologies Inc., BT-910). 3BDO was synthesized as described.Citation54 and dissolved in DMSO (0.1 M, Sigma-Aldrich, D2650) as a stock solution. DMSO used in the experiment was below 0.1% in culture medium (v/v) and did not affect cell viability.
RNA interference
The efficiency of TIA1.Citation19 and NUPR1.Citation25 silencing was previously described. Cells at 50% to 60% confluence were transfected with TIA1 (Santa Cruz Biotechnology, sc-29504), NUPR1 (Santa Cruz Biotechnology, sc-40792) or scrambled siRNA (Santa Cruz Biotechnology, sc-37007) for 24 h by use of RNAiFect Transfection Reagent according to the manufacturer's protocol (QIAGEN, 301605). The efficiency of RNA interference was determined by protein gel blot analysis.
Plasmid transfection
Cells were seeded onto 6-cm dishes at 1 × 106/mL and grown for 24 h. Cells at 70% to 80% confluence were transfected with the indicated expression vectors by use of Lipofectamine 2000 transfection reagent (Invitrogen, 11668019) according to the manufacturer's protocol. The pCMV6-TGFB2-OT1 vector was generated as previously reported.Citation19
Luciferase activity assay
A luciferase reporter vector (pmirGLO Dual-Luciferase miRNA Target Expression Vector; Promega) was used for the luciferase constructs. The TGFB2-OT1, 3′UTRs of CERS1, NAT8L and LARP1 were cloned by RT-PCR, then Luc-TGFB2-OT1-WT, mutants, Luc-CERS1-3′UTR, Luc-NAT8L-3′UTR and Luc-LARP1-3′UTR were constructed as previously reported.Citation19 HEK293 cells were seeded onto 96-well culture plates at 8000 cells per well in DMEM medium containing 10% fetal bovine serum and incubated overnight. Cells were cotransfected with Dual-Luciferase (containing Firefly and Renilla luciferase) reporter constructs and corresponding miRNA mimics or negative control by Lipofectamine 2000 transfection reagent (Invitrogen, 11668019). After transfection for 24 h, luciferase activity assays were determined by a dual-luciferase reporter system according to the protocol (Promega, E2920).
RT-PCR and quantitative real-time PCR (qPCR)
Total RNA was extracted from cells with use of TriZol Reagent (Sigma-Aldrich, T9424) following the manufacturer's protocol. An amount of 1 μg RNA was reverse-transcribed by use of the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, DRR047A). qPCR reactions involved use of the QuantiTect SYBR Green PCR kit (Takara, RR420) and LightCycler 2.0 system (Roche, Basel, Switzerland). Reactions were carried out in a 20 μl volume containing 10 μl 2×SYBR Green PCR Master Mix. The primer pair sequences were for TGFB2-OT1, forward 5'-GCAGTTTCACCTA AAGAGCAGC-3′, and reverse 5'- TTCCTTCCCACCTCCACCC-3′; and GAPDH, forward, 5′-ACCACAGTCCATGCCATCAC-3′, and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′ as a housekeeping gene. The rest of primer sequences are listed in Table S1. The fold changes in mRNA level were calculated by use of MxPro v4.00 (Stratagene). Relative gene expression was normalized to the GAPDH level.
Western blot analysis
Cell lysates were prepared in RIPA lysis buffer (Beyotime, P0013B) containing 1 mM PMSF. Equal amounts of proteins were run on SDS-polyacrylamide gel, then transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, IPFL00010), probed with primary antibodies, horseradish peroxidase-linked secondary antibodies (Santa Cruz Biotechnology, sc-2004 and sc-2302), and detected with use of an enhanced chemiluminesence detection kit (Thermo, 32209). The primary antibodies specific to GAPDH (sc-47724), TIA1 (sc-166247), CERS1 (sc-135033), LARP1 (sc-102006), and RELA (sc-372) were from Santa Cruz Biotechnology. Antibody for SQSTM1 (610833) was from BD Transduction Laboratories. Antibodies for ACTB (A5441), NUPR1 (SAB1104559) and NAT8L (HPA040677) were from Sigma-Aldrich. Antibodies for ATG3 (3415), ATG7 (8558) and CASP1 (2225) were from Cell Signaling Technology. The relative quantity of proteins was analyzed by ImageJ software and normalized to loading controls.
In situ hybridization
In situ hybridization to detect TGFB2-OT1 expression in HUVECs was performed as described by Roche Applied Science. Briefly, cells were fixed in 4% paraformaldehyde. Then the cells were rehydrated, digested and then refixed in 4% paraformaldehyde. After that, the cells were prehybridized with hybridization solution and then incubated with a digoxigenin-labeled TGFB2-OT1 probe at 42°C for 20 h. The next day, the cells were washed with 2× SSC, 1× SSC and 0.5× SSC and then incubated with a mouse anti-digoxin antibody conjugated with alkaline phosphatase (Sigma-Aldrich, A1054). For visualizing the positive signal, the sections were incubated in NBT/BCIP (Roche, 11681451001) and counterstained with nuclear fast red (Sigma-Aldrich, N8002). Staining of treated samples was performed for an equal length of time and experiments were repeated at least 3 times.
Immunofluorescence assay of cells
Immunofluorescence assay was performed as described.Citation19 In brief, after treatment, HUVECs were fixed with 4% paraformaldehyde for 15 min and blocked with 3% normal goat serum (ZSGB-BIO, ZLI-9026) or rabbit serum (ZSGB-BIO, ZLI-9026) for 20 min at room temperature. Then, the cells were incubated with primary antibody (1:100) at 4°C overnight and then corresponding secondary antibody (1:200) at 37°C for 1 h. Cells were rinsed 3 times with 0.1 M phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) to eliminate the uncombined secondary antibody. The samples were evaluated by Zeiss LSM700 (Carl Zeiss, Oberkochen, Germany).
Quantitative real-time PCR of miRNA expression
RNA was isolated with the Trizol reagent method. RNA samples were quantified by use of the NanoDrop. MiRNA was converted into cDNA by using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, PN4366596) and specific miRNA primers. The KAPA SYBR FAST qPCR Kits (Kapa Biosystems, KK4601) was used to quantify the expression of mature miRNA in HUVECs. Quantitative real-time PCR reactions involved use of the LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland). Following a single cycle at 95°C for 10 min, PCR involved 40 cycles at 95°C for 30 sec and 60°C for 1 min. MiRNA expression was calculated as relative to RNU6 expression, by independent triplicate measurements. The primer sequences used for MIR3960 (stem-loop primer): 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCA CTGGATACGACCCCCGCC-3′, (PCR primer): 5′-ATTTAAGGCGGCGGCGGA-3′; MIR4488 (stem-loop primer): 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCA CTGGATACGACGCCGGAG-3′, (PCR primer) 5′-TAATAAAGGGGGCGGGCT-3′; and RNU6, 5′-CTCGCTTCGGCAGCACATATACT-3′, 5′-ACGCTTCACGAATTTGCGTGTC-3′.
Transfection with miRNA mimics and inhibitor
RNA mimics and inhibitor for MIR3960, MIR4488, MIR4459 and negative control (NC) were designed and purchased from Invitrogen. The mimics and inhibitors were transfected into HUVECs or HEK293 cells with RNAiFect Transfection Reagent according to the manufacturer's protocol for 24 h. The final concentrations of the mimics or inhibitors were 25 or 50 nM.
Statistical analysis
All experiments were repeated independently for at least 3 times. Data are expressed as mean ± SE. SPSS 11.5 (SPSS Inc., Chicago, IL) was used for analysis by one-way ANOVA (followed by Scheffé F test for post-hoc analysis). A P < 0.05 was considered as statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary_material.zip
Download Zip (7.5 MB)Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by the National Natural Science Foundation of China (No. 91313303, 81321061, 91539105, 31270877, 20972088, 31070735, and 31501122) and the National 973 Research Project (No. 2011CB503906).
References
- Ronnau CG, Verhaegh GW, Luna-Velez MV, Schalken JA. Noncoding RNAs as novel biomarkers in prostate cancer. Biomed Res Internatl 2014; 2014:591703; http://dx.doi.org/10.1155/2014/591703
- Perkel JM. Visiting “noncodarnia.” Biotechniques 2013; 54:301, 3-4
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. The reality of pervasive transcription. PLoS Biol 2011; 9:e1000625; discussion e1102; http://dx.doi.org/10.1371/journal.pbio.1000625
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol 2010; 8:e1000371; PMID:20502517; http://dx.doi.org/10.1371/journal.pbio.1000371
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 2006; 20:1848-67; PMID:16847345; http://dx.doi.org/10.1101/gad.1422906
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Ann Rev Genet 2008; 42:733-72; PMID:18729722; http://dx.doi.org/10.1146/annurev.genet.42.110807.091711
- Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 2010; 7:582-5; PMID:20930520; http://dx.doi.org/10.4161/rna.7.5.13216
- Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 2010; 10:49; PMID:20459797; http://dx.doi.org/10.1186/1471-213X-10-49
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010; 39:925-38; PMID:20797886; http://dx.doi.org/10.1016/j.molcel.2010.08.011
- Moreno-Moya JM, Vilella F, Simon C. MicroRNA: key gene expression regulators. Fertility Sterility 2014; 101:1516-23; PMID:24314918; http://dx.doi.org/10.1016/j.fertnstert.2013.10.042
- Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Seminars Cell Dev Biol 2014; 34:9-14; http://dx.doi.org/10.1016/j.semcdb.2014.05.015
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47:648-55; PMID:22841487; http://dx.doi.org/10.1016/j.molcel.2012.06.027
- Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nature Commun 2013; 4:2939; http://dx.doi.org/10.1038/ncomms3939
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 2010; 11:R56; PMID:20507594; http://dx.doi.org/10.1186/gb-2010-11-5-r56
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011; 147:358-69; PMID:22000014; http://dx.doi.org/10.1016/j.cell.2011.09.028
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012; 14:659-65; PMID:22684254; http://dx.doi.org/10.1038/ncb2521
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146:353-8; PMID:21802130; http://dx.doi.org/10.1016/j.cell.2011.07.014
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465:1033-8; PMID:20577206; http://dx.doi.org/10.1038/nature09144
- Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z, Zhao J, Su L, Zhang S, Zhang Y, et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy 2014; 10:957-71; PMID:24879147; http://dx.doi.org/10.4161/auto.28363
- Peng N, Meng N, Wang S, Zhao F, Zhao J, Su L, Zhang S, Zhang Y, Zhao B, Miao J. An activator of mTOR inhibits oxLDL-induced autophagy and apoptosis in vascular endothelial cells and restricts atherosclerosis in apolipoprotein E(-)/(-) mice. Scientific Rep 2014; 4:5519
- Jones SA, Mills KH, Harris J. Autophagy and inflammatory diseases. Immunol Cell Biol 2013; 91:250-8; PMID:23318657; http://dx.doi.org/10.1038/icb.2012.82
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323-35; PMID:21248839; http://dx.doi.org/10.1038/nature09782
- Manley S, Williams JA, Ding WX. Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp Biol Med 2013; 238:525-38; http://dx.doi.org/10.1177/1535370213489446
- Choe JY, Jung HY, Park KY, Kim SK. Enhanced p62 expression through impaired proteasomal degradation is involved in caspase-1 activation in monosodium urate crystal-induced interleukin-1b expression. Rheumatology 2014; 53:1043-53; PMID:24587486; http://dx.doi.org/10.1093/rheumatology/ket474
- Meng N, Zhao J, Su L, Zhao B, Zhang Y, Zhang S, Miao J. A butyrolactone derivative suppressed lipopolysaccharide-induced autophagic injury through inhibiting the autoregulatory loop of p8 and p53 in vascular endothelial cells. Int J Biochem Cell Biol 2012; 44:311-9; PMID:22085531; http://dx.doi.org/10.1016/j.biocel.2011.11.001
- Jiang W, Ogretmen B. Ceramide stress in survival versus lethal autophagy paradox: ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Autophagy 2013; 9:258-9; PMID:23182807; http://dx.doi.org/10.4161/auto.22739
- Pessentheiner AR, Pelzmann HJ, Walenta E, Schweiger M, Groschner LN, Graier WF, Kolb D, Uno K, Miyazaki T, Nitta A, et al. NAT8L (N-acetyltransferase 8-like) accelerates lipid turnover and increases energy expenditure in brown adipocytes. J Biol Chem 2013; 288:36040-51; PMID:24155240; http://dx.doi.org/10.1074/jbc.M113.491324
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37:495-500; PMID:15806104; http://dx.doi.org/10.1038/ng1536
- Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol 2009; 334:186-97; PMID:19631203; http://dx.doi.org/10.1016/j.ydbio.2009.07.016
- Burrows C, Abd Latip N, Lam SJ, Carpenter L, Sawicka K, Tzolovsky G, Gabra H, Bushell M, Glover DM, Willis AE, et al. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res 2010; 38:5542-53; PMID:20430826; http://dx.doi.org/10.1093/nar/gkq294
- Pursiheimo JP, Rantanen K, Heikkinen PT, Johansen T, Jaakkola PM. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene 2009; 28:334-44; PMID:18931699; http://dx.doi.org/10.1038/onc.2008.392
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142:409-19; PMID:20673990; http://dx.doi.org/10.1016/j.cell.2010.06.040
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep 2012; 13:971-83; PMID:23070366; http://dx.doi.org/10.1038/embor.2012.145
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071-6; PMID:20393566; http://dx.doi.org/10.1038/nature08975
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013; 152:1308-23; PMID:23498939; http://dx.doi.org/10.1016/j.cell.2013.02.016
- Yan B, Liu J, Yao J, Li X, Wang X, Li Y, Tao Z, Song Y, Chen Q, Jiang Q. LncRNA-MIAT Regulates Microvascular Dysfunction by Functioning as a Competing Endogenous RNA. Circ Res 2015; 116(7):1143-56; PMID:25587098; http://dx.doi.org/10.1161/CIRCRESAHA.116.305510
- Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013; 341:789-92; PMID:23907535; http://dx.doi.org/10.1126/science.1240925
- Chen SS, Hu W, Wang Z, Lou XE, Zhou HJ. p8 attenuates the apoptosis induced by dihydroartemisinin in cancer cells through promoting autophagy. Cancer Biol Ther 2015; 16(5):1-10
- Weis S, Schlaich TC, Dehghani F, Carvalho T, Sommerer I, Fricke S, Kahlenberg F, Mossner J, Hoffmeister A. p8 Deficiency causes siderosis in spleens and lymphocyte apoptosis in acute pancreatitis. Pancreas 2014; 43:1277-85; PMID:25098696; http://dx.doi.org/10.1097/MPA.0000000000000172
- Weis S, Bielow T, Sommerer I, Iovanna J, Malicet C, Mossner J, Hoffmeister A. P8 deficiency increases cellular ROS and induces HO-1. Arch Biochem Biophy 2015; 565:89-94; http://dx.doi.org/10.1016/j.abb.2014.11.007
- Sanchez-Jimenez C, Ludena MD, Izquierdo JM. T-cell intracellular antigens function as tumor suppressor genes. Cell Death Dis 2015; 6:e1669; PMID:25741594; http://dx.doi.org/10.1038/cddis.2015.43
- Forch P, Valcarcel J. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis 2001; 6:463-8; http://dx.doi.org/10.1023/A:1012441824719
- Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 2002; 7:213-21; PMID:12380690; http://dx.doi.org/10.1379/1466-1268(2002)007<0213:VSTROE>2.0.CO;2
- Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY, Guo LJ, Xie H, Zhou HD, Wu XP, et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem 2011; 286:12328-39; PMID:21324897; http://dx.doi.org/10.1074/jbc.M110.176099
- Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem 2006; 281:25001-5; PMID:16793762; http://dx.doi.org/10.1074/jbc.R600010200
- Saddoughi SA, Garrett-Mayer E, Chaudhary U, O'Brien PE, Afrin LB, Day TA, Gillespie MB, Sharma AK, Wilhoit CS, Bostick R, et al. Results of a phase II trial of gemcitabine plus doxorubicin in patients with recurrent head and neck cancers: serum C(1)(8)-ceramide as a novel biomarker for monitoring response. Clin Cancer Res 2011; 17:6097-105; PMID:21791630; http://dx.doi.org/10.1158/1078-0432.CCR-11-0930
- Sentelle RD, Senkal CE, Jiang W, Ponnusamy S, Gencer S, Selvam SP, Ramshesh VK, Peterson YK, Lemasters JJ, Szulc ZM, et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol 2012; 8:831-8; PMID:22922758; http://dx.doi.org/10.1038/nchembio.1059
- Wiame E, Tyteca D, Pierrot N, Collard F, Amyere M, Noel G, Desmedt J, Nassogne MC, Vikkula M, Octave JN, et al. Molecular identification of aspartate N-acetyltransferase and its mutation in hypoacetylaspartia. Biochem J 2010; 425:127-36; http://dx.doi.org/10.1042/BJ20091024
- Ariyannur PS, Moffett JR, Manickam P, Pattabiraman N, Arun P, Nitta A, Nabeshima T, Madhavarao CN, Namboodiri AM. Methamphetamine-induced neuronal protein NAT8L is the NAA biosynthetic enzyme: implications for specialized acetyl coenzyme A metabolism in the CNS. Brain Res 2010; 1335:1-13; PMID:20385109; http://dx.doi.org/10.1016/j.brainres.2010.04.008
- Moscat J, Diaz-Meco MT. The atypical protein kinase Cs. Functional specificity mediated by specific protein adapters. EMBO Rep 2000; 1:399-403; PMID:11258478; http://dx.doi.org/10.1093/embo-reports/kvd098
- Dryden NH, Sperone A, Martin-Almedina S, Hannah RL, Birdsey GM, Khan ST, Layhadi JA, Mason JC, Haskard DO, Gottgens B, et al. The transcription factor Erg controls endothelial cell quiescence by repressing activity of nuclear factor (NF)-kappaB p65. J biol Chem 2012; 287:12331-42; PMID:22337883; http://dx.doi.org/10.1074/jbc.M112.346791
- Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA, Metallo CM, Diaz-Meco MT, et al. Metabolic Reprogramming of Stromal Fibroblasts through p62-mTORC1 Signaling Promotes Inflammation and Tumorigenesis. Cancer Cell 2014; 26:121-35; PMID:25002027; http://dx.doi.org/10.1016/j.ccr.2014.05.004
- Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 1973; 52:2745-56; PMID:4355998; http://dx.doi.org/10.1172/JCI107470
- Wang W, Liu X, Zhao J, Zhao B, Zhang S, Miao J. A novel butyrolactone derivative inhibited apoptosis and depressed integrin beta4 expression in vascular endothelial cells. Bioorganic Med Chem Lett 2007; 17:482-5; http://dx.doi.org/10.1016/j.bmcl.2006.10.023
