Abstract
GADD45A is a TP53-regulated and DNA damage-inducible tumor suppressor protein, which regulates cell cycle arrest, apoptosis, and DNA repair, and inhibits tumor growth and angiogenesis. However, the function of GADD45A in autophagy remains unknown. In this report, we demonstrate that GADD45A plays an important role in regulating the process of autophagy. GADD45A is able to decrease LC3-II expression and numbers of autophagosomes in mouse tissues and different cancer cell lines. Using bafilomycin A1 treatment, we have observed that GADD45A regulates autophagosome initiation. Likely, GADD45A inhibition of autophagy is through its influence on the interaction between BECN1 and PIK3C3. Immunoprecipitation and GST affinity isolation assays exhibit that GADD45A directly interacts with BECN1, and in turn dissociates the BECN1-PIK3C3 complex. Furthermore, we have mapped the 71 to 81 amino acids of the GADD45A protein that are necessary for the GADD45A interaction with BECN1. Knockdown of BECN1 can abolish autophagy alterations induced by GADD45A. Taken together, these findings provide the novel evidence that GADD45A inhibits autophagy via impairing the BECN1-PIK3C3 complex formation.
Abbreviations
| ACTB/β-actin | = | actin, beta |
| ATG | = | autophagy-related, Baf, bafilomycin A1 |
| BECN1 | = | Beclin 1, autophagy related |
| DAPI | = | 4′,6-diamidino-2-phenylindol dihydrochloride |
| DNA | = | deoxyribose nucleic acid |
| GADD45A | = | growth arrest and DNA-damage-inducible, alpha |
| GFP | = | green fluorescent protein |
| GST | = | glutathione s-transferase |
| LC3/MAP1LC3 | = | microtubule-associated protein 1 light chain 3 |
| MEFs | = | mouse embryonic fibroblasts |
| MTOR | = | mechanistic target of rapamycin (serine/threonine kinase) |
| PIK3C3 | = | phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| RFP | = | red fluorescent protein |
| SDS | = | sodium dodecyl sulfate |
| SQSTM1 | = | sequestosome 1 |
| UVR | = | ultraviolet radiation |
Introduction
Autophagy, a lysosome-dependent protein degradation pathway in eukaryocyte, is essential for cell survival, growth and homeostasis. Autophagy begins with a phagophore,Citation1,2 followed by the phagophore expansion to engulf intracellular cargo, such as protein aggregates, organelles and ribosomes, thereby sequestering the cargo in a double-membrane autophagosome.Citation3 The autophagosome eventually fuses with the lysosome, promoting the degradation of autophagosomal contents by lysosomal acid proteases.Citation3 It has also been reported that autophagy plays important roles in oncogenesis, which depends on cancer types and stages.Citation4-6 On the one hand, autophagy inhibits oncogenesis in certain types of cancer.Citation7-9 On the other hand, some cancer cells can utilize activated autophagy to ameliorate deficiencies in nutrition and oxygen, caused by rapid fission and ischemia.Citation10-12
BECN1/Beclin1, an ortholog of Vps30/Atg6 in yeast, is a central component in autophagy and also plays crucial roles in development, tumorigenesis and neurodegeneration.Citation7 Interestingly, BECN1 was initially discovered not as an autophagy protein but rather as an interaction partner for the antiapoptotic protein BCL2.Citation13 Autophagy and apoptosis, 2 self-destructive processes, have a complicated functional relationship.Citation14 It has been demonstrated that common upstream signals can induce either apoptosis or autophagy in a context-dependent fashion. One well-characterized regulatory factor in autophagy is BCL2, which interacts with BECN1 and disrupts the interaction of BECN1 with PIK3C3.Citation14,15 ROS favors proapoptotic mitochondrial outer membrane permeabilization (MOMP) as well as stimulates the proteolytic activity of ATG4, thereby stimulating autophagy.Citation16,17 Accumulating evidence suggests that TP53 (note that the mouse nomenclature is TRP53, but we use TP53 hereafter to refer to both the human and mouse genes/proteins for simplicity), a quintessential tumor suppressor and apoptosis inducer, regulates autophagy in a 2-faced fashion.Citation18-21 On the one hand, TP53 functions as a nuclear transcription factor and transactivates proapoptotic, cell cycle-arresting and proautophagic genes. On the other hand, cytoplasmic TP53 can operate at mitochondria to promote cell death and can repress autophagy via poorly characterized mechanisms.Citation19,22
GADD45A, a TP53-regulated and DNA-damage responsive protein, is induced by varieties of genotoxic stress agents, such as UV radiation (UVR), ionizing radiation (IR), alkylating agents and hydroxyurea.Citation23-28 A number of investigations indicate that GADD45A can induce cell cycle arrest,Citation29 apoptosis,Citation30 DNA damage repairCitation31 and angiogenesis inhibition.Citation32 Increasing evidence shows high frequency mutations or abnormal epigenetic modifications of GADD45A in pancreatic cancer, breast cancer and prostate cancer.Citation33-35 In vivo experiments have demonstrated that gadd45a knockout mice are susceptible to DNA-damage inducible tumors.Citation36 Thus GADD45A is an important tumor-suppressor gene. GADD45A is involved in the induction of apoptosis through inducing BCL2L11/Bim dissociation from the cytoskeleton and translocation to mitochondria. However, the possible functions and molecular mechanisms of GADD45A in autophagy are poorly understood.
In this report, we investigated the role of GADD45A in the regulation of autophagic system and found that GADD45A inhibited autophagy through impairing the formation of the BECN1-PIK3C3 complex. Furthermore, we observed that GADD45A interacted with BECN1 and disrupted the interaction between BECN1 and PIK3C3. The region harboring amino acids 71 to 81 of GADD45A was necessary and sufficient for interaction with BECN1. These findings demonstrate the negative role and the specific mechanism of GADD45A in autophagy.
Results
Deletion of GADD45A causes extensive autophagy
To explore the roles of GADD45A in the control of autophagy, gadd45a knockout mice (gadd45a−/−) were employed. We first examined the amounts of LC3 protein, an indicator of autophagic activity, in several organs including liver, lung and kidney using a LC3B-specific polyclonal antibody. As shown in , an approximately 18 kDa band representing LC3-I was observed in all preparations and there was a pronounced increase in the autophagosome-associated lipidation form LC3-II with apparent molecular mass of 16 kDa in the gadd45a−/− mouse organs, compared with Gadd45a+/+ mice (). The expression levels of SQSTM1, a cargo receptor for degradation of damaged or long-lived proteins that is often used as a protein marker of autophagy, were also evidently lower in gadd45a−/− mouse organs than that seen in Gadd45a+/+, suggesting that GADD45A might inhibit the autophagy process. Furthermore, we analyzed the numbers of autophagosomes by confocal microscopy assays in liver cells. There were large amounts of small bright green granular substances in gadd45a−/− mice liver cells, while only a few puncta were seen in the normal control ().
Figure 1. Elevated expression of LC3-II in different tissues of gadd45a−/− mice. (A) Western blot analysis of LC3-II levels in liver, lung and kidney tissues of mice. All tested tissues homogenates were loaded and analyzed by 15% SDS-PAGE. (B, F) The densities of signals were determined by densitometry and are shown relative to the Gadd45a+/+ group. Graphical data denote mean ± SD. (C) Confocal microscopy images and immunofluorescence analysis with anti-LC3 antibody. Scale bar: 10 μm. (D, H) The number of LC3 puncta per cell. Graphical data denote mean ± SD. (E) SQSTM1, LC3 and GADD45A expression variation were analyzed using western blots in MEF Gadd45a+/+ and gadd45a−/− cells. (G) MEF Gadd45a+/+, gadd45a−/− cells were seeded onto the cover slips and analyzed by immunofluorescence with anti-LC3 antibody. Scale bar: 10 μm. ***,P < 0.001; **,P < 0.01.
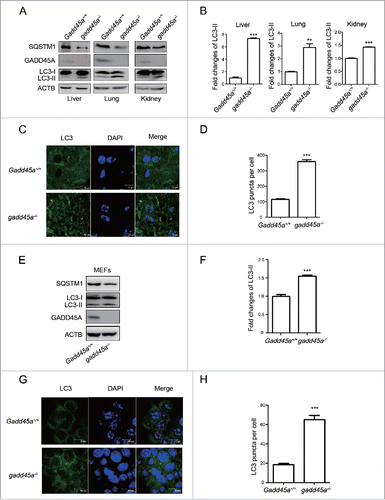
Next, we tested the expression of LC3 and SQSTM1 in Gadd45a+/+ and gadd45a−/− mouse embryonic fibroblasts (MEFs) and found that LC3-II-specific proteins dramatically increased and SQSTM1 decreased expression in the gadd45a−/− MEFs (). Similarly, when the numbers of LC3 puncta per cell were scored, bright green LC3 punctate signals were significantly observed in the cytoplasm of the gadd45a−/− MEFs (). These data clearly highlight a speculation that autophagosome formation is inhibited by GADD45A.
GADD45A levels negatively associates with the number of autophagosomes in tumor cells
To further investigate the association between the GADD45A level and autophagy, we generated GADD45A-stably-silenced clones with shRNA in KYSE30 cells and transiently knocked down GADD45A by RNA interference (RNAi) in KYSE150 cells. Real-time PCR and western blot results showed that the expression of GADD45A was significantly downregulated as shown in Figure S1. Downregulation of GADD45A resulted in an increased expression of LC3-II and a decreased expression of SQSTM1, compared with the control (). In addition, large amounts of small bright green granular substances were observed in the GADD45A knockdown cells, whereas evenly distributed green LC3 signals with a few puncta were seen in the control (, S2). These results indicate that the reduced expression of GADD45A leads to an increased number of autophagosomes.
Figure 2. GADD45A suppressed the expression of LC3-II in tumor cells. (A) GADD45A knockdown promotes the expression of LC3-II. KYSE30-H1 and KYSE30-shRNA cells were harvested at 70% to 80% confluence. KYSE30-shRNA cells were GADD45A-stably-silenced clones with specific shRNA and KYSE30-H1 was negative control cells. KYSE150 cells were harvested 48 h after transfection with siRNAs. SQSTM1, LC3, GADD45A and ACTB expression variation was analyzed using western blots. NC was the negative control. (B, F) The densities of signals were determined by densitometry and are shown relative to the control group. The data were presented in the form of mean ± SD. (C) KYSE30-H1 and KYSE30-shRNA cells were seeded onto the cover slips and analyzed by immunofluorescence with anti-LC3 antibody. Confocal microscopy images, scale bar: 10 μm. (D) The number of LC3 puncta per cell. Graphical data denote mean ± SD. (E) 293T, HeLa and EC9706 cells were transiently transfected with pCS2-MT or pCS2-MT-GADD45A vector. These cells were harvested at 48 h after transfection. A total of 50 μg of whole-cell protein was examined for SQSTM1, LC3, GADD45A and ACTB expression. (G, H) Cells were seeded onto the cover slips precoated with polylysine and analyzed by immunofluorescence with anti-LC3 antibody. Confocal microscopy images, scale bar: 10 μm. The number of LC3 puncta per cell. Graphical data denote mean ± SD. ***,P < 0.001; **,P < 0.01.
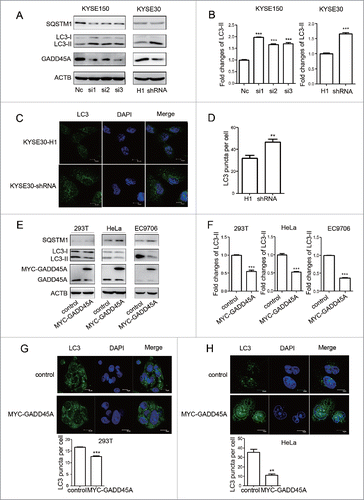
We also produced overexpression of GADD45A in 293T, HeLa and EC9706 cells transfected with the MYC-GADD45A vector. Compared with the control, overexpression of GADD45A suppressed LC3-II expression and induced SQSTM1 expression, which is an opposite effect to the GADD45A knockdown cells (). Confocal microscopy assays also showed that the numbers of autophagosomes in GADD45A-overexpressing cells were decreased (, S3), indicating that elevated expression of GADD45A inhibits the formation of autophagosomes. Taken together, these results corroborate that GADD45A is negatively associated with the formation of autophagosomes in tumor cells.
GADD45A blocks autophagic flux
Autophagy is a highly dynamic and multistep process, which can be modulated at several steps. An accumulation of autophagosomes could reflect the induction of autophagy or the reduction in autophagosome turnover.Citation37 Bafilomycin A1 (Baf), a small molecule inhibitor of autophagy, induces the accumulation of autophagosomes through blocking the fusion of autophagosomes and lysosomes. Baf treatment, which is a well-established method for monitoring autophagosome synthesis, can be used to reveal the mechanism of autophagosome accumulation. We then examined the expression of LC3-II in GADD45A knockdown cells treated with Baf and found that depletion of GADD45A resulted in increased LC3-II levels in KYSE30 and KYSE150 cells following Baf treatment, compared with the control cells treated with Baf (). Similar results were found in gadd45a−/− MEF cells (Fig. S4). Additionally, the level of LC3-II expression increased after exposure to Baf in cells transfected with control vector, but the decreased tendency of LC3-II expression caused by GADD45A was not changed in Baf-treated 293T, HeLa and EC9706 cells transfected with MYC-GADD45A vector (). These results indicate that GADD45A affects autophagosome formation.
Figure 3. The effect of GADD45A on autophagic flux. (A) KYSE30-H1 and KYSE30-shRNA cells were treated with Baf at 70% to 80% confluence. KYSE150 cells were treated with Baf after GADD45A was knocked down by siRNA. Cell lysates were prepared for analyzing the expression of LC3 and GADD45A. (C) 293T, HeLa and EC9706 cells were treated with Baf after transfection with pCS2-MT or pCS2-MT-GADD45A. LC3 and GADD45A were analyzed using western blots. (B, D) The densities of signals were determined by densitometry and are shown relative to the control group. Graphical data denote mean ± SD. (E, F) HeLa cells were transiently transfected with pCS2-MT, ptfLC3 or pCS2-MT-GADD45A, ptfLC3 vector. Cells were seeded onto the cover slips precoated with polylysine 24 h later after transfection and analyzed by immunofluorescence to detect the expression of RFP and GFP. KYSE30-H1 or KYSE30-shRNA cells were transfected with ptfLC3 vector and seeded onto the cover slips precoated polylysine. Immunofluorescence analyzed the expression of RFP and GFP. Scale bar: 10 μm. ***,P < 0.001.
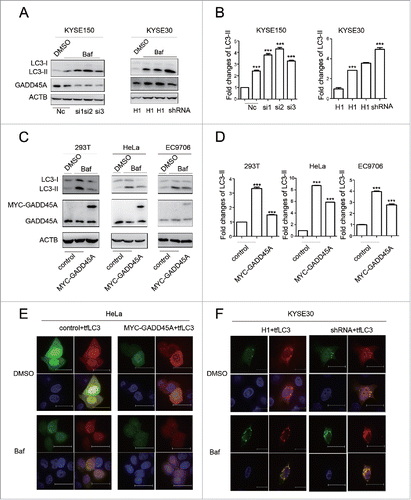
To further corroborate the above findings, tandem mRFP-GFP-LC3 reporter (tfLC3 plasmid) was transfected into tumor cell lines, in which autophagosomes trigger both RFP and GFP signals, whereas the autolysosomes emit only RFP signal because of quenching of the GFP in the acidic lysosomal environment. GADD45A overexpression compromised green (vesicles that appear yellow in the merged images) and red autophagosomes in HeLa cells. Similarly, after exposure to Baf, the number of both green and red autophagosomes still decreased in GADD45A-overexpressing cells compared with cells transfected with the control vectors (, S5A). In contrast, the number of green and red vesicles dramatically increased in GADD45A knockdown cells without or with Baf treatment (, S5B). These results suggest that GADD45A is involved in the inhibition of autophagosome formation at an early stage.
Next, we analyzed the inhibitory effect of GADD45A on autophagy flux in the same cell lines. The expression of LC3-II and the number of both green and red dots decreased in GADD45A-overexpressing KYSE30 cells without or with Baf treatment (Fig. S6A, S6C). Conversely, knockdown of GADD45A also led to enhancement of autophagic activities and the increased tendency was not changed after Baf treatment in GADD45A knockdown HeLa cells either through western blot or fluorescence analysis (Fig. S6B, S6D). These data further prove that the expression levels of GADD45A in cells are important for regulating autophagic flux.
GADD45A impairs the formation of the autophagy initiation complex
The MTOR pathway and the BECN1-PIK3C3 complex pathway are the most important negative and positive regulators for autophagy in mammalian cells, respectively.Citation38,39 To explore the possible alterations in those pathways, we measured the expression levels of MTOR and p-MTOR in gadd45a knockout MEFs and knockdown tumor cells using western blots. The results showed little effect of GADD45A on MTOR and p-MTOR expression (Fig. S7).
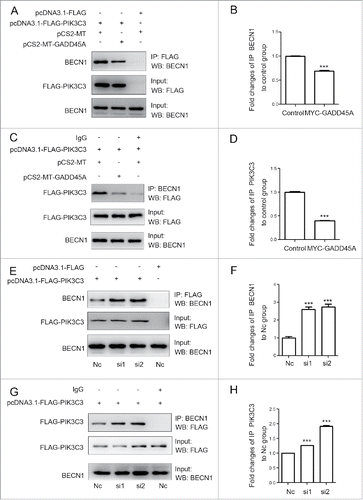
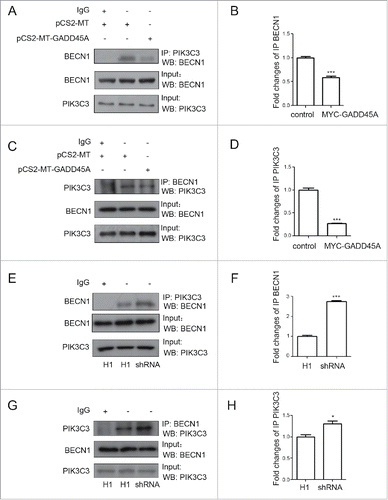
We then tested the expression of BECN1, which appears to act as a nexus point between autophagy and cell death pathways. The result showed that neither GADD45A overexpression nor deficiency changed the expression of BECN1. BECN1 is a part of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex and is required for the initiation of the autophagosome formation in autophagy. Human PIK3C3/VPS34 is the catalytic subunit of PtdIns3K. To test whether GADD45A is associated with the BECN1-PIK3C3 complex, 2 plasmids (pcDNA3.1-FLAG and pCS2-MT, pcDNA3.1-FLAG-PIK3C3 and pCS2-MT-GADD45A, pcDNA3.1-FLAG-PIK3C3 and pCS2-MT) were cotransfected into HeLa cells. Cells were lysed for immunoprecipitation (IP) with FLAG or BECN1 antibodies, and the immunocomplexes were subjected to western blot assays. As reported previously, BECN1 could interact with PIK3C3. Interestingly, we found that anti-FLAG antibody precipitated less BECN1 protein in GADD45A-overexpressing HeLa cells, compared with cells transfected with the control vectors (). There was also less coprecipitation of FLAG-PIK3C3 with BECN1 in the immunocomplexes precipitated by anti-BECN1 antibody (). In contrast, in GADD45A knockdown cells, there were more amount of copurifying BECN1 protein precipitated by FLAG magnetic beeds and BECN1 also precipitated more FLAG-PIK3C3 protein compared with the negative control cells (). Collectively, these data imply that GADD45A is able to disrupt the formation of the BECN1-PIK3C3 complex that is essential for autophagy initiation.
Figure 4. GADD45A impaired the formation of the BECN1-PIK3C3 complex. (A, B, C, D) HeLa cells were harvested after cotransfection with pCS2-MT and pcDNA3.1-FLAG-PIK3C3 or pCS2-MT-GADD45A and pcDNA3.1-FLAG-PIK3C3 or pCS2-MT and pcDNA3.1-FLAG. (A) Immunoprecipitation was performed with anti-FLAG M2 magnetic beads and the precipitated complexes were analyzed by immunoblots with anti-BECN1 antibody. (C) Immunoprecipitation was performed with BECN1 antibody. Precipitated complexes were analyzed by immunoblots with anti-FLAG antibody. (E, F, G, H) KYSE150 cells were harvested after cotransfection with negative control and pcDNA3.1-FLAG-PIK3C3 or RNAi and pcDNA3.1-FLAG -PIK3C3 or negative control and pcDNA3.1-FLAG. (E) Immunoprecipitation was performed with anti-FLAG M2 magnetic beads and the precipitated complexes were analyzed by immunoblots with anti-BECN1 antibody. (G) Immunoprecipitation was performed with BECN1 antibody. FLAG-PIK3C3 was analyzed using western blots in immunoprecipitated complexes. (B, D, F, H) The densities of signals were determined by densitometry and are shown relative to the control group or the H1 group. Graphical data denote mean ± SD. ***,P < 0.001.
To analyze the influence of GADD45A on the endogenous BECN1 and PIK3C3 interaction, coimmunoprecipitation was used to test the interaction in GADD45A overexpression or deficiency cells. Clearly, anti-PIK3C3 antibody precipitated less endogenous BECN1 in GADD45A-overexpressing cells but it did more in GADD45A knockdown cells compared with the control cells (). Likewise, the levels of endogenous PIK3C3 precipitated by BECN1 were affected by expression of GADD45A (). These data confirm the function of GADD45A in the process of autophagy, which was shown in the above transfection experiments.
Figure 5. GADD45A disrupted the endogenous BECN1 and PIK3C3 interaction. (A, B, C, D) HeLa cells were harvested after transfection with pCS2-MT or pCS2-MT-GADD45A. (A, E) Immunoprecipitation was performed with PIK3C3 antibody. BECN1 was analyzed using western blots in immunoprecipitated complexes. (C, G) Immunoprecipitation was performed with BECN1 antibody. PIK3C3 was analyzed using western blots in immunoprecipitated complexes. (B, D, F, H) The densities of signals were determined by densitometry and are shown relative to the control group or the H1 group. Graphical data denote mean ± SD. *,P < 0.05; ***,P < 0.001.
Two distinct PtdIns3-kinase complexes are well known: The ATG14 complex (PIK3C3, PIK3R4/VPS15, BECN1, ATG14 and NRBF2) is required for autophagosome formation,Citation40 and the UVRAG complex (PIK3C3, PIK3R4, BECN1 and UVRAG) has a unique activity in the endocytic pathway.Citation41,42 ATG14 can interact with BECN1 and PIK3C3. Thus, we checked the influence of GADD45A on the interaction between ATG14 and BECN1. But as shown in the Figure S8, there was no significant effect of GADD45A on the interaction between BECN1 and ATG14.
GADD45A interacts with BECN1
To investigate the underlying mechanism by which GADD45A interferes with the interaction between BECN1 and PIK3C3, we examined the physical associations among GADD45A, BECN1 and PIK3C3. Cellular lysates from HeLa cells cotransfected with MYC-GADD45A and FLAG-BECN1 or FLAG control vectors were immunoprecipitated with anti-FLAG M2 magnetic beads. After immunoblotting analysis with MYC antibodies, MYC-GADD45A was detected in the HeLa cells transfected with FLAG-BECN1 but not in cells transfected with FLAG control vectors (). Cells were also lysed for IP with GADD45A antibodies, and BECN1 was detected in the immunocomplexes by western blot assays (). Similar results were also found in HeLa cells cotransfected with MYC-GADD45A and FLAG-PIK3C3 or FLAG control vectors (). These data indicate that GADD45A interacts with the BECN1-PIK3C3 complex. Moreover, we performed a glutathione S-transferase (GST) affinity isolation experiment to determine which components of the BECN1-PIK3C3 complex were associated with GADD45A. To do this, GST-GADD45A fusion protein was incubated with cellular lysates isolated from HeLa cells, followed by western blots. Clearly, GST-GADD45A was able to pull down BECN1 compared with GST alone (), but PIK3C3 did not interact with GST-GADD45A (results not shown). To further confirm the interaction between GADD45A and BECN1, we examined their subcellular localizations in HeLa cells by staining with MYC and BECN1 antibodies, respectively. Confocal microscopy assays exhibited a significant colocalization between BECN1 (bright green particles) and GADD45A (bright red) (), but the colocalization between GADD45A and PIK3C3 was not clearly seen (Fig. S9). Furthermore, the endogenous BECN1 and GADD45A interaction was verified in HeLa cells by coimmunoprecipitation (). Taken together, these results indicate a physical association between GADD45A and BECN1.
Figure 6. The interaction between GADD45A and the BECN1-PIK3C3 complex. (A, B) HeLa cells were harvested after cotransfection with pCS2-MT-GADD45A and pcDNA3.1-FLAG or pCS2-MT-GADD45A and pcDNA3.1-FLAG-PIK3C3 or pCS2-MT-GADD45A and pcDNA3.1-FLAG-BECN1. (A) Immunoprecipitation assays were performed with anti-FLAG M2 magnetic beads and the precipitated complexes were analyzed by immunoblots with anti-MYC antibody. (B) Immunoprecipitation was performed with GADD45A antibody. Precipitated complexes were analyzed by immunoblots with anti-FLAG antibody. (C) GST affinity isolation assay was performed with GST-GADD45A and cell lysates from exponentially growing HeLa cells. GST tag alone was used as internal control. The binding proteins were detected with specified antibodies. (D) HeLa cells were harvested after transfection with pCS2-MT-GADD45A. Double immunofluorescence staining for MYC and BECN1 analyzed colocalization. Scale bar: 10 μm. (E) Immunoprecipitation was performed with GADD45A or BECN1 antibody and the precipitated complexes were analyzed by immunoblots with the other antibody in HeLa cells. (F) A series of MYC-tagged-GADD45A deletion mutants were introduced into HeLa cells via transient transfection. Immunoprecipitation were performed with BECN1 antibody. Precipitated complexes were analyzed by immunoblots with MYC and PIK3C3 antibodies.
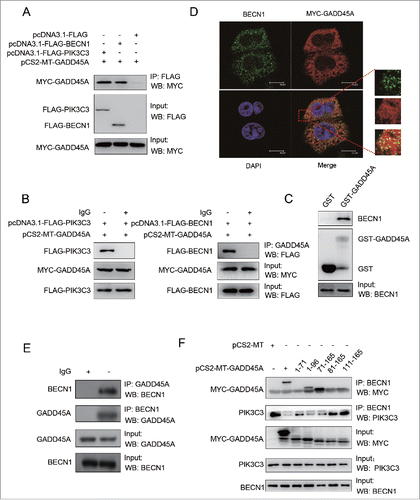
Next, we sought to identify the regions of GADD45A that are required for interaction with BECN1 using a series of MYC-tagged GADD45A deletion protein expression vectors. Cell lysates from HeLa cells transfected with MYC-tagged GADD45A deletion vectors were immunoprecipitated with antibody to BECN1 and blotted with anti-MYC antibody. As shown in , MYC-GADD45A deletion was found in transfected HeLa cells. After incubation with BECN1 antibody, full-length MYC-GADD45A protein was apparently detected in immunocomplexes, while deletion of the carboxyl terminus of GADD45A in (1-96) GADD45A and deletion of the N-terminus of GADD45A in (71-165) GADD45A still retained their binding activity for the interaction of GADD45A with BECN1. However, (1-71) GADD45A, (81-165) GADD45A and (111-165) GADD45A were deficient for BECN1 binding activity. These observations suggest that the central region of the GADD45A protein (amino acids between 71 and 81) is essential for its interaction with BECN1. Along with these observations, interaction between BECN1 and PIK3C3 was declined in immunocomplexes containing GADD45A (1-96, 71-165), but GADD45A (1-71, 81-165 and 111-165) did not interfere the interaction. Collectively, these data demonstrate that GADD45A central region (aa 71 to 81) is necessary and sufficient for the interaction between GADD45A and BECN1.
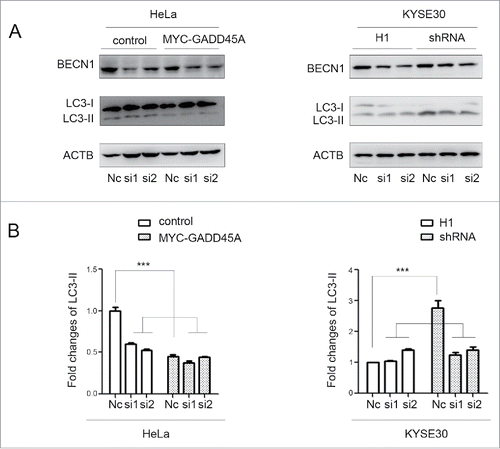
In order to further demonstrate whether the interaction between GADD45A and BECN1 mediates the role of GADD45A in autophagy, we knocked down BECN1 in GADD45A knockdown cells (KYSE30) and overexpressing cells (HeLa). As shown in and 7B, the overexpression of GADD45A in HeLa cells substantially suppressed autophagy, while knockdown of BECN1 resulted in negligible decrease of LC3-II, indicating that BECN1 mediates GADD45A-induced autophagy inhibition. The similar result was detected in GADD45A knockdown KYSE30 cells. Taken together, the interaction between GADD45A and BECN1 is required, to a great extent, for the role of GADD45A in the regulation of autophagy.
Figure 7. The downward trend of LC3-II expression after cells silencing BECN1. HeLa cells were harvested 48 h after cotransfection with pCS2-MT and siRNA or pCS2-MT-GADD45A and siRNA. KYSE30-H1 and KYSE30-shRNA cells were harvested after BECN1 was knocked down by siRNA. (A) Cell lysates were prepared for analyzing the expression of LC3 and BECN1. (B) The densities of signals were determined by densitometry and are shown relative to the Nc group. Graphical data denote mean ± SD. ***,P < 0.001.
Discussion
The GADD45 (growth arrest and DNA-damage-inducible) proteins have been implicated in regulation of many cellular functions including DNA repair, cell cycle control, senescence and genotoxic stress. The GADD45 gene family encodes 3 related GADD45 protein including GADD45A, GADD45B and GADD45G. Functions of GADD45 family proteins are similar, but not identical, and their induction differs under diverse physiological conditions or in different cell types.Citation43 GADD45A, one of the most critical downstream targets of tumor suppressor TP53 and BRCA1,Citation44 can regulate a range of cellular processes, including DNA damage repair, proliferation, cell cycle checkpoint, apoptosis and tumor angiogenesis. Autophagy is an important lysosomal degradation pathway in tumorigenesis and development. Many molecular components of autophagy have been identified, but little is known about the regulatory mechanisms underlying their effector functions. Recent research demonstrated that GADD45B inhibited autophagy through regulating phosphorylation of MAPK14, but GADD45A could not induce MAPK14 phosphorylation.Citation45,46 Hence, these findings prompted us to study whether GADD45A can induce an autophagy response and what is the underlying mechanism. In this study, through western blot and immunofluorescence assays for specific markers of the autophagic system, we found that the number of autophagosomes in GADD45A knockout mice and MEF cells significantly increased as a result of GADD45A deletion. Further, GADD45A overexpression resulted in a significant reduction of LC3-II expression and autophagosomes amount in multiple tumor cell lines. Thus, GADD45A, an important tumor suppressor gene, may regulate cancer malignant development through autophagy.
Autophagy is a highly dynamic and multi-step process. The steady state levels of LC3-II are affected by both synthesis and degradation. One approach to assess the effects on autophagy by clamping LC3-II and autophagosome degradation is to use Baf, a small molecule inhibitor of autophagy.Citation37 In our study, we found that the expression of LC3-II increased in GADD45A knockdown cells and decreased in GADD45A overexpressing cells compared with the control after cells treated with Baf. The tendency is in agreement with the observations in cells without Baf treatment. In addition, we also obtained similar results in examining RFP-GFP-LC3 protein that is used to detect autolysosomes. Therefore, these findings demonstrate that GADD45A may impair autophagy at early stage of autophagosome formation.
With regard to the mechanism by which GADD45A regulates autophagy, we found that the regulatory effect of GADD45A on autophagy is closely related to the formation of the BECN1-PIK3C3 autophagy initiation complex. GADD45A is able to physically associate with BECN1, but not PIK3C3. The interaction between GADD45A and BECN1 dissociates the BECN1 and PIK3C3 complex. Further evidence in the study shows that the central region of GADD45A (aa 71 to 81) is critical for GADD45A interaction with BECN1. Thus, it can be concluded that GADD45A directly interacts with BECN1 to destabilize the BECN1-PIK3C3 complex, and in turn abrogates the autophagy process (). But we found that there was no significant effect of GADD45A on the interaction between BECN1 and ATG14. BECN1 forms multiple complexes with PIK3C3: ATG14-BECN1-PIK3C3-PIK3R4, UVRAG-BECN1-PIK3C3-PIK3R4, BECN1-PIK3C3. BECN1 and PIK3C3 are the core components of these complexes. Thus GADD45A disruption of the interaction between BECN1 and PIK3C3 can also affect the whole complex integrity, that could be the ATG14 complex or it could be the UVRAG complex.
Figure 8. Regulation of GADD45A in autophagy in cancer cells. (A) In normal conditions, GADD45A directly interacts with BECN1 to destabilize the BECN1-PIK3C3 complex, and in turn abrogates the autophagy process. GADD45A is an important regulator of autophagy. (B) GADD45A can be induced by varieties of genotoxic stress agents, such as UVR, IR, alkylating agents and hydroxyurea. After radiotherapy and chemotherapy, GADD45A induced by stresses in cancer cells may dramatically inhibit autophagy to reduce tumor recurrence. Thus, GADD45A may be an important target in anticancer therapy.
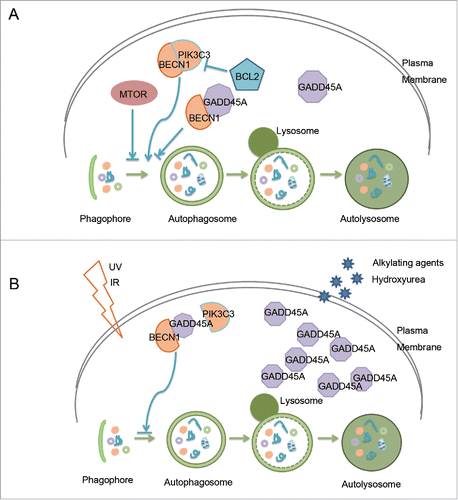
Apoptosis and autophagy are both tightly regulated biological processes that play a central role in tissue homeostasis, development and disease. Apoptosis occurs spontaneously in malignant tumors, often markedly retarding their growth, and it is increased in tumors responding to irradiation, cytotoxic chemotherapy, heating and hormone ablation.Citation47,48 Autophagy is a double-edged sword which can either kill cancer cells or protect them from unfavorable conditions. At the early stage of tumorigenesis, autophagy functions as a tumor suppressor, but at advanced stages of tumor development, autophagy promotes tumor progression.Citation49 Multiple studies suggest that autophagy is a protective response through which cancer cells acquire resistance to radiotherapy and chemotherapy. Cancer stem cells are highly resistant to ionizing radiation and other anticancer treatments with increased autophagy.Citation49 Meanwhile, GADD45A is induced by varieties of genotoxic stress agents including ionizing radiation and alkylating agents. In our previous report, we demonstrated that GADD45A can induce apoptosis, which is coupled with BCL2L11 release from the microtubule associated dynein motor complex and translocation to the mitochondria.Citation30 In this study, we show that GADD45A inhibits autophagy through the interaction with BECN1. Thus, as a tumor suppressor, GADD45A may induce cancer cell death through 2 different pathways during the process of tumorigenesis and development. We hypothesize the possible role of GADD45A in cancer treatment that the increased expression of GADD45A after radiotherapy and chemotherapy in cancer cells induces apoptosis and inhibits autophagy to reduce tumor recurrence. Thus, GADD45A may be an important target in anticancer therapy ().
In summary, our study demonstrated that GADD45A may function as a negative regulator in autophagy via affecting the interaction between BECN1 and PIK3C3. These findings explore a new function of GADD45A and unveil the mechanisms of GADD45A in regulating autophagy. Therefore, GADD45A is able to induce cancer cells death through 2 important manners: apoptosis and autophagy, and may act as an important potential target for clinical anticancer treatment, especially after radiotherapy and chemotherapy.
Materials and Methods
Mice
Gadd45a knockout mice were a kind gift from Professor Albert J. Fornace (Georgetown University), and were maintained in a C57BL/6 (CD45.2) background. The Animal Care and Ethics Committee at Cancer Institute and Hospital, Chinese Academy of Medical Sciences approved all animal experiments in our study. Animal housing and experimental protocols were in accordance with the Chinese Regulations for the Administration of Affairs Concerning Experimental Animals.
Cell culture and treatment
Mouse embryonic fibroblasts that were Gadd45a+/+ and gadd45a−/− transformed by Ras/E1A were kindly provided by Professor Fornance. The 2 MEFs, KYSE150, KYSE30, 293T, HeLa and EC9706 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, 11995065), containing 10% fetal bovine serum (Invitrogen, 10099141), 100 mg/ml streptomycin and 100 U/ml penicillin (Invitrogen, 15140122), at 37°C with 5% CO2. KYSE30 cells (human esophageal squamous cell carcinoma; provided by Dr. Yutaka Shimada, Kyoto University, Japan) were transfected with pSIH-H1 vector encoding scrambled sequences or pSIH-shRNA vector encoding the target sequence against GADD45A. Forty-eight hours after transfection, stably silenced clones were selected by 1 μg/ml puromycin. Twenty percent to 30% of confluent cells were transfected with 50 nM of siRNAs and 60% to 70% of confluent cells were transfected with 2 µg plasmids using Lipofectamine 2000 (Invitrogen, 11668019) following the manufacturer’s instructions. For transient transfections, cells were exposed to 400 nM Baf (Sigma, B1793) 4 h after transfected 44 h. MEFs Gadd45a+/+, gadd45a−/− and KYSE30 (H1, shRNA) cells were harvested after treated with 400 nM Baf for 4 h.
Reagents and plasmids
Bafilomycin A1 (Baf) was purchased from Sigma (Sigma, B1793). SiRNAs for GADD45A and BECN1 were synthesized by Invitrogen. Antibodies against GADD45A, BECN1 were ordered from Cell Signaling Technology (4632, 3738). Antibody against GADD45A (Santa Cruz Biotechnology, sc-797) was used to detect the expression of GADD45A in mouse tissues and cell lines. Commercial rabbit polyclonal antibodies anti-LC3 and anti-SQSTM1 were from Sigma-Aldrich (L8918, P0067). Rabbit polyclonal antibody anti-ATG14 was from MBL (PD062). Antibody against PIK3C3 was ordered from Invitrogen (382100). Mouse anti-ACTB, anti-FLAG monoclonal antibodies and anti-FLAG M2 magnetic beads were also from Sigma Aldrich (A1719, F3165, M8823). Mouse anti-MYC monoclonal antibody was purchased from Millipore (M047). MYC-GADD45A vector was described in our previous study.Citation30 Plasmid ptfLC3, pcDNA3.1-FLAG, pcDNA3.1-FLAG-BECN1, pcDNA3.1-FLAG-PIK3C3 were kindly provided by Dr. Yin Xu (Chinese center for Disease Control and Prevention).
Western blot assay
After transfection with plasmid or siRNA for 48 h, cells were harvested in RIPA buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100 [Amresco, 0694], 0.1% Na-deoxycholate [Sigma, T0875], 0.1% SDS [Amresco, 0227] and 5 mM EDTA [ABI, 15576028]) containing 1% Nonidet P-40 (Amresco, J619), 1 × protease inhibitors cocktail (Roche, P9599) and 50 mg/ml phenylmethylsulphonyl fluoride. A total of 30 µg cellular proteins was subjected to 15% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane. Membranes were blocked for 1 h at room temperature in 2% BSA and incubated with the indicated antibodies overnight at 4°C. Membranes were washed 3 times in 1 × TBS (Amresco, J640) with 0.1% Tween 20 (Amresco, 0777), followed by incubation with anti-mouse (CST, 7076) or anti-rabbit (Abcam, 6721) secondary antibodies conjugated to horseradish peroxidase (HRP). After incubation with HRP substrate (Pierce, 32209), the chemiluminescence signal was detected with a Luminescent Image Analyzer LAS-4000 (Fujifilm, Tokyo, Japan) and analyzed by the Multi Gauge V3.2 software.
Immunofluorescent staining
Cells transfected with siRNA or plasmids for 24 h, were placed in glass slides in 6-well microplates. After 24 h, cells were subjected to fixation with cold methanol at −20°C and washed with phosphate-buffered saline (PBS; Amresco, 0780). Cells were blocked in 1% normal goat serum (Santa Cruz Biotechnology, SC2043) and then incubated with the 1:200-diluted, indicated antibody at 4°C for overnight. The slides were subsequently incubated with Alexa Fluor 488-labeled or Alexa Fluor 568-labeled secondary antibody (Invitrogen, A-11034, A-11004) for 1 h. Finally, nuclei were detected by 1 μg/ml DAPI. The slides were then mounted in permount and analyzed by confocal microscopy Leica ST2 (Leica, Mannheim, Germany). The numbers of LC3 puncta were counted according to previous guidelines.Citation37
GST affinity isolation and immunoprecipitation
GST-GADD45A fusion protein was induced in E.coli with 0.2 mM IPTG (isopropyl-β-D-thiogalactopyranoside; Amresco, 0487) at 31°C overnight. Bacteria were harvested by centrifugation at 12,000 g for 10 min, washed with PBS and resuspended in cold STE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl). After the addition of 100 µg/ml lysozyme solution (Millipore, 71110) and 10 nM DTT (Amresco, 0281), pellets were broken by sonication for 1 min. After centrifugation at 12,000 g for 20 min, the supernatant fractions were transferred to fresh tubes, and Triton X-100 was included at a final concentration of 2%. The mixtures were incubated at room temperature for 30 min prior to the incubation with the glutathione-agarose beads (BD, 554780) at 4°C overnight. The glutathione-agarose beads conjugated to GST fusion proteins were washed 3 times with PBS and mixed with cellular lysates (1 mg) at 4°C for 6 h. At last, beads was washed 5-7 times with lysis buffer and boiled in loading buffer. The binding proteins were analyzed by western blot assay.
For immunoprecipitation, cellular lysates were incubated with 2 µg antibody and 20 µl protein A/G agarose beads (Santa Cruz Biotechnology, sc2003) or Anti-FLAG M2 magnetic beads at 4°C overnight. Beads with immunocomplexes were treated as for the GST affinity isolation assay. Rabbit or mouse isotype antibody (Santa Cruz Biotechnology, sc2025 or sc2027) was used as negative controls.
Statistical analysis
Statistical analysis was performed using the SPSS17.0 statistical package. Quantitative analysis of immunoblotting images was performed using ImageJ software. The values of each target blot were evaluated. Data were presented as mean ± SD. Means of 2 groups were compared using the Student t test, with a 2-tailed P value. P values less than 0.05 were considered to be statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
KAUP_A_1112484_Supplemental.zip
Download Zip (6.6 MB)Acknowledgments
Plasmid ptfLC3, pcDNA3.1-FLAG, pcDNA3.1-FLAG-BECN1, pcDNA3.1-FLAG-PIK3C3 were kindly provided by Dr. Yin Xu from Chinese Center for Disease Control and Prevention. The ESCC cell lines (KYSE30, KYSE150) were provided by Dr. Yutaka Shimada of Kyoto University. Gadd45a−/− mouse and Mouse embryonic fibroblasts (MEFs) Gadd45a+/+, gadd45a−/− transformed by Ras/E1A were kindly provided by Professor Albert J Fornance from Goregetown University.
Supplemental Material
Supplemental data for this article can be accessed on the publisher’s website.
Funding
This work was supported by funding from the National Natural Science Foundation of China (81021061 and 81230047) and the 973 National Fundamental Research Program of China (2015CB553904).
References
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; PMID:18725538; http://dx.doi.org/10.1083/jcb.200803137
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 2009; 186:773-82; PMID:19797076; http://dx.doi.org/10.1083/jcb.200907014
- Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861-73; PMID:18006683; http://dx.doi.org/10.1101/gad.1599207
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27-42; PMID:18191218; http://dx.doi.org/10.1016/j.cell.2007.12.018
- Levine B. Cell biology: autophagy and cancer. Nature 2007; 446:745-7; PMID:17429391; http://dx.doi.org/10.1038/446745a
- Eskelinen EL. The dual role of autophagy in cancer. Curr Opin Pharmacol 2011; 11:294-300; PMID:21498118; http://dx.doi.org/10.1016/j.coph.2011.03.009
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672-6; PMID:10604474; http://dx.doi.org/10.1038/45257
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003; 100:15077-82; PMID:14657337; http://dx.doi.org/10.1073/pnas.2436255100
- Morselli E, Galluzzi L, Kepp O, Marino G, Michaud M, Vitale I, Maiuri MC, Kroemer G. Oncosuppressive functions of autophagy. Antioxid Redox Signal 2011; 14:2251-69; PMID:20712403; http://dx.doi.org/10.1089/ars.2010.3478
- Ma XH, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, Li LZ, Amaravadi RK. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res 2011; 17:3478-89; PMID:21325076; http://dx.doi.org/10.1158/1078-0432.CCR-10-2372
- Romano S, D'Angelillo A, Pacelli R, Staibano S, De Luna E, Bisogni R, Eskelinen EL, Mascolo M, Cali G, Arra C, et al. Role of FK506-binding protein 51 in the control of apoptosis of irradiated melanoma cells. Cell Death Differ 2010; 17:145-57; PMID:19696786; http://dx.doi.org/10.1038/cdd.2009.115
- Marino ML, Fais S, Djavaheri-Mergny M, Villa A, Meschini S, Lozupone F, Venturi G, Della Mina P, Pattingre S, Rivoltini L, et al. Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis 2010; 1:e87; PMID:21368860; http://dx.doi.org/10.1038/cddis.2010.67
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 1998; 72:8586-96; PMID:9765397
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927-39; PMID:16179260; http://dx.doi.org/10.1016/j.cell.2005.07.002
- Pattingre S, Levine B. Bcl-2 inhibition of autophagy: a new route to cancer? Cancer Res 2006; 66:2885-8; PMID:16540632; http://dx.doi.org/10.1158/0008-5472.CAN-05-4412
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007; 26:1749-60; PMID:17347651; http://dx.doi.org/10.1038/sj.emboj.7601623
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8:741-52; PMID:17717517; http://dx.doi.org/10.1038/nrm2239
- Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol 2008; 10:637-9; PMID:18521069; http://dx.doi.org/10.1038/ncb0608-637
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008; 10:676-87; PMID:18454141; http://dx.doi.org/10.1038/ncb1730
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A 2005; 102:8204-9; PMID:15928081; http://dx.doi.org/10.1073/pnas.0502857102
- Abida WM, Gu W. p53-Dependent and p53-independent activation of autophagy by ARF. Cancer Res 2008; 68:352-7; PMID:18199527; http://dx.doi.org/10.1158/0008-5472.CAN-07-2069
- Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol 2010; 22:181-5; PMID:20044243; http://dx.doi.org/10.1016/j.ceb.2009.12.001
- Fornace AJ, Jr., Alamo I, Jr., Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A 1988; 85:8800-4; PMID:3194391; http://dx.doi.org/10.1073/pnas.85.23.8800
- Papathanasiou MA, Kerr NC, Robbins JH, McBride OW, Alamo I, Jr., Barrett SF, Hickson ID, Fornace AJ, Jr. Induction by ionizing radiation of the gadd45 gene in cultured human cells: lack of mediation by protein kinase C. Mol Cell Biol 1991; 11:1009-16; PMID:1990262; http://dx.doi.org/10.1128/MCB.11.2.1009
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ, Jr. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 1992; 71:587-97; PMID:1423616; http://dx.doi.org/10.1016/0092-8674(92)90593-2
- Zhan Q, Bae I, Kastan MB, Fornace AJ, Jr. The p53-dependent gamma-ray response of GADD45. Cancer Res 1994; 54:2755-60; PMID:8168107
- Zhan Q, Chen IT, Antinore MJ, Fornace AJ, Jr. Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol Cell Biol 1998; 18:2768-78; PMID:9566896; http://dx.doi.org/10.1128/MCB.18.5.2768
- Zhan Q, Fan S, Smith ML, Bae I, Yu K, Alamo I, Jr., O'Connor PM, Fornace AJ, Jr. Abrogation of p53 function affects gadd gene responses to DNA base-damaging agents and starvation. DNA Cell Biol 1996; 15:805-15; PMID:8892753; http://dx.doi.org/10.1089/dna.1996.15.805
- Jin S, Tong T, Fan W, Fan F, Antinore MJ, Zhu X, Mazzacurati L, Li X, Petrik KL, Rajasekaran B, et al. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene 2002; 21:8696-704; PMID:12483522; http://dx.doi.org/10.1038/sj.onc.1206034
- Tong T, Ji J, Jin S, Li X, Fan W, Song Y, Wang M, Liu Z, Wu M, Zhan Q. Gadd45a expression induces Bim dissociation from the cytoskeleton and translocation to mitochondria. Mol Cell Biol 2005; 25:4488-500; PMID:15899854; http://dx.doi.org/10.1128/MCB.25.11.4488-4500.2005
- Carrier F, Georgel PT, Pourquier P, Blake M, Kontny HU, Antinore MJ, Gariboldi M, Myers TG, Weinstein JN, Pommier Y, et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol 1999; 19:1673-85; PMID:10022855; http://dx.doi.org/10.1128/MCB.19.3.1673
- Yang F, Zhang W, Li D, Zhan Q. Gadd45a suppresses tumor angiogenesis via inhibition of the mTOR/STAT3 protein pathway. J Biol Chem 2013; 288:6552-60; PMID:23329839; http://dx.doi.org/10.1074/jbc.M112.418335
- Wang W, Huper G, Guo Y, Murphy SK, Olson JA, Jr., Marks JR. Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene 2005; 24:2705-14; PMID:15735726; http://dx.doi.org/10.1038/sj.onc.1208464
- Yamasawa K, Nio Y, Dong M, Yamaguchi K, Itakura M. Clinicopathological significance of abnormalities in Gadd45 expression and its relationship to p53 in human pancreatic cancer. Clin Cancer Res 2002; 8:2563-9; PMID:12171884
- Ramachandran K, Gopisetty G, Gordian E, Navarro L, Hader C, Reis IM, Schulz WA, Singal R. Methylation-mediated repression of GADD45alpha in prostate cancer and its role as a potential therapeutic target. Cancer Res 2009; 69:1527-35; PMID:19190346; http://dx.doi.org/10.1158/0008-5472.CAN-08-3609
- Hollander MC, Kovalsky O, Salvador JM, Kim KE, Patterson AD, Haines DC, Fornace AJ, Jr. Dimethylbenzanthracene carcinogenesis in Gadd45a-null mice is associated with decreased DNA repair and increased mutation frequency. Cancer Res 2001; 61:2487-91; PMID:11289119
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/10.4161/auto.19496
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43:67-93; PMID:19653858; http://dx.doi.org/10.1146/annurev-genet-102808-114910
- Xu Y, Tian C, Wang SB, Xie WL, Guo Y, Zhang J, Shi Q, Chen C, Dong XP. Activation of the macroautophagic system in scrapie-infected experimental animals and human genetic prion diseases. Autophagy 2012; 8:1604-20; PMID:22874564; http://dx.doi.org/10.4161/auto.21482
- Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem J 2014; 461:315-22; PMID:24785657; http://dx.doi.org/10.1042/BJ20140515
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360-72; PMID:18843052; http://dx.doi.org/10.1091/mbc.E08-01-0080
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20:355-62; PMID:20356743; http://dx.doi.org/10.1016/j.tcb.2010.03.002
- Tamura RE, de Vasconcellos JF, Sarkar D, Libermann TA, Fisher PB, Zerbini LF. GADD45 proteins: central players in tumorigenesis. Curr Mol Med 2012; 12:634-51; PMID:22515981; http://dx.doi.org/10.2174/156652412800619978
- Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, et al. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell 1999; 97:575-86; PMID:10367887; http://dx.doi.org/10.1016/S0092-8674(00)80769-2
- Hocker R, Walker A, Schmitz I. Inhibition of autophagy through MAPK14-mediated phosphorylation of ATG5. Autophagy 2013; 9:426-8; PMID:23235332; http://dx.doi.org/10.4161/auto.22924
- Keil E, Hocker R, Schuster M, Essmann F, Ueffing N, Hoffman B, Liebermann DA, Pfeffer K, Schulze-Osthoff K, Schmitz I. Phosphorylation of Atg5 by the Gadd45beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ 2013; 20:321-32; PMID:23059785; http://dx.doi.org/10.1038/cdd.2012.129
- Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature 2001; 411:342-8; PMID:11357141; http://dx.doi.org/10.1038/35077213
- Kerr JF, Winterford CM, Harmon BV. Apoptosis. Its significance in cancer and cancer therapy. Cancer 1994; 73:2013-26; PMID:8156506; http://dx.doi.org/10.1002/1097-0142(19940415)73:8%3c2013::AID-CNCR2820730802%3e3.0.CO;2-J
- Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 2005; 5:726-34; PMID:16148885; http://dx.doi.org/10.1038/nrc1692
