Abstract
Microtubules, the major components of cytoskeleton, are involved in various fundamental biological processes in plants. Recent studies in mammalian cells have revealed the importance of microtubule cytoskeleton in autophagy. However, little is known about the roles of microtubules in plant autophagy. Here, we found that ATG6 interacts with TUB8/β-tubulin 8 and colocalizes with microtubules in Nicotiana benthamiana. Disruption of microtubules by either silencing of tubulin genes or treatment with microtubule-depolymerizing agents in N. benthamiana reduces autophagosome formation during upregulation of nocturnal or oxidation-induced macroautophagy. Furthermore, a blockage of leaf starch degradation occurred in microtubule-disrupted cells and triggered a distinct ATG6-, ATG5- and ATG7-independent autophagic pathway termed starch excess-associated chloroplast autophagy (SEX chlorophagy) for clearance of dysfunctional chloroplasts. Our findings reveal that an intact microtubule network is important for efficient macroautophagy and leaf starch degradation.
Abbreviations
| AD | = | activation domain |
| AGPase | = | ADP-glucose pyrophosphorylase |
| APM | = | amiprophos-methyl |
| APS1/ADG1 | = | small subunit 1 of AGPase |
| At | = | Arabidopsis thaliana |
| ATG | = | autophagy related |
| BD | = | binding domain |
| CFP | = | cyan fluorescent protein |
| chlorophagy | = | chloroplast autophagy |
| CLSM | = | confocal laser scanning microscopy |
| cLUC | = | C-terminal domain of firefly luciferase |
| CoIP | = | coimmunoprecipitation |
| DMSO | = | dimethyl sulfoxide |
| DPE | = | disproportionating enzyme |
| GST | = | glutathione S-transferase |
| IB | = | immunoblotting |
| IP | = | immunoprecipitation |
| LCI | = | firefly luciferase complementation imaging |
| MAPs | = | microtubule associated proteins |
| MBD | = | microtubule binding domain |
| MEX1 | = | maltose excess 1 |
| MV | = | methyl viologen |
| Nb | = | Nicotiana benthamiana; |
| nLUC | = | N-terminal domain of firefly luciferase |
| Nt | = | Nicotiana tabacum |
| PtdIns3K | = | phosphatidylinositol 3-kinase |
| ROS | = | reactive oxygen species |
| SEX | = | starch-excess |
| Sl | = | Solanum lycopersicum; |
| TEM | = | transmission electron microscopy |
| TRV | = | tobacco rattle virus |
| TUB | = | β-tubulin |
| TUA | = | α-tubulin |
| VIGS | = | virus-induced gene silencing |
| wpi | = | weeks post-agroinfiltration |
| X-gal | = | 5-bromo-4-chloro-3-indolyl-D-galactoside |
| Y2H | = | yeast 2-hybrid |
Introduction
Autophagy is an evolutionarily conserved degradation pathway in eukaryotes that mainly contributes to the clearance of intracellular proteins and organelles and sometimes targets lipids, carbohydrates, nucleic acids and invading pathogens as well.Citation1,2 Two types of autophagy, microautophagy and macroautophagy, have been described in plants.Citation3 In canonical macroautophagy (hereafter referred to as autophagy unless otherwise specified), substrates are wrapped in the cytoplasm by cup-shaped membranes called phagophores, which expand to form double-membrane vesicles known as autophagosomes, and are ultimately delivered to the vacuoles or lysosomes for degradation.Citation4 This process occurs constitutively at low basal levels under normal conditions to renovate cytoplasmic components and maintain cellular homeostasis (basal autophagy) and can also be massively induced by internal or external stresses to cope with diverse physiological and pathological situations (induced autophagy).Citation5,6 In plants, autophagy plays roles in diverse biological processes, including adaption to various abiotic stresses, such as oxidation, drought and high salinity,Citation7,8 immune defense against virus, bacterial and fungal pathogens,Citation9-12 development of roots, flowers and xylem,Citation13-16 chloroplast recycling,Citation17 peroxisome degradationCitation18-20 and nighttime metabolism of amino acids and leaf starch.Citation21,22
As the most remarkable feature of autophagy, the biogenesis of autophagosomes requires the involvement of at least 18 key autophagy–related genes, including ATG1 to ATG10, ATG12 to ATG14, ATG16 to ATG18, ATG29, and ATG31.Citation1,23 Orthologs of most of these genes have also been identified in plants except for ATG14, ATG29 and ATG31.Citation24,25 Yeast Vps30, whose mammalian and Arabidopsis orthologs are BECN1 and ATG6 respectively, is a major component of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex localized at the phagophore assembly sites and is important for the construction of phagophores through recruiting other effector proteins.Citation23 In addition, ATG6 participates in multiple interactions with other proteins to regulate both autophagic and nonautophagic process in mammals.Citation26,27 However, few ATG6-binding proteins have been reported in plants.
Microtubules are polar cytoskeletal filaments assembled from heterodimers of α- and β-tubulins and exhibit dynamic behaviors characterized by stochastic switching between periods of growing and shrinking.Citation28,29 Plant microtubules are involved in numerous fundamental biological processes such as cell division, polarity of growth, cell wall deposition, and stress sensing.Citation30,31 Evidence supporting the involvement of microtubules in endomembrane organization and trafficking has emerged recently in plants.Citation32 Investigations of the relationship between cytoskeleton and autophagy in mammalian cells reveal the importance of microtubule cytoskeleton in various aspects of autophagy including autophagosome biogenesis, movement and autolysosome formation.Citation6,33 In plants, Arabidopsis ATG8 and JOKA2, a selective autophagy receptor in tobacco, colocalize with microtubules,Citation34,35 which indicates a possible link between microtubules and autophagy. However, little is known about the exact role of microtubules in plant autophagy.
In this study, we report that ATG6 interacts with microtubules, and microtubule disorganization reduces autophagosome formation upon rapid induction of macroautophagy and severely blocks leaf starch degradation. Massive starch accumulation in microtubule-disrupted cells triggers malformation and degradation of chloroplast in the central vacuole through an ATG6-, ATG5- and ATG7-independent pathway termed starch excess-associated chloroplast autophagy. Our study reveals roles of intact microtubule cytoskeletons in macroautophagy and leaf starch degradation.
Results
Autophagy-related protein 6 interacts with β tubulin 8
To better understand the role of ATG6 in plant autophagy, we performed a yeast 2-hybrid (Y2H) screen of a tomato cDNA libraryCitation36 using Nicotiana tabacum ATG6 (NtATG6) as bait to identify ATG6-binding host proteins. One clone encoding partial β-tubulin 8 was identified as a putative candidate for ATG6-interacting factor. We cloned its N. tabacum homolog gene and named it NtTUB8. Sequence alignment analysis showed that NtTUB8 shares 96% amino acid identity with TUB8/β-tubulin 8 in Arabidopsis thaliana (AtTUB8) and 98.9% identity with TUB8 in Solanum lycopersicum (SlTUB8) (Fig. S1). To test whether NtTUB8 interacts with NtATG6, we introduced TUB8 into the activation domain (AD) vector and cotransformed it into yeast with a bait vector carrying ATG6 fused with LexA DNA binding domain (BD). As indicated by the galactose (Gal) dependent growth of yeast on Leu-deficient plates and β-galactosidase assays on 5-bromo-4-chloro-3-indolyl-D-galactoside (X-gal) containing plates, NtTUB8 interacts with NtATG6 in yeast ().
Figure 1. ATG6 interacts with TUB8 in yeast and N. benthamiana. (A) Interaction of NtATG6 with NtTUB8 in yeast. Yeast cells harboring both BD-ATG6 and AD-TUB8 were able to turn blue on X-Gal-containing plate and grow on Leu-deficient medium in the presence of galactose and raffinose. Yeast harboring other control constructs showed no growth or color-changes, on corresponding induction plates. (B) NtTUB8 coimmunoprecipitates with NtATG6. Immunoprecipitation (IP) by anti-HA antibody was performed on total protein extracts from N. benthamiana leaves transiently expressing HA-TUB8 or other control groups. Precipitates were then analyzed by immunoblotting (IB) using anti-MYC (top panel) or anti-HA (middle panel) antibodies. Expression of MYC-tagged proteins was checked with anti-MYC (bottom panel). (C) Firefly luciferase complementation imaging (LCI) assay shows the interaction of ATG6 with TUB8. Coexpression of cLUC-TUB8 with ATG6-nLUC, but not other negative controls, reconstituted LUC activities and generated luminescence in the presence of luciferin. Infiltration areas of various combinations of constructs are indicated by the dashed circle.
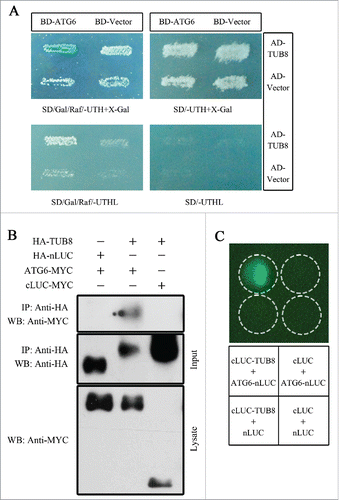
To test TUB8-ATG6 interaction in plants, coimmunoprecipitation (CoIP) assay was performed. MYC-tagged ATG6 (ATG6-MYC) or MYC-tagged C-terminal domain of firefly luciferase (cLUC-MYC) were coexpressed transiently with HA-tagged TUB8 (HA-TUB8) or HA-tagged N-terminal domain of firefly luciferase (HA-nLUC) in N. benthamiana leaves for about 60 h. CoIP using anti-HA antibodies was subsequently performed on the total proteins extracted from infiltrated leaves. ATG6-MYC was specifically detected in the immunoprecipitates of HA-TUB8 but not of HA-nLUC (). Similarly, cLUC-MYC, another negative control, did not coimmunoprecipitate with HA-TUB8 (). These results suggest an interaction of TUB8 with ATG6 in vivo.
The in vivo TUB8-ATG6 interaction was further confirmed by firefly luciferase complementation imaging (LCI) assays.Citation37 Immunoblot analysis showed expression of nLUC- and cLUC-fusion constructs in leaves (Fig. S2B). The positive luminescence signals were only detected at sites where cLUC-TUB8 and ATG6-nLUC were coexpressed but not at sites for the combinations of controls (). These data, along with the CoIP and Y2H results, demonstrate that ATG6 interacts with TUB8.
The tubulin family consists of multiple evolutionarily conserved proteins in plants (Fig. S3 and S4). In order to test the interaction of ATG6 with other tubulin proteins, we cloned TUB1, TUB6 and TUA6 in tobacco and confirmed the corresponding expression of cLUC-fusion proteins in inoculated leaves (Fig. S2B). LCI assay showed that these tubulin proteins also interact with ATG6 (Fig. S2A).
ATG6 colocalizes with microtubules in vivo
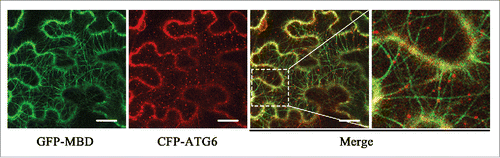
Since ATG6 interacts with tubulins, the major components of microtubules, we hypothesize that a putative interaction exists between ATG6 and microtubules. To test this, we performed a colocalization assay in plant cells. MAP4 is one of the well-studied microtubule-associated proteins (MAPs) in mammalian cells and GFP-tagged MAP4 or its microtubule binding domain (MBD) has been used as a microtubule reporter in mammals and plants.Citation38,39 Here, we constructed a GFP-tagged MBD domain of HsMAP4, GFP-MBD, to label the cortical microtubule networks in leaf cells of N. benthamiana. Cyan fluorescent protein (CFP)-tagged ATG6 (CFP-ATG6) was transiently coexpressed with GFP-MBD in leaves for 48 h, followed by microscopic observation under confocal laser scanning microscopy (CLSM). Interestingly, CFP-ATG6 decorated a number of punctate structures and all of them displayed localizations on or adjacent to cortical microtubules (). Detailed observation of movement of CFP-ATG6 by time-lapse microscopy showed that some dots of CFP-ATG6 could move along microtubules for short distances (Fig. S5B and C; Movie S1). We also captured one CFP-ATG6-decorated structure showing synchronous movement with the shrinking microtubule (Fig. S5C; Movie S1). Similar colocalization of ATG6 with microtubules was observed when CFP-ATG6 was coexpressed with YFP-MBD (Fig. S5A). Taken together, these data suggest that ATG6 binds to microtubules in vivo.
Figure 2. ATG6 colocalizes with microtubules in vivo. Images were taken when CFP-ATG6 and the microtubule reporter, GFP-MBD, were transiently coexpressed in N. benthamiana leaves for 48 h. GFP-MBD-labeled microtubules appear green and CFP-ATG6-labeled punctate structures are pseudocolored red. Scale bars: 20 μm.
Silencing of tubulin genes results in disordered cortical microtubule arrays and aberrant plant development
To investigate the function of tubulin genes, we used a Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) systemCitation40 to silence tubulin genes. TUB8-silenced plants developed a leaf curling and crinkling phenotype at 2 wk post-agroinfiltration () and showed more severe developmental aberrations including stunted top and root growth, leaf chlorosis and flower defects at later stages of silencing (; Fig. S6A and B). Similar phenotypes were also observed in TUA6/α-tubulin 6 (TUBULIN ALPHA-6)-silenced plants (Fig. S6C). Real-time RT-PCR showed that silencing of TUB8 or TUA6 in N. benthamiana greatly reduces the mRNA level of its respective target gene but has no significant effect on mRNA abundance of Actin (ACT7) and other nontargeted tubulin genes such as TUB1 or TUA5 ( and Fig. S7).
Figure 3. Silencing of TUB8 affects plant development and disorganizes microtubule arrays. (A) Leaf curling and crinkling phenotype in TUB8-silenced plants. Photos were taken at 2 wk post-agroinfiltration (wpi) for VIGS. (B) Realtime RT-PCR shows relative mRNA levels of TUB8 in silenced or nonsilenced plants. ACT7 was used as the internal control. Values are means ± SE of 5 replicate samples. The Student t test was applied to determine statistically significant differences (**P <0.01). (C and D) Developmental defects in TUB8-silenced plants at later stages of silencing, including defective flower development in (C) and leaf chlorosis in (D). Photos were taken at 7 wpi for VIGS. (E) Disordered cortical microtubule arrays in TUB8-silenced leaves. Various aberrations of GFP-MBD-labeled microtubules, including segmentation (magenta dashed oval), bending (white dashed square) and parallel distribution (yellow dashed rectangle) are shown in the pavement cells of TUB8-silenced plants. Scale bars: 10 μm.
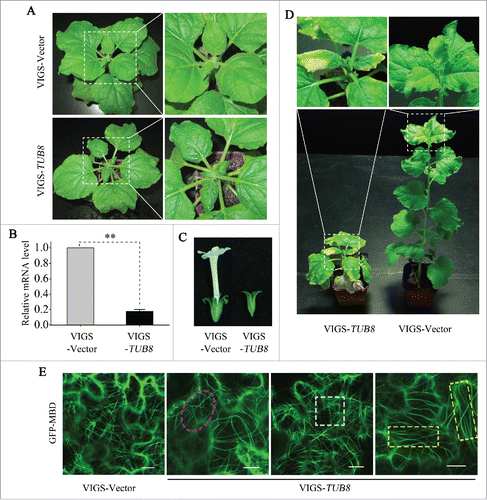
To test the effect of silencing TUB8 on microtubule cytoskeletons, we visualized GFP-MBD-labeled cortical microtubule arrays in pavement cells of TUB8-silenced N. benthamiana. Unlike the intact, dense and randomly oriented arrays in the nonsilenced TRV control leaf cells, microtubule networks in TUB8-silenced plants appear sparse and disorganized (). Some microtubules were bent, parallel or broken into short segments and detached from cell membrane (). Similar disordered arrays were found in the TUA6-silenced plants (Fig. S6D). These results show that silencing of tubulin genes results in disorganized cortical microtubule arrays and aberrant plant development.
Disruption of microtubule cytoskeleton reduces autophagosome formation during upregulation of macroautophagy
We have previously shown that nocturnal autophagy in N. benthamiana leaves undergoes an upregulation before midnight and a downregulation by dawn.Citation21 Using this as a model system, we tested the role of microtubule in macroautophagy. CFP-tagged ATG8f (CFP-ATG8f) was used to label autophagic structures in mesophyll cells as described previously.Citation21 As expected, a lot of punctate structures labeled by CFP-ATG8f appeared in mesophyll cells of nonsilenced plants after 4 h of exposure to darkness (, upper panel). However, many fewer autophagic structures were detected in the TUB8-silenced leaves (, lower panel). Quantitative analysis of autophagic structures showed that a 66% reduction of autophagic activities occurred in TUB8-silenced leaves at midnight (). Similar reduction of autophagic activity was observed in TUA6-silenced leaves (Fig. S8C and D). We also tested the basal autophagic activity occurring in plants exposed to 0 h or 8 h of darkness. No obvious differences, however, were detected between TUB8-silenced and nonsilenced plants (Fig. S8A and B). These results indicate that microtubules may not be required but are still important for efficient autophagosome biogenesis during upregulation of nocturnal autophagy.
Figure 4. Disruption of microtubules suppresses upregulation of nocturnal autophagy. (A and B) Suppressed nocturnal autophagy in TUB8-silenced leaves at midnight. (A) Representative images of CFP-ATG8f-labeled autophagic structures in leaves. CFP-ATG8f is in cyan, and chloroplasts are in red. White arrows indicate CFP-ATG8f-labeled autophagic structures. Scale bars: 20 μm. (B) Relative autophagic activity in TUB8-silenced plants. The autophagic activity in nonsilenced plants was set to 1. This experiment was repeated 6 times. Values represent means ± SE. The Student t test was used to determine significant differences (**, P < 0.01). (C and D) Suppressed nocturnal autophagy in microtubule inhibitor-treated leaves at midnight. (C) Relative autophagic activity in microtubule inhibitor-treated leaves. The autophagic activity in leaves infiltrated with 0.1% dimethyl sulfoxide (Mock) was set to 1. This experiment was repeated 10 times. Values represent means ± SE. The Student t test was used to determine significant differences (**, P < 0.01). (D) Representative images of autophagic structures and cortical microtubule arrays in leaves after 4 h of treatment with microtubule inhibitors. Magenta arrowheads indicate short segments of depolymerized microtubules. Scale bars: 20 μm.
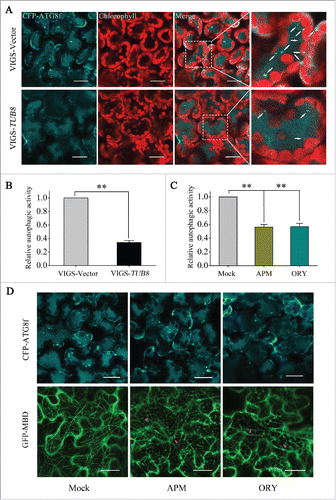
We further confirmed the involvement of microtubules in the nocturnal autophagic process by antimicrotubule herbicides. Amiprophos-methyl (APM) and oryzalin have been well-studied as microtubule-disrupting agents in plants, both of which cause microtubule depolymerization through their binding to tubulins.Citation41,42 We infiltrated 10 μM APM or oryzalin into leaves to depolymerize microtubules at the end of day to allow 4-h drug treatment, and we visualized microtubule organizations and autophagic activities, respectively, at midnight. As indicated by GFP-MBD, most cortical microtubules were disrupted, leaving only some short segments of depolymerized microtubules in pavement and mesophyll cells after the application of APM or oryzalin (; Fig. S9). Concomitantly, autophagosome formation was markedly decreased and fewer CFP-ATG8-labeled structures appeared when the microtubules were depolymerized (). Quantitative analysis revealed that treatment with either APM or oryzalin resulted in an approximately 50% reduction in nocturnal autophagic activities compared to that in mock treatment ().
We also investigated roles of microtubules in oxidation-induced autophagy. Methyl viologen (MV), an inducer of reactive oxygen species (ROS), could induce macroautophagy in plants.Citation7,43 Similarly, 1-d treatment with 10 μM MV in nonsilenced N. benthamiana plants caused a 3.25-fold increase in basal activity of autophagy as indicated by quantitative analysis of CFP-ATG8f-labeled structures. However, silencing of either TUB8 or TUA6 significantly blocked MV-triggered formation of autophagosomes (Fig. S8E).
Taken together, microtubule integrity is important for efficient autophagosome formation when macroautophagy is induced by either internal signals or external environment changes.
Silencing of TUB8 severely blocks leaf starch degradation
Since macroautophagy contributes to leaf starch degradationCitation44 and silencing of TUB8 blocks the upregulation of nocturnal autophagy, we reasoned that TUB8 silencing could affect the breakdown of transitory starch in leaves. Indeed, we detected large amounts of starch reserves in TUB8-silenced leaves at the end of night using iodine staining (). Strikingly, these reserves were conspicuously higher than those found in ATG6-silenced plants (Fig. S10) and could not be consumed efficiently even when the time of dark treatment was prolonged to 120 h (). This extremely strong starch-excess (SEX) phenotype in TUB8-silenced plants implies that leaf starch degradation is largely blocked when microtubule cytoskeletons are disorganized.
Figure 5. Silencing of TUB8 in N. benthamiana blocks leaf starch degradation. (A) Iodine staining of leaf discs taken from plants subjected to prolonged dark treatment. The plants were kept in darkness for up to 120 h of treatment until all the samples of each time point were collected. At each time point, 5 to 6 leaf discs were punched from the plants and incubated in ethanol to remove leaf pigments. Iodine staining was carried out when all the samples were harvested. Representative results are presented. (B to D) The SEX phenotype in TUB8-silenced leaves is caused by a blockage of starch degradation. (B) Iodine staining of leaves detached from silenced and nonsilenced plants at the indicated time. These results were reproduced in 2 experiments using more than 3 leaves in each experiment. Representative results are presented. (C) Real-time RT-PCR shows successful silencing of target genes in either individually silenced or cosilenced plants. ACT7 was used as the internal control. Values are means ± SE of 2 replicate samples. (D) Quantitative analysis of leaf starch content in silenced and nonsilenced plants at the indicated time. Values are means ± SE of 2 or 3 replicate samples.
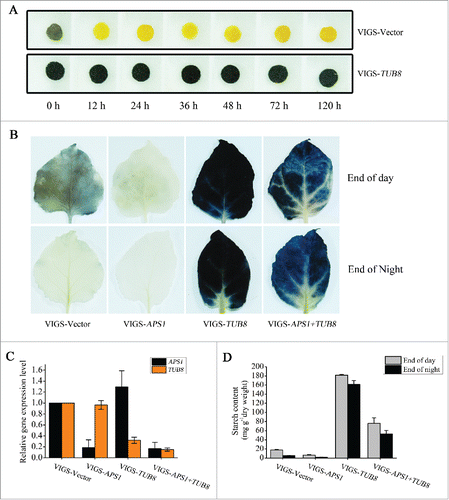
To exclude the possibility that the SEX phenotype in TUB8-silenced plants is caused by enhanced starch synthesis, we studied the mobilization process of starch when starch synthesis was inhibited in TUB8-silenced plants. ADP-glucose pyrophosphorylase (AGPase) catalyzes the first rate-limiting step in starch biosynthesis in plants,Citation45 and mutations in the gene encoding its small subunit 1 (APS1/ADG1) lead to a significant reduction in starch synthesis.Citation46 Here, we successfully silenced APS1 and TUB8 individually or simultaneously in N. benthamiana and determined starch content of leaves harvested at the end of day or night by iodine staining and quantitative measurement (). Only 35.8% of leaf starch content of control plants was detected in APS1-silenced plants after a 16-h illumination period (), suggesting silencing of APS1 decreases starch synthesis in N. benthamiana as well. We noticed that there was lower starch accumulation in APS1- and TUB8-cosilenced leaves than TUB8-silenced leaves at either time point (). However, the APS1- and TUB8-cosilenced leaves still had a much higher level of starch reserves than nonsilenced leaves (). These data clearly demonstrate that silencing of TUB8 severely blocks leaf starch degradation.
Starch overaccumulation in TUB8-silenced leaves triggers starch excess-associated chloroplast autophagy
To better understand the extreme SEX phenotype of TUB8-silenced plants, transmission electron microscopy (TEM) was used to analyze the cellular ultrastructure of TUB8-silenced mesophyll cells when the nocturnal metabolism was finished. Large oval-shaped granules accumulated in the chloroplasts of TUB8-silenced plants, whereas no visible starch granules remained in the nonsilenced plants at the end of night (), consistent with the results of starch assays (; Fig. S10). Additionally, we visualized cells with variant degrees of starch accumulation in TUB8-silenced leaves at different stages of silencing (). At 3 wk post-agroinfiltration, some chloroplasts of TUB8-silenced leaves accumulated starch at levels of illuminated wild-type cells (, panel 2), whereas more chloroplasts accumulated large granules, which severely exceeded the capacity of stroma (, panel 3). The former contained well-organized thylakoid membranes and grana stacks, had normal shapes and were located in an orderly fashion in the thin layer of cytoplasm (, panel 2), while the latter contained fewer visible thylakoid membrane systems, became swollen, enlarged and globular in shape and sometimes were separated from the cytosol and taken into the vacuole (, panel 3). Measurements of ultrastructural characteristics of chloroplast (the length/width ratio and cross-sectional area) further supported the changes in chloroplast morphology of TUB8-silenced leaves (Fig. S11). The misshapen phenotypes of chloroplasts were more evident in chlorotic leaves of TUB8-silenced plants at about 5 wk post-agroinfiltration ( panels 4 and 5, and ). Nearly all the chloroplasts were fully filled with massive starch granules, appearing as if specific storage organelles of starch, and an increasing number of chloroplasts swarmed into vacuoles ( panels 4 and 5). In addition to whole chloroplasts with several granules, we sometimes observed vacuolar localization of smaller chloroplastic structures with one starch granule bounded by extremely thin thylakoid membrane (, yellow arrows) or small spherical structures with only aberrant thylakoids but no granules (, cyan arrows). These smaller chloroplastic structures are likely products of a budding process from whole, large chloroplasts (, yellow arrowhead). Vesicles were occasionally found to be close to, but never engulf, starchy chloroplast or chloroplastic structures (, red arrowhead). In a few cases, starch granules were released from the extremely disrupted chloroplasts into vacuoles (, red asterisk). Other remnants of chloroplasts, like fragmented thylakoids (, black arrows), were observed in vacuoles. In chlorotic leaves, collapsed cells were occasionally observed, in which massive starchy chloroplasts and other organelles were taken into the central vacuole ( panel 5). We called the above self-eating process for clearing disorganized starchy chloroplast ‘starch excess-associated chloroplast autophagy’ (hereafter short for SEX chlorophagy). Remarkably, the ultrastructural characteristics of aberrant chloroplasts were well matched with some special findings in chloroplast morphologies of TUB8-silenced leaves under CLSM, including lower or hollow chlorophyll fluorescence (Fig. S12) and vacuolar localization of chloroplast or its derivative structures (Movies S2 to S4).
Figure 6. Starch overaccumulation in TUB8-silenced leaves triggers chloroplast malformation and SEX chlorophagy. (A) Ultrastructural analysis shows varying degrees of starch accumulation in mesophyll cells of TUB8-silenced plants at different stages of silencing. Blue arrows refer to the vacuole-localized SEX chloroplast. (B) Ultrastructure of misshapen chloroplasts in chlorotic leaves of TUB8-silenced plants at 5 wk post-agroinfiltration (wpi). Red arrowheads indicate vesicles adjacent to starchy chloroplast or chloroplastic structures. Yellow and cyan arrows indicate smaller chloroplastic structures with or without starch granules, which were probably generated by a budding-like process (yellow arrowhead) from the whole, large chloroplasts. Occasionally, remnants of disrupted chloroplast, like starch granule (red asterisk) and segments of thylakoids (black arrows), were observed in the vacuole. (C) Ultrastructural analysis of starch accumulation and SEX chlorophagy in mesophyll cells of TUB8-silenced and APS1- and TUB8-cosilenced plants at 3 wk post-agroinfiltration (wpi). (D) Quantification of vacuole-localized SEX chloroplast count per cell in TEM images of TUB8-silenced, as well as APS1- and TUB8-cosilenced leaves. Values are means ± SE from more than 50 cells. Two asterisks indicate a highly significant difference (P < 0.01; the Student t test). All the leaf samples used for TEM sectioning were taken from plants that had just finished nocturnal metabolism at the indicated weeks post-agroinfiltration (wpi) for VIGS. S, starch; V, vacuole; Cp, chloroplast; CW, cell wall. Black scale bars: 5 μm; white scale bars: 2 μm.
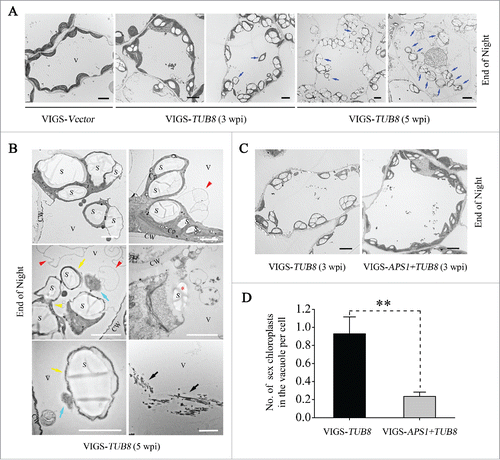
To study the influence of starch content on SEX chlorophagy, we further analyzed ultrastructure of mesophyll cells in APS1- and TUB8-cosilenced plants. In agreement with previous results (), starch granules deposited in chloroplasts of dark-treated APS1-and TUB8-cosilenced leaves were fewer and smaller than that of TUB8-silenced leaves (). Furthermore, quantitative analysis of sex chloroplasts in vacuoles showed that APS1 and TUB8 cosilencing resulted in significantly reduced SEX chlorophagy compared to individual silencing of TUB8 (). This result shows that the occurrence of sex chlorophagy is closely related to leaf starch content in chloroplasts.
SEX chlorophagy is independent of ATG6, ATG5 and ATG7
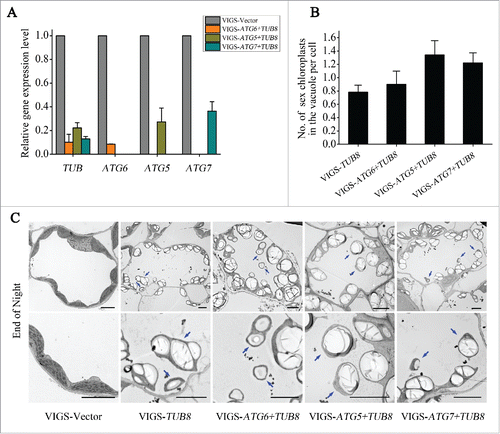
ATG-dependent autophagic pathway plays a role in the degradation of whole or partial chloroplast during senescence in individually darkened leaves.Citation47 The partially suppressed autophagosome-mediated autophagic activities in TUB8-silenced plants indicate that the degradation of SEX chloroplast may be independent of macroautophagy. To further test this hypothesis, we cosilenced TUB8 together with several key ATG genes, including ATG6, ATG5 and ATG7 (), and investigated the involvement of these ATG genes in SEX chlorophagy. TEM observations showed that the aberrant phenotypes of starch overaccumulation and chloroplast degradation in vacuole were still developed when TUB8 was cosilenced together with ATG6, ATG5 or ATG7 (). Quantitative analysis of vacuole-localized SEX chloroplasts demonstrated that no obvious difference existed in the level of SEX chlorophagy between the TUB8 individually silenced and ATG/TUB8 cosilenced plants (). These results suggest that SEX chlorophagy is ATG6-,ATG5- and ATG7-independent.
Figure 7. The occurrence of SEX chlorophagy Is independent of ATG6, ATG5 and ATG7. (A) Real-time RT-PCR suggests silencing of the corresponding target genes in either individually silenced or cosilenced plants. ACT7 was used as the internal control. Values are means ± SE of 2 replicate samples. (B) Quantification of the number of vacuole-localized SEX chloroplast per cell in TEM images of TUB8-silenced, as well as ATG and TUB8-cosilenced plants. Values are means ± SE from more than 30 cells. (C) Ultrastructural analysis shows occurrence of SEX chlorophagy in ATG and TUB8-cosilenced plants. Leaf samples used for TEM sectioning were taken from plants that had just finished nocturnal metabolism at 25 d post-agroinfiltration (3 or 4 wpi) for VIGS. Blue arrows refer to the vacuole-localized SEX chloroplasts. Scale bars: 5 μm.
Microtubule-depolymerizing drugs phenocopy TUB8-silenced plants
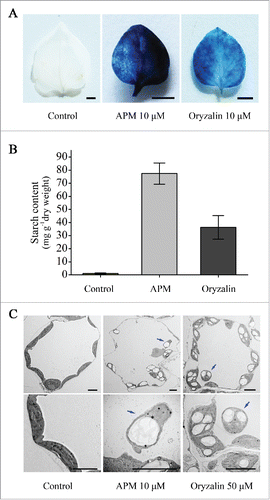
To further confirm whether the aberrant phenotypes of TUB8-silenced plants, including developmental defects, blocked starch depletion and concomitant chloroplast degradation, were caused by a dysfunctional microtubule cytoskeleton, we tested effects of microtubule-depolymerizing drugs, APM and oryzalin, on these biological processes. Two approaches were adopted for the application of microtubule inhibitors: one was germination of N. benthamiana seeds directly on drug-containing MS plates for 2 to 3-weeks growth (hereafter referred to as germination assay); the other one was transplanting seedlings to the drug-containing MS plates for a 4-wk treatment (hereafter referred to as transplanting assay). Normally, N. benthamiana seedlings grown on MS media for 3 wk were 15 to 20 mm in height and had 4 to 7 leaves and elongated hypocotyls and roots. However, seedlings germinated on MS plates containing either 10 μM APM or 10 μM oryzalin stopped growing after cotyledon opening and displayed defects in hypocotyl elongation and root development (Fig. S13A). Similar growth retardation was observed in N. benthamiana plants from the transplanting assay (Fig. S13D). Additionally, chlorosis phenotype was evident in the transplanted plants treated with either 10 μM APM, 50 μM APM or 50 μM oryzalin (Fig. S13D). Iodine staining and quantitative measurement of starch contents revealed that large amounts of starch remained in drug treated-seedlings or leaves that had finished nocturnal metabolism ( and B; Fig. S13B and C). Furthermore, TEM observations shows that SEX chlorophagy occurred in mesophyll cells of transplanted plants treated with APM or oryzalin (). Taken together, these results suggest that functional microtubules are required for leaf starch degradation and normal plant development.
Figure 8. Starch accumulation and SEX chlorophagy in microtubule depolymerizing agent-treated plants. (A) Iodine staining of leaves indicates massive starch reserves in the microtubule depolymerizing agent-treated plants from transplanting assay. Leaves used for determination of starch content were harvested from the plants treated with 10 μM APM or oryzalin at the end of night. These results were reproduced in 2 experiments using 6 to 8 leaves in each experiment. Representative results are presented. Scale bars: 0.5 cm. (B) Quantitative analysis of leaf starch content. Values are means ± SE of 3 replicate samples. (C) Ultrastructural analysis shows occurrence of starchy chloroplast autophagy in microtubule depolymerizing agent-treated plants from the transplanting assay. Blue arrows refer to the vacuole-localized starchy chloroplasts. Scale bars: 5 μm. Leaf samples used here for starch assay and TEM sectioning were all collected at the end of night.
Discussion
Role of microtubules in macroautophagy
Microtubules play important roles in macroautophagic process in mammalian cells but not yeast cells.Citation6 Their participation in different steps of mammalian autophagy is well investigated through pharmacological approaches.Citation33 Whereas an agreement has been reached on the role of microtubules in autophagosome movement,Citation48-51 the involvement of microtubules in autolysosome formation has been under debate.Citation6,33 Likewise, microtubules have initially been believed to have little or no effects on autophagic sequestration in early studies,Citation52,53 but are proven to have a facilitative role in autophagosome formation during starvation or mitosis.Citation49,51,54,55 For example, microtubule disruption reduces the number of starvation-induced autophagosomes in primary rat hepatocytes or Chinese hamster ovary cells,Citation51,54 and microtubule dynamics and acetylation state of tubulins can affect autophagosome biogenesis.Citation49,55 Nevertheless, interruption of microtubules seldom affects the basal levels of autophagy.Citation51,54,55
In contrast to the comprehensive studies in mammals, little is known about the involvement of microtubules in plant autophagy. In this study, we investigated the autophagic response to microtubule disassembly in N. benthamiana via silencing of tubulin genes or treatment with microtubule-depolymerizing agents. Both approaches interrupted cortical microtubules arrays successfully but to different extents (, Figs. S6 and S9). This discrepancy might be caused by the difference in availability of cellular pools of tubulin dimers. Local application of APM or oryzalin in leaves transiently releases too much herbicide for cells to keep dynamic equilibrium between tubulin dimers and microtubules, while silencing of TUB8 or TUA6 has less or no effect on the nontargeted tubulins, which are also always able to reach a new appropriate equilibrium. We observed CFP-ATG8f-labeled autophagosomes not only in cells with intact microtubules but also in cells with disorganized microtubules (, Fig. S8). Moreover, there was no obvious difference between the number of autophagosomes formed in normal cells and microtubule-disorganized cells at the end of day or night when the level of autophagy was supposed to be basal and low (Fig. S8A, B and E), implying that plant microtubules are not crucial elements involved in autophagosome formation at baseline. However, a significant suppression of autophagosome biogenesis occurred in microtubule-disrupted cells during the upregulation process of macroautophagy at midnight or under oxidative stress (, Fig. S8), demonstrating that intact microtubules are important for efficient biogenesis of autophagosomes induced by either internal signals or external environment changes. These findings are consistent with reports on mammalian cells that suggest regular microtubules function as facilitators of autophagosome formation rather than essential elements.Citation51,54,55
Microtubule involvement in macroautophagy has also been supported by biochemical and molecular findings in mammalian cells. Mammalian orthologs and paralogs of yeast Atg8 can be associated with microtubules.Citation49,51,56 A subset of ATG proteins engaged in phagophore formation, including BECN1/Vps30/ATG6, WIPI1 and the ATG12–ATG5 complex, are enriched in the labile microtubule fractions upon starvation, suggesting that dynamic microtubules are important for the assembly of preautophagosomal structures and subsequent autophagosome formation.Citation49 Additionally, BECN1/Vps30/ATG6 can be sequestered on microtubules by 2 complexes comprising dynein light chain1 (DYNLL1/LC8) and AMBRA1 or BCL2L11/Bim to regulate the onset of autophagy.Citation57,58
In this study, we detected interactions of ATG6 with tubulin proteins (; Fig. S2) and its colocalizations with microtubules in plants (; Fig. S5). The dynamic short movement of some CFP-ATG6-decorated structures on microtubules clearly shows the binding of ATG6 to microtubules (Fig. S5; Movie S1). A similar movement of WIPI1, an ortholog of yeast Atg18 engaged in early stage of autophagosome formation, along microtubules has been reported in mammalian cells.Citation49 The PtdIns3K complex containing BECN1/Vps30/ATG6 is essential for the vesicle nucleation step by producing PtdIns3P and recruiting PtdIns3P-binding effectors, such as WIPI1/Atg18 and ZFYVE1, to autophagosome formation sites: the phagophore assembly site in yeast and omegasome in mammals.Citation23,59,60 Moreover, 2 ubiquitin-like conjugation systems, the ATG12–ATG5-ATG16L1 complex and ATG8–PE, turn out to be downstream of the PtdIns3K complex in the hierarchical model of ATG proteins in autophagosome formation.Citation60-62 Considering the positive role of microtubules in autophagosome formation, we propose that the localization of ATG6 on microtubules may facilitate efficient recruitment of other ATG proteins to assemble scaffolds for autophagosome biogenesis when cellular autophagic activities need to be upregulated. On the other hand, when autophagy occurs at basal level, a low level of pre-existing, available ATG proteins is probably sufficient to meet the requirement for autophagosome biogenesis without further microtubule-dependent recruiting.
A functional microtubule cytoskeleton is necessary for leaf starch degradation
Leaf starch accumulates temporarily in chloroplast as primary products of photosynthesis during the day and is mobilized at night to provide neutral sugars, mainly in the form of maltose and glucose, for the continued metabolism of plants.Citation63 Any aberrations in starch synthesis or degradation would lead to altered starch levels in leaves either at the end of day or night.Citation46,64 SEX mutants identified so far in Arabidopsis are mostly deficient in key enzymes involved in leaf starch degradation, like α-glucan water dikinase, phosphoglucan phosphatase, β-amylase and starch debranching enzyme.Citation65 In addition, mutations in genes involved in transporting starch breakdown products out of the chloroplast, like maltose transporter MEX1 (maltose excess 1), or metabolizing them in the chloroplast and cytosol, like chloroplastic or cytosolic disproportionating enzyme DPE1 and DPE2, cause feedback inhibition of starch degradation and generate SEX phenotypes as well.Citation66-70 Unexpected or pleiotropic SEX phenotypes are sometimes reported.Citation71 In N. benthamiana, higher starch reserves are also detected when autophagy is blocked, suggesting autophagy is involved in leaf starch degradation.Citation21
Here, we observed an extremely severe SEX phenotype in N. benthamiana plants lacking intact microtubules ( and Fig. S13). Large amounts of starch reserves were detected even when the TUB8-silenced plants were subjected to dark treatment as long as 120 h (). Moreover, APS1- and TUB8-cosilenced leaves still developed SEX phenotype, suggesting the excess starch accumulation is due to defects in mobilization process rather than enhanced starch synthesis (). In our previous studies, we report that the macroautophagic pathway, distinct from the major, classical plastidial route, is responsible for the extra-chloroplast degradation of small starch granule-like structures.Citation21,44 A blockage of the macroautophagic pathway in leaf starch degradation would partially account for the impaired starch turnover in microtubule-disrupted plants since the nocturnal autophagic activity was significantly suppressed when the microtubules were disorganized ( and Fig. S8C and D). However, this is obviously not the whole reason for the SEX phenotype, because much higher levels of starch accumulated in TUB8-silenced leaves than ATG6-silenced leaves (Fig. S10). The classic plastidial starch degradation must have been inhibited in the microtubule-disrupted cells, although the mechanism linking microtubule cytoskeleton and leaf starch degradation in chloroplasts is completely unknown. Microtubule cytoskeletons might exert influence on starch degradation through unknown, direct or indirect connections to SEX phenotype-generating factors. Further studies are required to clarify how the severe SEX phenotype is generated in microtubule-disassembled plants.
ATG5, ATG6 and ATG7 are not required for SEX chlorophagy
Organelles become substrates of autophagy when they are damaged or not needed in some special cases.Citation72 As a specialized organelle in plants and photoautotrophic algae, the chloroplast is no exception. Vacuolar degradation of the whole chloroplast in a phagocytic-type way was proposed decades ago in pioneering ultrastructural studies of dark-induced senescing primary wheat leaves.Citation73 Other studies in naturally or induced senescent leaves report 2 pathways for degradation of chloroplastic structures: ATG-dependent internalization of the whole chloroplast or Rubisco-containing bodies into central vacuole and ATG-independent degradation of stromal proteins in senescence-associated vacuoles.Citation74-77 It is thought that the vacuolar degradation of chloroplastic structures play essential roles in remobilization and recycling of nutrients in leaves during plant senescence.Citation17 A recent study shows a distinct autophagy-independent pathway for stress-induced degradation of thylakoid and stromal proteins, which is mediated by the formation of chloroplast vesiculation-containing vesicles.Citation78 In other cases, chloroplast damages due to either virus infection or gene mutations in chloroplast translocon or maltose transporter also caused vacuolar degradation of chloroplasts,Citation67,79,80 suggesting that chloroplast-targeted autophagy plays an important role in organelle quality control.
In this study, misshapen chloroplasts bearing large starch granules were delivered into vacuole of mesophyll cells with disrupted microtubules through a process termed SEX chlorophagy (). The aberrations in chloroplast morphology and the occurrence of SEX chlorophagy were more evident when considerable starch granules accumulated in chlorotic leaves of TUB8-silenced plants (). Apart from whole chloroplasts, chloroplast remnants like thylakoid membranes were occasionally observed in vacuoles (), suggesting that cells might be undergoing vacuolar degradation of chloroplasts. These extreme phenotypes of chloroplast degeneration probably contribute to leaf chlorosis occurrance at later growth stages of microtubule-disrupted plants (; Fig. S6B and C, as well as S13D). Indeed, defective metabolism of starch breakdown products in Arabidopsis mex1 and the dpe1 mex1 double mutant triggers chloroplast degradation and leads to chlorosis.Citation67 The abnormal vacuolar deposition of chloroplasts in TUB8-silenced cells was further confirmed by time-lapse observations of mesophyll cells under CLSM, which showed Brownian motion of chloroplasts or chloroplastic structures within the central vacuole (Movies S2 and S3). To demonstrate the vacuolar localization of SEX chloroplasts, we have also made attempts to isolate vacuoles from TUB8-silenced N. benthamiana protoplasts as described.Citation81 Whereas vacuoles were successfully released from nonsilenced protoplasts by osmotic and thermal shock, few vacuoles were observed to release from TUB8-silenced protoplast for unknown reason at the corresponding step (data not shown). Strikingly, the phenotypic aberrations in chloroplast morphologies and SEX chlorophagy in TUB8-silenced leaves were alleviated by TUB8 and APS1 cosilencing which markedly reduced starch accumulation (), revealing a positive correlation between starch content, malformations of chloroplasts and vacuolar degradation in TUB8-silenced plants.
The concurrence of SEX chlorophagy and compromised macroautophagy in microtubule-disrupted plants indicates that SEX chlorophagy may be macroautophagy-independent. This hypothesis was further confirmed by cosilencing experiments. The number of vacuole-localized SEX chloroplasts did not decrease when TUB8 was cosilenced with other ATG genes (ATG6, ATG5 or ATG7) (). Moreover, no vesicle embedding chloroplast was observed either in the cytoplasm or the vacuole (). All evidence supported a dispensable role of macroautophagy in SEX chlorophagy. In fact, the size of normal mature chloroplasts, about 5 to 10 µm across, far exceeds the reported capability of classic double-membrane autophagosomes (around 1.5 µm),Citation17 not to mention the size of swollen chloroplasts resulting from massive starch accumulation. Cargo selectivity seems to exist during the degradation of SEX chloroplast, as not all the chloroplasts swarmed into the vacuole at the same time unless the cell collapsed (). How SEX chloroplasts enter vacuoles is still unknown and requires further investigation in the future.
Materials and Methods
Plant materials and plasmids
N. benthamiana plants were cultivated in growth rooms at 25°C under a 16-h light/8-h dark cycle. The Bait vector BD-ATG6 and Prey vector AD-TUB8 used in the yeast 2-hybrid were generated by inserting the full-length gene ATG6 and TUB8 into pYL302 and pJG4-5 respectively.Citation82 Constructs for LCI assay were generated by cloning target genes into 35S-nLUC or 35S-cLUC respectively.Citation37 Expression Vectors used in coIP and confocal assays were generated by cloning target genes into pCAMBIA1300-based T-DNA vectors with respective tags.Citation82 The VIGS vector TRV1 has been described;Citation40 TRV2-TUB8, TRV2-TUA6 were constructed by cloning the respective cDNA fragments into pTRV2-LIC as described previously.Citation83 Other VIGS vectors used in cosilencing assays, including TRV2-APS1-Luc, TRV2-TUB8-Luc, TRV2-TUB8-APS1, TRV2-TUB8-ATG6, TRV2-TUB8-ATG5, TRV2-TUB8-ATG7, were generated by inserting a 900-bp overlapping PCR product of a 450-bp sequence of a target gene with another 450-bp fragment of a target gene or a nontarget gene encoding luciferase. Autophagy marker CFP-ATG8f was described.Citation21 Fluorescent markers for microtubules were generated by cloning MBD domains of MAP4 into A pCAMBIA1300-based T-DNA vector with GFP or YFP. Primers used for gene cloning are listed in Table S1.
Transient expression by agroinfiltration and VIGS
Agroinfiltration-mediated transient expression in N. benthamiana leaves was performed as described.Citation82 Leaf samples were detached at 40 to 60 h postinfiltration for the corresponding assays. For VIGS assay, mixed Agrobacterium cultures containing TRV1 or TRV2-derivative plasmids (ratio 1:1) were infiltrated into the leaves of 6-leaf stage N. benthamiana plants as described previously.Citation21
Protein-protein interaction assay
A yeast 2-hybrid system was used to screen and confirm protein-protein interactions as described.Citation82
For the firefly luciferase complementation imaging (LCI) assay, Agrobacterium cultures containing nLUC- and cLUC-fusion constructs were infiltrated into N. benthamiana. Leaves were detached at 40 h post-infiltration and sprayed with 1 mM luciferin (Gold Biotechnology, LUCK-1G). Luminescence was captured with a low-light cooled Andor iXon CCD camera (Andor Technology, South Windsor, CT, USA) after 10 min of exposure. Anti-firefly LUC antibodies (Sigma, L0159) were used to detect the expression of nLUC- and cLUC-fusion constructs.
For coimmunoprecipitation (CoIP) assay, total proteins were extracted in buffer containing 10% (v/v) glycerol (Sinopharm Chemical Reagent, 10010692), 25 mM Tris-HCl (pH 7.5; CAMYCAL, T41571), 150 mM NaCl (Sinopharm Chemical Reagent, 10019318), 1 mM EDTA (Sinopharm Chemical Reagent, 10009717), 2 mM DTT (Sigma-Aldrich, D0632), 0.1% (v/v) Triton X-100 (Sigma-Aldrich, T8787), 1 mM PMSF (Sigma-Aldrich, P7626) and protease inhibitor cocktail (1 x; Roche, 04693116001). Protein extracts were incubated with the anti-HA antibody (Santa Cruz Biotechnology, sc-7392) for 4 h at 4°C followed by incubation with protein A/G plus agarose beads (Santa Cruz Biotechnology, sc-2003) for 1 h to overnight. The beads were collected and washed 3 times with the extraction buffer. Immunoprecipitated samples were then used for western blot analysis using the anti-MYC antibody (Santa Cruz Biotechnology, sc-40).
Microtubule colocalization assay in vivo
For colocalization assay in vivo, CFP-ATG6 and microtubule reporter GFP-MBD or YFP-MBD were transiently coexpressed in leaves of 6-leaf stage N. benthamiana for 48 h. Their colocalizations were then detected with a Zeiss LSM 710 laser scanning microscope (Carl Zeiss, Jena, Germany) by sequential scanning of different fluorescences in 2 tracks.
Real-time RT-PCR
Real-time RT PCR was used to test the silencing efficiency and target-specificity of VIGS. The upper expanded leaves of N. benthamiana plants silenced for about 3 wk were detached for RNA extraction and reverse transcription as described.Citation21 Real-time PCR analysis was performed on a Bio-Rad CFX96™ Real-Time PCR Detection System (Bio-Rad, Gladesville, NSW, Australia) using Power SYBR→ Green PCR Master Mix (Applied Biosystems, 4367659). ACT7 and EF1A were used as the internal controls. Primers used for RT-PCR analysis are listed in Table S1.
Live-cell imaging of microtubules and autophagic structures
Microtubule reporter GFP-MBD was transiently expressed in N. benthamiana leaves for 48 h and detached for confocal imaging under an inverted Zeiss LSM 710 laser scanning microscope. The GFP-derived fluorescence was monitored by a 488-nm excitation laser.
Autophagy marker CFP-ATG8f was imaged at 48 to 60 h postinfiltration using a Zeiss LSM 710 microscope with an excitation light of 405 nm and chloroplast autofluorescence was excited at 543 nm. Nocturnal autophagy was examined at time points of 0 h (at the end of day), 4 h (at midnight), 8 h (at the end of night) after onset of darkness. For oxidation-induced autophagy, N. benthamiana leaves were infiltrated with 10 µM methyl viologen (Sigma-Aldrich, 856177), at the area where CFP-ATG8f had been transiently expressed for about 36 h, to keep the 24-h treatment under normal light/dark cycle before autophagy examination at the next end of night. All image processing was performed using ZEN 2009 Light Edition. For quantitative analysis of autophagic activity, the number of cells and CFP-ATG8f-labeled structures in a field of view were counted separately to calculate the average number of autophagic structure per cell. More than 100 mesophyll cells from several views of one independent experiment were used for quantification. Relative autophagic activity was obtained by normalizing the number of autophagic structure per cell in tested samples to that of the control. Three or more independent experiments were performed to calculate the final relative autophagic activity.
Iodine staining and starch content measurement
Leaves or seedlings were harvested at the indicated time and decolorized by ethanol for subsequent qualitative and quantitative determination of starch contents as described.Citation21
Transient and long-term application of microtubule inhibitors
For transient application of microtubule inhibitors, amiprophos-methyl (APM; Sigma-Aldrich, 03992) and oryzalin (Sigma-Aldrich, 36182) were used at the concentration of 10 µM, and 0.1% (V/V) Dimethyl sulfoxide (DMSO; Amresco, 0231) was used as solvent control. Using a needless syringe, reagents were infiltrated into expanded N. benthamiana leaves transiently overexpressing CFP-ATG8f or GFP-MBD at the end of night. After a 4-h treatment, leaves were detached for examination of drug effects on autophagic structures or cortical microtubule arrays.
For long-term application of microtubule inhibitors, MS media containing APM or oryzalin at the concentration of 10 µM or 50 µM was used. In the germination assay, surface-sterilized seeds were plated directly on the inhibitor-containing MS plates and grown at 25°C under under a 16-h light/8-h dark cycle for about 2 or 3 wk. In the transplanting assay, N. benthamiana was germinated and cultured on normal MS media for about 5 or 6 wk and transplanted to new culture bottles with inhibitor-containing MS media for another 4-wk treatment under regular light-dark cycles. Sample collection for starch assay and TEM was performed at the end of night.
TEM
The general procedures used to prepare TEM samples and sections have been described previously.Citation21 Sections were stained with uranyl acetate and lead citrate and then examined with a Hitachi H-7650 electron microscope (Hitachi High-Technologies, Nishi-Shimbashi, Minato-Ku, Tokyo, Japan).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: NtTUB8 (KP316398), NtTUB6 (KP316399), NtTUB1 (KP316400), NbTUA6 (KP316402) and NtATG6 (KP316403).
Statistical analysis
Statistical analyses were performed with a 2-tailed unpaired Student t test. P < 0.05 was considered statistically significant (shown as *); P < 0.01 was considered highly statistically significant (shown as **).
Disclosure of Potential Conflicts of Interest
There were no potential conflicts of interest that needed to be disclosed.
Acknowledgments
We thank Professor Tonglin Mao (China Agricultural University) for helpful suggestions in the visualization of microtubule.
Supplemental material
Supplemental data for this article can be accessed on the publisher's website.
Supplemental_Files.zip
Download Zip (95.8 MB)Funding
This work was supported by the National Basic Research Program of China (2011CB910100), the National Natural Science Foundation of China (3142100007, 31470254, 31270182), China Postdoctoral Science Foundation (2014M550048), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20120002130005) and the National Transgenic Program of China (2014ZX08009-003, 2014ZX08005-001). Y. W. was supported in part by the Postdoctoral Fellowship of Tsinghua-Peking Joint Center for Life Sciences.
References
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/10.1146/annurev-cellbio-092910-154005
- Rabinowitz JD, White E. Autophagy and metabolism. Science 2010; 330:1344-8; PMID:21127245; http://dx.doi.org/10.1126/science.1193497
- Bassham DC. Plant autophagy–more than a starvation response. Curr Opin Plant Biol 2007; 10:587-93; PMID:17702643; http://dx.doi.org/10.1016/j.pbi.2007.06.006
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/10.1038/nature06639
- Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ 2005; 12 Suppl 2:1535-41; PMID:16247501; http://dx.doi.org/10.1038/sj.cdd.4401728
- Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc 2009; 84:431-48; PMID:19659885; http://dx.doi.org/10.1111/j.1469-185X.2009.00082.x
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 2007; 143:291-9; PMID:17098847; http://dx.doi.org/10.1104/pp.106.092106
- Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009; 5:954-63; PMID:19587533; http://dx.doi.org/10.4161/auto.5.7.9290
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005; 121:567-77; PMID:15907470; http://dx.doi.org/10.1016/j.cell.2005.03.007
- Lai Z, Wang F, Zheng Z, Fan B, Chen Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J 2011; 66:953-68; PMID:21395886; http://dx.doi.org/10.1111/j.1365-313X.2011.04553.x
- Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, Bassham DC, Vierstra RD, Parker JE, Bautor J, et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 2011; 66:818-30; PMID:21332848; http://dx.doi.org/10.1111/j.1365-313X.2011.04546.x
- Wang Y, Nishimura MT, Zhao T, Tang D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 2011; 68:74-87; PMID:21645148; http://dx.doi.org/10.1111/j.1365-313X.2011.04669.x
- Yano K, Suzuki T, Moriyasu Y. Constitutive autophagy in plant root cells. Autophagy 2007; 3:360-2; PMID:17426438; http://dx.doi.org/10.4161/auto.4158
- Chen CN, Chen HR, Yeh SY, Vittore G, Ho TH. Autophagy is enhanced and floral development is impaired in AtHVA22d RNA interference Arabidopsis. Plant Physiol 2009; 149:1679-89; PMID:19151132; http://dx.doi.org/10.1104/pp.108.131490
- Ghiglione HO, Gonzalez FG, Serrago R, Maldonado SB, Chilcott C, Cura JA, Miralles DJ, Zhu T, Casal JJ. Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J 2008; 55:1010-24; PMID:18547393; http://dx.doi.org/10.1111/j.1365-313X.2008.03570.x
- Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J 2010; 64:151-64; PMID:20659276
- Ishida H, Izumi M, Wada S, Makino A. Roles of autophagy in chloroplast recycling. Biochim Biophys Acta 2014; 1837:512-21; PMID:24269172; http://dx.doi.org/10.1016/j.bbabio.2013.11.009
- Yoshimoto K, Shibata M, Kondo M, Oikawa K, Sato M, Toyooka K, Shirasu K, Nishimura M, Ohsumi Y. Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci 2014; 127:1161-8; PMID:24463818; http://dx.doi.org/10.1242/jcs.139709
- Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B. Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 2013; 25:4085-100; PMID:24179123; http://dx.doi.org/10.1105/tpc.113.113407
- Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, Hayashi M, Sakamoto W, Ohsumi Y, Nishimura M. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 2013; 25:4967-83; PMID:24368788; http://dx.doi.org/10.1105/tpc.113.116947
- Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al. Autophagy contributes to leaf starch degradation. Plant Cell 2013; 25:1383-99; PMID:23564204; http://dx.doi.org/10.1105/tpc.112.108993
- Izumi M, Hidema J, Makino A, Ishida H. Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol 2013; 161:1682-93; PMID:23457226; http://dx.doi.org/10.1104/pp.113.215632
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458-67; PMID:19491929; http://dx.doi.org/10.1038/nrm2708
- Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell 2013; 26:9-18; PMID:23810512; http://dx.doi.org/10.1016/j.devcel.2013.05.024
- Yoshimoto K. Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 2012; 53:1355-65; PMID:22764279; http://dx.doi.org/10.1093/pcp/pcs099
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22:140-9; PMID:20097051; http://dx.doi.org/10.1016/j.ceb.2010.01.001
- Wirawan E, Lippens S, Vanden Berghe T, Romagnoli A, Fimia GM, Piacentini M, Vandenabeele P. Beclin1: a role in membrane dynamics and beyond. Autophagy 2012; 8:6-17; PMID:22170155; http://dx.doi.org/10.4161/auto.8.1.16645
- Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol 2008; 20:71-6; PMID:18226514; http://dx.doi.org/10.1016/j.ceb.2007.11.010
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 2008; 9:309-22; PMID:18322465; http://dx.doi.org/10.1038/nrm2369
- Nick P. Microtubules, signalling and abiotic stress. Plant J 2013; 75:309-23; PMID:23311499; http://dx.doi.org/10.1111/tpj.12102
- Breviario D, Giani S, Morello L. Multiple tubulins: evolutionary aspects and biological implications. Plant J 2013; 75:202-18; PMID:23662651; http://dx.doi.org/10.1111/tpj.12243
- Brandizzi F, Wasteneys GO. Cytoskeleton-dependent endomembrane organization in plant cells: an emerging role for microtubules. Plant J 2013; 75:339-49; PMID:23647215; http://dx.doi.org/10.1111/tpj.12227
- Mackeh R, Perdiz D, Lorin S, Codogno P, Pous C. Autophagy and microtubules - new story, old players. J Cell Sci 2013; 126:1071-80; PMID:23620510; http://dx.doi.org/10.1242/jcs.115626
- Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett 2004; 567:302-6; PMID:15178341; http://dx.doi.org/10.1016/j.febslet.2004.04.088
- Zientara-Rytter K, Sirko A. Selective autophagy receptor Joka2 co-localizes with cytoskeleton in plant cells. Plant Signal Behav 2014; 9:e28523; PMID:24705105; http://dx.doi.org/10.4161/psb.28523
- Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 2002; 14:1483-96; PMID:12119369; http://dx.doi.org/10.1105/tpc.002493
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 2008; 146:368-76; PMID:18065554; http://dx.doi.org/10.1104/pp.107.111740
- Olson KR, McIntosh JR, Olmsted JB. Analysis of MAP 4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol 1995; 130:639-50; PMID:7622564; http://dx.doi.org/10.1083/jcb.130.3.639
- Marc J, Granger CL, Brincat J, Fisher DD, Kao T, McCubbin AG, Cyr RJ. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 1998; 10:1927-40; PMID:9811799
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 2002; 30:415-29; PMID:12028572; http://dx.doi.org/10.1046/j.1365-313X.2002.01297.x
- Falconer MM, Seagull RW. Amiprophos-methyl (APM): A rapid, reversible, anti-microtuble agent for plant cell cultures. Protoplasma 1987; 136:118-24; http://dx.doi.org/10.1007/BF01276360
- Morejohn LC, Bureau TE, Mole-Bajer J, Bajer AS, Fosket DE. Oryzalin, a dinitroaniline herbicide, binds to plant tubulin and inhibits microtubule polymerization in vitro. Planta 1987; 172:252-64; PMID: 24225878; http://dx.doi.org/10.1007/BF00394595
- Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y, Liu Y. Cytoplastic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with ATG3 to Negatively Regulate Autophagy and Immunity in Nicotiana benthamiana. Plant Cell 2015; 27:1316-31; PMID:25829441; http://dx.doi.org/10.1105/tpc.114.134692
- Wang Y, Liu Y. Autophagic degradation of leaf starch in plants. Autophagy 2013; 9:1247-8; PMID:23722252; http://dx.doi.org/10.4161/auto.25176
- Jin X, Ballicora MA, Preiss J, Geiger JH. Crystal structure of potato tuber ADP-glucose pyrophosphorylase. EMBO J 2005; 24:694-704; PMID:15692569; http://dx.doi.org/10.1038/sj.emboj.7600551
- Lin TP, Caspar T, Somerville C, Preiss J. Isolation and Characterization of a Starchless Mutant of Arabidopsis thaliana (L.) Heynh Lacking ADPglucose Pyrophosphorylase Activity. Plant Physiology 1988; 86:1131-5; PMID:16666044; http://dx.doi.org/10.1104/pp.86.4.1131
- Ishida H, Wada S. Autophagy of whole and partial chloroplasts in individually darkened leaves: a unique system in plants? Autophagy 2009; 5:736-7; PMID:19395861; http://dx.doi.org/10.4161/auto.5.5.8568
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 2010; 188:253-69; PMID:20100911; http://dx.doi.org/10.1083/jcb.200907015
- Geeraert C, Ratier A, Pfisterer SG, Perdiz D, Cantaloube I, Rouault A, Pattingre S, Proikas-Cezanne T, Codogno P, Poüs C. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J Biol Chem 2010; 285:24184-94; PMID:20484055; http://dx.doi.org/10.1074/jbc.M109.091553
- Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic 2008; 9:574-87; PMID:18182013; http://dx.doi.org/10.1111/j.1600-0854.2008.00701.x
- Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 2006; 281:36303-16; PMID:16963441; http://dx.doi.org/10.1074/jbc.M607031200
- Seglen PO, Berg TO, Blankson H, Fengsrud M, Holen I, Stromhaug PE. Structural aspects of autophagy. Adv Exp Med Biol 1996; 389:103-11; PMID:8860999; http://dx.doi.org/10.1007/978-1-4613-0335-0_12
- Aplin A, Jasionowski T, Tuttle DL, Lenk SE, Dunn WA, Jr. Cytoskeletal elements are required for the formation and maturation of autophagic vacuoles. J Cell Physiol 1992; 152:458-66; PMID:1506410; http://dx.doi.org/10.1002/jcp.1041520304
- Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 2006; 7:129-45; PMID:16420522; http://dx.doi.org/10.1111/j.1600-0854.2005.00368.x
- Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol 2010; 11:89; PMID:21092184; http://dx.doi.org/10.1186/1471-2121-11-89
- Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem 1994; 269:11492-7; PMID:7908909
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 2010; 191:155-68; PMID:20921139; http://dx.doi.org/10.1083/jcb.201002100
- Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O, Rubinsztein DC. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell 2012; 47:359-70; PMID:22742832; http://dx.doi.org/10.1016/j.molcel.2012.05.040
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 2013; 14:759-74; PMID:24201109; http://dx.doi.org/10.1038/nrm3696
- Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764-76; PMID:20639694; http://dx.doi.org/10.4161/auto.6.6.12709
- Ohsumi Y. Historical landmarks of autophagy research. Cell Res 2014; 24:9-23; PMID:24366340; http://dx.doi.org/10.1038/cr.2013.169
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007; 12:209-18; PMID:17295840; http://dx.doi.org/10.1111/j.1365-2443.2007.01050.x
- Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochem J 2007; 401:13-28; PMID:17150041; http://dx.doi.org/10.1042/BJ20061393
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, Somerville C. Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol 1991; 95:1181-8; PMID:16668109; http://dx.doi.org/10.1104/pp.95.4.1181
- Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 2010; 61:209-34; PMID:20192737; http://dx.doi.org/10.1146/annurev-arplant-042809-112301
- Niittyla T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC. A previously unknown maltose transporter essential for starch degradation in leaves. Science 2004; 303:87-9; PMID:14704427; http://dx.doi.org/10.1126/science.1091811
- Stettler M, Eicke S, Mettler T, Messerli G, Hortensteiner S, Zeeman SC. Blocking the metabolism of starch breakdown products in Arabidopsis leaves triggers chloroplast degradation. Mol Plant 2009; 2:1233-46; PMID:19946617; http://dx.doi.org/10.1093/mp/ssp093
- Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman SC, Smith SM, Smith AM. A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant Journal 2004; 37:853-63; PMID:14996213; http://dx.doi.org/10.1111/j.1365-313X.2003.02012.x
- Lu Y, Sharkey TD. The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 2004; 218:466-73; PMID:14593480; http://dx.doi.org/10.1007/s00425-003-1127-z
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM. A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 2001; 26:89-100; PMID:11359613; http://dx.doi.org/10.1046/j.1365-313x.2001.01012.x
- Streb S, Zeeman SC. Starch metabolism in Arabidopsis. Arabidopsis Book 2012; 10:e0160; PMID:23393426; http://dx.doi.org/10.1199/tab.0160
- Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol 2014; 205:435-45; PMID:24862571; http://dx.doi.org/10.1083/jcb.201402054
- Wittenbach VA, Lin W, Hebert RR. Vacuolar localization of proteases and degradation of chloroplasts in mesophyll protoplasts from senescing primary wheat leaves. Plant Physiol 1982; 69:98-102; PMID:16662193; http://dx.doi.org/10.1104/pp.69.1.98
- Minamikawa T, Toyooka K, Okamoto T, Hara-Nishimura I, Nishimura M. Degradation of ribulose-bisphosphate carboxylase by vacuolar enzymes of senescing French bean leaves: immunocytochemical and ultrastructural observations. Protoplasma 2001; 218:144-53; PMID:11770431; http://dx.doi.org/10.1007/BF01306604
- Chiba A, Ishida H, Nishizawa NK, Makino A, Mae T. Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol 2003; 44:914-21; PMID:14519773; http://dx.doi.org/10.1093/pcp/pcg118
- Otegui MS, Noh YS, Martinez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 2005; 41:831-44; PMID:15743448; http://dx.doi.org/10.1111/j.1365-313X.2005.02346.x
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 2009; 149:885-93; PMID:19074627; http://dx.doi.org/10.1104/pp.108.130013
- Wang S, Blumwald E. Stress-Induced Chloroplast Degradation in Arabidopsis Is Regulated via a Process Independent of Autophagy and Senescence-Associated Vacuoles. Plant Cell 2014; 26(12):4875-88; PMID: 25538186
- Niwa Y, Kato T, Tabata S, Seki M, Kobayashi M, Shinozaki K, Moriyasu Y. Disposal of chloroplasts with abnormal function into the vacuole in Arabidopsis thaliana cotyledon cells. Protoplasma 2004; 223:229-32; PMID:15221529; http://dx.doi.org/10.1007/s00709-004-0037-7
- Seay M, Hayward AP, Tsao J, Dinesh-Kumar SP. Something old, something new: plant innate immunity and autophagy. Curr Top Microbiol Immunol 2009; 335:287-306; PMID:19802571
- Robert S, Zouhar J, Carter C, Raikhel N. Isolation of intact vacuoles from Arabidopsis rosette leaf-derived protoplasts. Nat Protoc 2007; 2:259-62; PMID:17406583; http://dx.doi.org/10.1038/nprot.2007.26
- Zhao J, Liu Q, Zhang H, Jia Q, Hong Y, Liu Y. The rubisco small subunit is involved in tobamovirus movement and Tm-2(2)-mediated extreme resistance. Plant Physiol 2013; 161:374-83; PMID:23148080; http://dx.doi.org/10.1104/pp.112.209213
- Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP. A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 2007; 145:1161-70; PMID:17932306; http://dx.doi.org/10.1104/pp.107.107391
