Abstract
Aneuploidy, defined as an alteration in chromosome number that is not a multiple of the haploid complement, severely affects cellular physiology. Changes in chromosome number lead to imbalances in cellular protein composition, thus disrupting cellular processes and causing proteins to misfold and aggregate. We recently reported that in mammalian cells protein aggregates are readily encapsulated within autophagosomes but are not degraded by lysosomes. This leads to a lysosomal stress response in which the transcription factor TFEB induces expression of factors needed for macroautophagy-mediated protein degradation. Our studies uncover lysosomal degradation defects as a feature of the aneuploid state, and a role for the transcription factor TFEB in the response thereto.
The changes in gene copy number brought about by aneuploidy translate into changes in protein expression. This leads to dramatic alterations in the cell’s protein composition. It has been proposed that the altered protein composition of aneuploid cells causes increased protein misfolding and aggregation, and proteotoxic stress ensues. To gain insights into how increased protein misfolding affects the protein quality control pathways of aneuploid cells we investigated the consequences of chromosome mis-segregation and aneuploidy on one of the cell’s protein quality control pathways, macroautophagy (hereafter autophagy). We found that following chromosome mis-segregation, autophagosomal constituents accumulate within lysosomes. Analysis of lysosomal function did not reveal any substantive defects. Lysosomal pH and catalytic activity of cathepsins does not appear impaired in aneuploid cells raising the possibility that autophagosomal content is not effectively cleared because lysosomal capacity is limiting rather than compromised. Our studies further suggested that it is protein aggregate-containing autophagosomes that accumulate in the lysosomal compartment. We found SQSTM1/p62, which serves as a protein aggregate receptor in autophagy, to accumulate in lysosomes. Electron microscopy analysis further showed that autolysosomes were filled with electron-dense material. Together, our findings indicate that aneuploidy has a dramatic impact on the degradative capacity of the lysosomal compartment. It will be interesting to define which proteins aggregate in aneuploid cells and whether it is specific protein aggregates that accumulate within lysosomes. Whether autophagic cargo other than protein aggregates such as mitochondria accumulates in the lysosomal compartment in aneuploid cells also remains to be determined.
Cells respond to the accumulation of autophagic cargo in lysosomes. Upon chromosome mis-segregation we observed translocation of the transcription factor TFEB into the nucleus and a concomitant upregulation of TFEB-responsive genes. Our data further showed that inhibition of proteasomal degradation also leads to TFEB activation indicating that aggregated/misfolded proteins can activate TFEB. A key question that remains to be addressed is how aneuploidy controls TFEB activity. Previous studies showed that proteasome inhibition induces autophagy because internal amino acid pools are depleted when protein recycling by the proteasome is inhibited. TFEB could be regulated by this same mechanism. Changes in TFEB’s subcellular localization are accompanied by changes in TFEB phosphorylation, indicating that this posttranslational modification regulates TFEB’s localization in the cell. Furthermore TORC1, the protein kinase complex that is regulated by amino acid availability, controls TFEB phosphorylation and localization. Whether TFEB is activated by this mechanism in aneuploid cells is not known. Our preliminary data indicate that TORC1 is not responsible for TFEB activation in response to chromosome mis-segregation, which leads us to speculate that other signals are responsible for controlling TFEB activity in response to lysosome saturation in aneuploid cells. Identifying the protein kinase(s) that targets TFEB in response to cargo accumulation in the lysosome will be essential in elucidating the mechanism whereby aneuploidy activates the transcription factor.
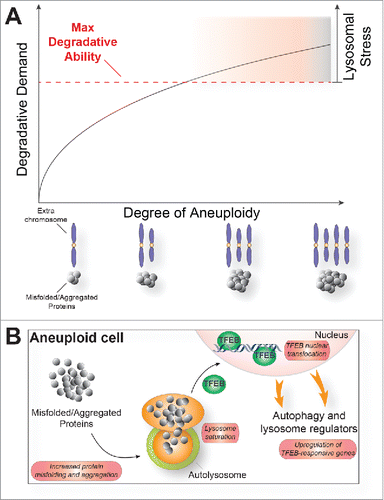
In summary, our study provides evidence that, in mammalian cells, aneuploidy limits lysosomal capacity. This defect does not manifest itself immediately after chromosome mis-segregation, but requires 2–3 cell divisions to become apparent. This finding suggests that lysosomal defects are not an immediate consequence of aneuploidy, but require the continuous presence of an aneuploid karyotype and the proteomic imbalances associated with it. We propose that cells have a maximal degradative capacity that is reached by the persistence of misfolded and/or aggregated proteins (). When this maximal degradative capacity is reached, a lysosomal stress response ensues that is aimed at producing more autophagosomes and lysosomes ().
Figure 1. A model as to how proteotoxic load induces a lysosomal stress response. An increasing load of misfolded and/or aggregated proteins brought about by aneuploidy causes lysosome-mediated protein degradation to become limiting (A). This in turn triggers a lysosomal stress response, which lead to translocation of the transcription factor TFEB from the cytoplasm into the nucleus where it stimulates the transcription of genes involved in autophagic degradation (B).
Our analysis focused on the effects of aneuploidy on autophagy in primary cells. It will be interesting to determine whether chromosome mis-segregation and aneuploidy cause lysosomal saturation in cancer cells. Cancer cells harbor high levels of aneuploidy and are genomically unstable with karyotypes changing continuously. This places a profound pressure on the protein quality control machinery of aneuploid cells. Indeed, cancer cells have previously been shown to not only depend on protein quality control mechanisms for their survival but to also upregulate their protein folding machinery. It is thus not unlikely that other protein quality control pathways such as autophagy are also more active in aneuploid cancer cells. Indeed our preliminary studies suggest that many aneuploid cancer cell lines do not experience lysosomal saturation following chromosome mis-segregation. This result raises the interesting possibility that one aspect of malignant transformation is the development of mechanisms that allow cells to upregulate lysosomal capacity, perhaps by upregulating TFEB activity. Addressing the importance of autophagic degradation for tumorigenesis and determining whether and how autophagic activity is altered in aneuploid cells will be an important future question. The observation that RAS-driven and BRAF-driven cancers upregulate autophagy and are dependent on autophagic degradation for survival already points toward this protein quality control process as playing a critical role in tumorigenesis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institute of Health GM056800 to A.A and the Kathy and the Curt Marble Cancer Research Fund. A.A is also an investigator of the Howard Hughes Medical Institute and the Glenn Foundation for Biomedical Research. S. S. was supported by the American Italian Cancer Foundation (AICF) and by a Fellowship in Cancer Research from Marie Curie Actions and the Italian Association for Cancer Research (AIRC).
