ABSTRACT
An evolutionarily conserved gene network regulates the expression of genes involved in lysosome biogenesis, autophagy, and lipid metabolism. In mammals, TFEB and other members of the MiTF-TFE family of transcription factors control this network. Here we report that the lysosomal-autophagy pathway is controlled by Mitf gene in Drosophila melanogaster. Mitf is the single MiTF-TFE family member in Drosophila and prior to this work was known only for its function in eye development. We show that Mitf regulates the expression of genes encoding V-ATPase subunits as well as many additional genes involved in the lysosomal-autophagy pathway. Reduction of Mitf function leads to abnormal lysosomes and impairs autophagosome fusion and lipid breakdown during the response to starvation. In contrast, elevated Mitf levels increase the number of lysosomes, autophagosomes and autolysosomes, and decrease the size of lipid droplets. Inhibition of Drosophila MTORC1 induces Mitf translocation to the nucleus, underscoring conserved regulatory mechanisms between Drosophila and mammalian systems. Furthermore, we show Mitf-mediated clearance of cytosolic and nuclear expanded ATXN1 (ataxin 1) in a cellular model of spinocerebellar ataxia type 1 (SCA1). This remarkable observation illustrates the potential of the lysosomal-autophagy system to prevent toxic protein aggregation in both the cytoplasmic and nuclear compartments. We anticipate that the genetics of the Drosophila model and the absence of redundant MIT transcription factors will be exploited to investigate the regulation and function of the lysosomal-autophagy gene network.
Introduction
The lysosomal-autophagy system is key to ensuring the continuous turnover of macromolecules and exhausted organelles, the removal of abnormal proteins, and the maintenance of a healthy cellular environment. Autophagy is the main mechanism contributing to the turnover of cellular components, playing diverse physiological roles in normal and pathological conditions.Citation1,2 Lysosomes are key organelles in this process, providing the acidic environment and hydrolases required for the degradation of autophagosomal content after lysosome-autophagosome fusion.Citation3
In mammals, the basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor EB (TFEB) functions as a master regulator of lysosomal biogenesis and autophagy,Citation4,5 and plays critical roles in cellular clearance and lipid metabolism.Citation6,7 TFEB coordinates the expression of genes belonging to the coordinated lysosomal expression and regulation (CLEAR) network, by direct binding to specific CLEAR-box sequences located near their promoters.Citation4,8 Through the positive regulation of the CLEAR network, TFEB controls several aspects of cellular clearance, including the biogenesis of lysosomes and autophagosomes, and their fusion during autophagy.Citation4,5 TFEB also regulates lysosomal exocytosis by inducing the release of intracellular Ca2+ and increasing the population of lysosomes in the close proximity to the cell surface.Citation6
TFEB function is particularly required in response to several pathological conditions, such as starvation, infection, and accumulation of lysosomal substrates,Citation5,9,4 and its regulation is finely tuned. In general, when nutrients are available TFEB is mainly located in the cytoplasm where it interacts with the mechanistic target of rapamycin (serine/threonine kinase) complex 1 (MTORC1) on the lysosomal membrane. MTORC1-mediated phosphorylation prevents TFEB nuclear translocation. During starvation, MTORC1 is released from the lysosomal membrane and TFEB translocates to the nucleus, where it induces target gene transcription.Citation10-12 The vacuolar-type H+-ATPase (V-ATPase) complex, a well-known proton pump that creates and maintains the lysosomal acidic environment,Citation13 also plays a crucial role in the MTORC1-mediated activation of cell growth. The presence of amino acids in the lysosomal lumen promotes the interaction of V-ATPase with the scaffolding complex Ragulator, which in turn activates MTORC1 translocation to the lysosomal membrane.Citation14,15
TFEB is a member of the MiTF-TFE subfamily (also known as the MIT family), together with TFEC, TFE3 and MITF. These transcription factors recognize similar target sequences (3′-CANNTG-5′) and bind DNA as homodimers or heterodimers in combination with other family members.Citation16,17 Recent data indicates that TFE3 and MITF are regulated by MTORC1 and drive transcriptional programs that are similar to those regulated by TFEB,Citation18,19 suggesting that in mammals such overlapping roles give rise to functional redundancy and compensation effects among family members. However, the invertebrate Caenorhabditis elegans has only one gene showing meaningful sequence similarity to the MiTF-TFE family, namely hlh-30. Notably, the HLH-30 protein works similarly to mammalian TFEB,Citation20,21 suggesting that regulation of lysosomal-autophagic pathways are evolutionarily conserved.
These findings in mammals and nematodes raised the question whether lysosomal-autophagic pathways are transcriptionally regulated in Drosophila as well.Citation22 The sole Drosophila MIT transcription factor, Mitf, functions in eye development in a similar manner to the mammalian MITF.Citation23-25 Since there are no other members of the MiTF-TFE family in the fruit fly genome, we investigated whether Mitf has additional functions modulating lysosomal biogenesis and autophagy. Our results indicate that the regulation of lysosomal biogenesis, autophagy and lipid metabolism is evolutionarily conserved and coordinated by the same family of evolutionarily conserved transcription factors in different species.
Results
Comparison of Drosophila Mitf and MiTF-TFE family members
To evaluate the possible functional relationships between Mitf and its human and worm homologs, we compared their protein functional domains. Amino acid sequence alignment of the MiTF-TFE human family members with Drosophila Mitf and C. elegans HLH-30 showed conservation of the basic, helix-loop-helix, and leucine zipper functional domains (). This suggests similar DNA binding specificities of the Drosophila and human proteins, which are known to bind the CLEAR box.
Figure 1. Drosophila Mitf shows sequence similarity to other MiTF-TFE family members and regulates the expression of V-ATPase and other target genes. (A) Amino acid sequence alignment of bHLH-Zip functional domains of human TFEB, TFEC, TFE3, MITF, C. elegans HLH-30, and D. melanogaster Mitf. Amino acids are color-coded based on side chain properties. (B) Phylogenetic tree depicting the distance between human members of MiTF-TFE family, C. elegans HLH-30 and D. melanogaster Mitf; 2 other human bHLH transcription factors are also shown as controls. (C) Heat map of the scores associated with the coexpression analysis of Drosophila lysosomal genes. A cluster of genes encoding V-ATPase subunits with strongly associated expressions is indicated (red box). (D) Logo representation of the dCLEAR element. The height of nucleotide symbols at each position is proportional to the conservation of nucleotides at that position. Graph shows the distribution of dCLEAR sites at the promoters of analyzed genes. (E) qRT-PCR analysis of gene expression of TFEB-network homologs in fat body samples isolated from larvae in which Mitf was overexpressed or silenced (Mitf KD) using the fat body driver (lsp2-GAL4). White bars show the fold change of the mRNA levels of target genes in Mitf-overexpressing versus control larvae. Black bars show the fold change of mRNA levels in Mitf-silenced larvae vs. control larvae. Gene expression was normalized relative to Act5C gene. Data are mean of replicates (n=3) ± SEM. *, P < 0.05; **, P < 0.005 by the Student t test.
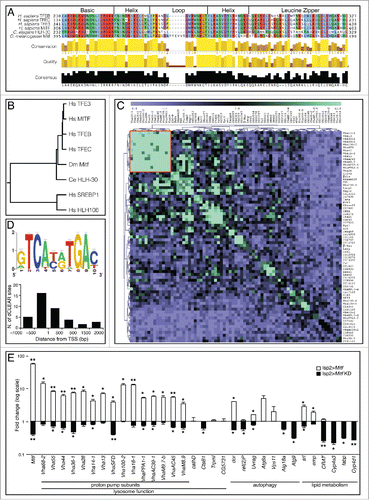
Phylogenetic analysis of the protein sequences indicated that human MiTF-TFE family members, the Drosophila Mitf and the C. elegans HLH-30 proteins belong to the same branch of the evolutionary tree (). Interestingly, Mitf is not closer to human MITF than to TFEB. The common ancestor gene underwent multiple rounds of duplication after the separation of the vertebrate and invertebrate lineages. Because gene duplication is often accompanied by specialization in gene function, we hypothesized that the distinct functions that have been described for the mammalian proteins—eye development for MITF, lysosome-autophagy regulation for TFEB and TFE3—may coexist in the same protein, Mitf, in Drosophila.
Coexpression analysis of Drosophila lysosomal genes reveals regulation of proton pump subunits
Gene expression analysis can be used to identify groups of genes that are coexpressed and therefore may have related functions.Citation26 To investigate the expression relationships among Drosophila lysosomal genes, we performed hierarchical clustering based on their coexpression scores, which were calculated using a recently described procedure.Citation4,27 The results showed a cluster of strongly coexpressed genes, which was entirely composed of the genes encoding the subunits of V-ATPase (). To examine whether the V-ATPase genes share any regulatory motif in common, we performed a de novo motif analysis looking for over-represented motifs in their putative regulatory regions (promoters, 5′ untranslated regions and first introns).Citation28 The analysis resulted in the identification of a palindromic 10-base pair (bp) motif (GTCA[CG][TA]TGAC) (), which is essentially the same as the mammalian TFEB target sequence; i.e., the CLEAR motif.Citation4 Together, these data suggest that the expression of genes encoding V-ATPase subunits is coordinated via the CLEAR motif and implicate Mitf as a prime candidate to orchestrate such coregulation.
We then examined the distribution of Mitf CLEAR sites (dCLEAR) in the sequence of Drosophila genes involved in lysosomal biogenesis, autophagy and lipid metabolism (listed in ). We focused our analysis on the regions spanning from –1000 to +2000 bp from all annotated transcription start sites (TSS) of this subset of genes that are homologs of human TFEB targets. Pattern-matching analysis showed that dCLEAR elements are highly enriched in this gene set, displaying a tight association (i.e., within 500 bp) with TSS (). Among the 33 genes analyzed, 60% (20 of 33) were found to have dCLEAR elements in the genomic region that was investigated, either as a single sequence or as multiple sites. In 45% of the cases (9 of 20) 2 or more dCLEAR sites were detected. Furthermore, we found that 80% of dCLEAR sequences (16 of 20) display a tendency to be concentrated in the region between –500 bp and the TSS. Interestingly, 86% of proton pump subunit genes (13 of 15) have at least one dCLEAR element.
Table 1. Distribution of dCLEAR elements near the promoters of Mitf target genes with a known role in lysosomal function, autophagy or lipid metabolism.
Mitf transcriptionally regulates genes involved in lysosomal biogenesis, autophagy and lipid metabolism
Reducing or increasing mammalian TFEB expression leads to downregulation and upregulation of its target genes, respectively.Citation4 In order to elucidate the role of Mitf in lysosomal biogenesis, autophagy and lipid metabolism, we studied animals in which the expression of Mitf was either increased or decreased. Animals overexpressing wild-type MitfCitation23 or a hairpin RNA (hpRNA) against Mitf mRNA were used. In both cases, transgenes were inserted downstream of yeast UAS sequences, in order to use the GAL4-UAS system and allow targeted, tissue-specific gene expression.Citation29 Using the fat-body-specific driver (lsp2-GAL4), expression analyses revealed a 60-fold increase of Mitf gene in Mitf-overexpressing larval fat body and a 60% reduction of Mitf gene expression in Mitf-knockdown samples (). We then investigated the expression of genes encoding V-ATPase subunits and other lysosome, autophagy and lipid metabolism genes—homologs of TFEB targets. V-ATPase subunit-encoding genes displayed significant upregulation in Mitf-overexpressing larvae and downregulation in Mitf-knockdown animals (). Notable examples of other Mitf-regulated genes include: 1) the ortholog of MCOLN1 (Trpml), which mediates TFEB activation of lysosomal exocytosis;Citation6 2) the orthologs of mammalian UVRAG, GABARAP, WIPI1 and ATG9A (uvrag, Atg8a, Atg18a and Atg9, respectively), which are involved in the main steps of autophagosome formation and maturation;Citation30-33 3) and the homolog of the polyubiquitin-binding protein SQSTM/p62 (ref(2)P), which is involved in the targeting of polyubiquitinated proteins to autophagosomes and is itself selectively degraded via the autophagic pathway.Citation34,35 Consistent with the mammalian TFEB gene network, Mitf also regulates genes involved in Drosophila lipid metabolism. Interestingly, one of the genes whose expression was significantly upregulated following Mitf overexpression was PPARGC1A (srl) (), a known key regulator of liver lipid metabolism that is transcriptionally induced during starvation.Citation7 A similar downregulation of the same target genes was obtained using a dominant negative form of Mitf (Mitf DN) (Fig. S1A).Citation23 These data suggest that Mitf transcriptionally regulates genes belonging to the lysosomal-autophagic pathway and lipid metabolism in Drosophila, similarly to its mammalian homolog TFEB.
Reduction of Mitf function leads to accumulation of autophagy substrates in Drosophila
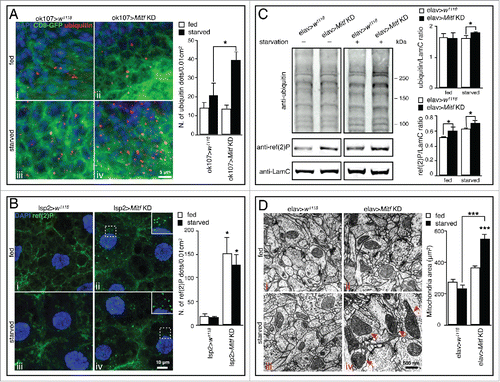
To test the hypothesis that Mitf has other roles beyond its reported function in eye development,Citation23 we investigated potential cellular phenotypes elicited by its loss of function. In the brain of adult flies in which Mitf was silenced using neuronal drivers (ok107-GAL4 and elav-GAL4) we observed a slight but significant accumulation of polyubiquitinated protein aggregates upon starvation ( iv and ). These aggregates are also observed in neurons in the context of lysosomal storage disorders (LSDs),Citation36,37 as well as in several other neurodegenerative diseases in which the lysosomal-autophagic pathway is impaired.Citation38-41 Numerous progressive neurological diseases that are accompanied by dysregulation of autophagy are also associated with the formation of intracellular SQSTM1-positive aggregates.Citation42 Similarly, the Drosophila ortholog of SQSTM1, ref(2)P, accumulates in the fat body and in the brain of adult Mitf-knockdown flies in the fed condition as well as upon starvation ( ii, 2B iv, and 2C), and these observations are consistent with impaired autophagy. Studies using transmission electron microscopy (TEM) revealed the presence of giant mitochondria in adult brain cells of starved Mitf-knockdown flies ( iv, arrowheads), further suggesting impaired autophagic flux. In conclusion, the accumulation of autophagic substrates, such as polyubiquitinated proteins, ref(2)P-positive aggregates, and mitochondria are consistent with a block of autophagy caused by Mitf knockdown.
Figure 2. Cellular phenotypes induced by Mitf knockdown. (A) Orthogonal projection of z-stacks confocal microscopy images of adult brain from fed and starved control flies or from flies in which Mitf was silenced using the mushroom body specific driver (ok107-GAL4). Tissues were stained with a polyubiquitin antibody (red) and mounted in DAPI (blue); CD8-GFP (green) labels cell membranes. Graph shows means of number of polyubiquitinated dots per 0.01 cm2 area. (B) Orthogonal projection of z-stacks confocal microscopy images of fat body from fed and starved control larvae or from larvae in which Mitf was silenced using the fat body specific driver (lsp2-GAL4). Tissues were stained with a ref(2)P antibody (green) and mounted in DAPI (blue). The regions within the dotted box are magnified in the insets (7x). Graph shows means of number of ref(2)P dots per 0.01 cm2 area. (C) Immunoblot against polyubiquitin and ref(2)P in fed and starved head extracts taken from control flies and from flies in which Mitf was silenced using the pan-neuronal driver (elav-GAL4). LamC was used as a loading control. Graph shows ubiquitin/LamC and ref(2)P/LamC quantification ratios. Experiments were performed in triplicate and band intensities were quantified using ImageJ software. (D) TEM images of fed and starved adult brains from control flies, and from flies in which Mitf was silenced using the pan-neuronal driver (elav-GAL4). Arrowheads show giant mitochondria detected in starved Mitf-knockdown animals. Graph shows the mean area of mitochondria. Measurement of mitochondrial area was performed using ImageJ software and at least 100 mitochondria/group were analyzed. Experiments were performed in triplicate and error bars represent SEM. *, P < 0.05; ***, P < 0.0005 by the Student t test.
Mitf regulates the cellular response to starvation
In mammals, TFEB mediates the transcriptional response to amino acid starvation and to lysosomal stress.Citation5,10 To investigate whether Mitf performs similar functions in Drosophila, we examined the starvation response in live tissues using the pH-sensitive fluorescent dye LysoTracker Green. Fat body from control fed animals displayed faint and diffuse LysoTracker Green staining, while starved control animals showed a typical punctate staining, indicating an increased number of acidic vesicles, including lysosomes and autolysosomes ( iv). Fat body overexpressing Mitf showed similar punctate LysoTracker Green staining in both fed and starved conditions ( ii and 3A v) revealing a higher rate of lysosome/autolysosomes production even in the fed condition. When Mitf was downregulated in the fed condition, the tissue displayed a diffuse staining, similar to control tissue ( iii). However, upon starvation we observed impaired activation of LysoTracker Green staining ( vi) suggesting either that acidification is impaired, or that the number of lysosomes and autolysosomes in starved Mitf-knockdown fat body is significantly reduced compared to starved control animals. This result is consistent with the idea that Mitf controls lysosomal acidic pH through regulation of the expression of genes encoding V-ATPase subunits as suggested by coexpression and RT-PCR data ().
Figure 3. Mitf regulates starvation-induced lysosomal biogenesis. (A) Orthogonal projection of z-stacks confocal microscopy images of LysoTracker Green staining (green) and DAPI (blue) on fed and starved fat body isolated from control larvae and larvae in which Mitf is either overexpressed or downregulated using the fat body driver (lsp2-GAL4). Graph shows means of number of vesicles per 0.01 cm2 area. (B) Confocal microscopy images of LAMP1-GFP (green) and DAPI (blue) in fed and starved fat body isolated from control larvae and larvae in which Mitf is either overexpressed or downregulated using the fat body driver (lsp2-GAL4). The regions within the dotted box are magnified in the insets (7x). Graph shows means of number of lysosomes per 0.01 cm2 area. (C) TEM images of adult brains from fed and starved control flies and flies in which Mitf is either overexpressed or downregulated using the pan-neuronal driver (elav-GAL4). Lysosomes (L) and lamellar bodies (LB) are indicated. Animals were raised at 23°C. Experiments were performed in triplicate and error bars represent SEM. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 by the Student t test.
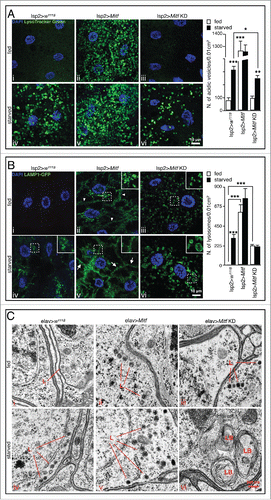
LAMP1-GFP transgenic flies were used to independently evaluate the status of the lysosomal compartment in fed and starved animals with normal and altered Mitf activities. As expected, we observed expansion of the lysosomal compartment in fat bodies of starved control larvae compared to fed controls indicating starvation-mediated activation of lysosomal biogenesis ( iv). Overexpression of Mitf in fat bodies significantly increased the number of LAMP1-GFP-positive structures, some of which were located near the cell surface ( ii, arrowheads) suggesting a potential induction of lysosomal exocytosis. Interestingly, upon nutrient starvation we could not detect a significant additional increase in the number of lysosomes, but the migration of lysosomes to the plasma membrane was clearly boosted (). These observations were confirmed by TEM experiments, which showed increased number of lysosome-like structures in the adult brain of starved Mitf-overexpressing flies ( ii and 3C v, marked as “L”). We observed a slight increase in the number of LAMP1-GFP-positive structures in fed Mitf-knockdown tissue ( iii), compared to control counterparts. Such structures were not stained by the pH sensitive LysoTracker Green dye ( iii) suggesting that they represent immature lysosomes in which lysosomal acidification is abnormal. Upon amino acid starvation vesicle size was increased in Mitf-knockdown larvae when compared to LAMP1-GFP-positive structures detected in starved control animals ( vi, magnified in the insets) suggesting lysosomal burden following downregulation of Mitf. Moreover, starving Mitf-knockdown flies caused lysosomal accumulation of undigested membranes, as implied by the observation of lamellar bodies ( vi, marked as “LB”). Taken together these results suggest that starvation-induced activation of autophagy increased the flow of membranes and cytoplasmic material to the lysosome, but Mitf silencing impairs the normal activation of lysosomal biogenesis as a response to starvation.
In mammalian cells, TFEB overexpression significantly increases the number of autophagosomes and enhances lysosome-autophagosome fusion.Citation5 Accordingly, using the UAS-Atg8a-mCherry transgene we found that Mitf-overexpressing adult flies display a higher number of autophagosomes in presence of nutrients, compared to control fed animals ( ii). Upon nutrient starvation we observed a further increase of autophagosomes ( v) suggesting an additive effect on autophagosome biogenesis.
Figure 4. Mitf regulates autophagy activation and lipid breakdown. (A) Orthogonal projection of z-stacks confocal microscopy images of UAS-Atg8a-mcherry (red) and DAPI (blue) in fed and starved adult brain isolated from control flies and flies in which Mitf is either overexpressed or downregulated using the pan-neuronal driver (elav-GAL4). Graph shows means of number of autophagosomes per 0.01 cm2 area. (B) Confocal microscopy images of Nile red staining (red) and DAPI (blue) on fat body tissue isolated from fed and starved control larvae and larvae in which Mitf is either overexpressed or downregulated using the fat body driver (lsp2-GAL4). Graph shows means of lipid droplet areas. Experiments were performed in triplicate and error bars represent SEM. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 by the Student t test.
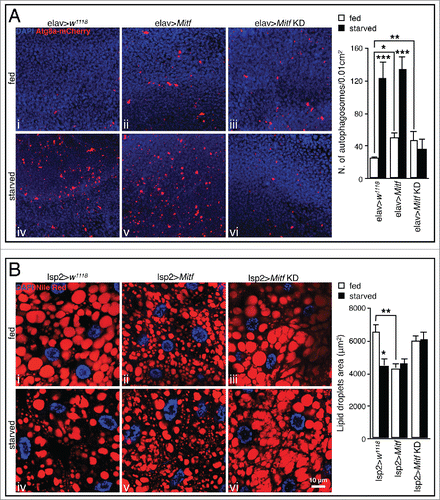
In conditions of reduced/impaired activity of lysosomal enzymes the ability of lysosomes to fuse with autophagosomes is reduced,Citation43 eventually leading to accumulation of autophagosomes.Citation36 Consistent with this, we observed a slight but significant accumulation of autophagosomes in Mitf-knockdown adult brain ( iii). Upon nutrient deprivation no significant change in the number of autophagosomes was detected ( vi), suggesting that in Mitf-knockdown animals the downregulation of autophagy-related genes () affects autophagosome biogenesis during the cellular response to starvation. Supporting this hypothesis, we observed that several genes involved in autophagy and especially those implicated in autophagosome formation and maturation (i.e., uvrag, Atg8a, Atg18a and Atg9) were significantly less upregulated after starvation in Mitf-knockdown larvae, compared to starved control animals (Fig. S1B). These data are consistent with the observation that in mammals the downregulation of TFEB leads to decreased levels of LC3-II both in normal and starved conditions.Citation5 Together, these data highlight the similarities between the roles of Drosophila Mitf and mammalian TFEB in the cellular response to starvation and the regulation of autophagy pathway.
Mitf regulates lipid breakdown in the fat body
Since TFEB controls lipid metabolism in mouse liver,Citation5 we investigated the possibility that Mitf performs a similar function in Drosophila. Using the lipophilic dye Nile red to stain the fat body we observed a significant reduction of lipid droplets in Mitf-overexpressing tissues compared to control animals ( ii). Together with previous results on lipid metabolism gene expression (), these data suggest a role for Mitf in lipid breakdown. Unlike autophagy activation, lipid degradation was not significantly increased upon starvation treatment of Mitf-overexpressing larvae ( v). We could not detect significant differences in Nile red staining between fat body samples of Mitf-knockdown larvae and controls ( iii). However, after a 4 h fast, the fat body in Mitf-knockdown specimens did not show the starvation-mediated lipid degradation observed in fasted control samples, suggesting a defect in intracellular lipid degradation ( vi). These data highlight the importance of Mitf in the control of lipid metabolism in Drosophila. Together, the data described above support a role of Drosophila Mitf in lysosome biogenesis, starvation-induced autophagy, and lipid breakdown, recapitulating the main features and functions of mammalian TFEB.
Mitf is required for starvation-induced fusion of lysosomes and autophagosomes
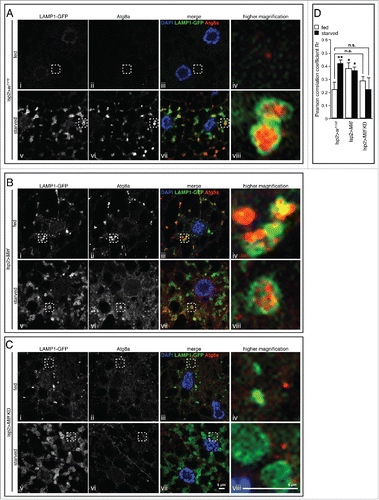
In mammalian cells TFEB enhances the autophagic flux as indicated by the increase in the number of autolysosomes in TFEB-overexpressing cells.Citation5 We asked whether Mitf overexpression and downregulation in Drosophila affect the formation of autolysosomes, as a consequence of lysosome-autophagosome fusion. To this end, we examined the colocalization of Atg8a and LAMP1 and quantified the fusion process in control, Mitf-overexpressing and -knockdown animals and in fed or starved conditions. As a measure of colocalization we calculated the correlation coefficient (Pearson Rr). Upon induction of autophagy by starvation we observed Atg8a-positive structures surrounded by LAMP1 (i.e., autolysosomes) in starved control animals ( vii and 5A viii) and the correlation coefficient was significantly higher than fed controls, indicating vesicle fusion. Significant Atg8a and LAMP1 colocalization was observed under both fed and starved conditions in Mitf-overexpressing tissue ( iii, 5B iv, 5B vii and 5B viii), consistent with the accumulation of LysoTracker Green-positive acidic vesicles, lysosomes and autophagosomes detected in the same conditions ( ii, 3A v, 3B ii, 3B v, 4A ii and 4A v). Interestingly, we observed a block of vesicle fusion upon starvation in Mitf-knockdown animals ( vii and 5C viii), suggesting that downregulation of genes encoding the entire proton pump subunit complex together with other lysosome- and autophagy-related proteins profoundly affects the autophagosome-lysosome fusion. In conclusion, these data demonstrate that Mitf induces autolysosome formation even in nutrient-rich conditions and they also indicate that Mitf functions are necessary for starvation-induced fusion of lysosomes and autophagosomes.
Figure 5. Mitf is required for starvation-induced fusion of lysosomes and autophagosomes. Confocal microscopy images of LAMP1-GFP (green), Atg8a (red) in fed and starved fat body isolated from control larvae (A) and larvae in which Mitf is either overexpressed (B) or downregulated (C) using the fat body driver (lsp2-GAL4). Tissues were stained with an Atg8a antibody (red) and mounted in DAPI (blue). The regions within the dotted boxes are magnified in the right column (5x). Autolysosomes are identified as enlarged Atg8a-positive vesicles decorated by a LAMP1-GFP-positive ring. (D) Graph shows quantification of LAMP1-GFP and Atg8 colocalization using ImageJ software to determine Pearson correlation coefficient Rr; at least 10 images/group were analyzed. Animals were raised at 25°C. Experiments were performed in triplicate and error bars represent SEM. *, P < 0.05; **, P < 0.005 by Student t test. Nonsignificant data are indicated (n.s.).
Inhibition of Drosophila MTORC1 leads to Mitf nuclear translocation and activation of target genes
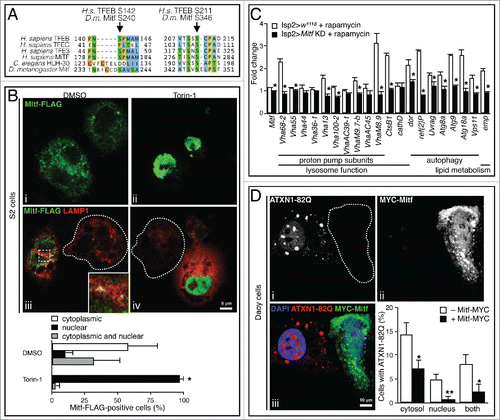
The nutrient content of the lysosome is sensed by a complex machinery, which includes MTORC1, V-ATPase subunits and additional complexes.Citation44 TFEB protein is phosphorylated by MTORC1 at several serine residues, including Ser142 and Ser211, the latter being crucial for the interaction of TFEB with the cytosolic chaperone YWHA/14-3-3.Citation12 MTORC1-mediated phosphorylation leads to TFEB sequestration in the cytosol.Citation5,10,11 Amino acid sequence comparison indicates conservation of Ser142 and Ser211 in TFEB, which correspond to Ser240 and Ser346 in the Drosophila protein () suggesting a similar regulatory mechanism. Therefore, we tested whether treating Schneider 2 (S2) Drosophila cells with the MTORC1 inhibitor Torin-1 induced Mitf nuclear translocation. S2 cells were transiently cotransfected with pUAST-Mitf-FLAG and pACT-GAL4. In control cells (DMSO) Mitf-FLAG localization was often diffuse throughout the cytoplasm with occasional colocalization with lysosomes (labeled with LAMP1 antibody) ( i and 6B iii, magnified in the inset). This indicates retention of Mitf outside of the nucleus, and is consistent with its transient interaction with the lysosomal surface. Upon Torin-1 treatment, Mitf-FLAG translocates to the nucleus ( ii and 6B iv) implying MTORC1-mediated regulation of Mitf activity in Drosophila. Interestingly, following Torin-1-mediated nuclear translocation of Mitf we detected expansion of the lysosomal compartment compared to nontransfected cells ( iv, dashed lines highlight nontransfected cells). Furthermore, we monitored the expression of several target genes in fat bodies upon rapamycin treatment of control and Mitf-knockdown Drosophila larvae. While the expression of Mitf target genes was upregulated in fat bodies from control larvae after treatment with rapamycin, this upregulation was significantly reduced for the majority of tested genes in Mitf-knockdown larvae (). We also treated S2 cells with rapamycin and observed Mitf nuclear translocation (Fig. S1C) in agreement with the observations in rapamycin-treated larvae. These results indicate that Drosophila MTORC1 inhibits Mitf nuclear translocation in a similar manner as mammalian MTORC1 acts on TFEB. Additionally, Mitf is required to activate an appropriate transcriptional response following the inhibition of Drosophila MTOR pathway.
Figure 6. Mitf regulation by MTORC1 and clearance of expanded ATXN1. (A) Amino acid sequence alignment of region containing MTORC1 target serines of human TFEB, TFEC, TFE3 and MITF, C. elegans HLH-30, and D. melanogaster Mitf. Amino acids are color-coded based on side chain properties. Arrows indicate target serine residues. (B) Confocal microscopy images of S2 Drosophila cells treated for 1 h with Torin-1 (250 nM) or DMSO and stained with anti-FLAG (green) to label Mitf-FLAG, anti-LAMP1 (red) to label lysosomes. The region within the dotted box is magnified in the inset. Dashed lines indicate nontransfected cells. The region within the dotted box is magnified in the inset (9x). Graph shows percentage of Mitf-FLAG-positive cells displaying Mitf localization in the cytoplasm, in the nucleus or in both compartments. Error bars represent SEM of 3 different experiments, at least 50 cells/group were counted. (C) qRT-PCR analysis of target gene expression in fat body samples isolated from control larvae and from larvae in which Mitf was silenced using the fat body driver (lsp2-GAL4). Animals were treated with rapamycin (1 μM) or with DMSO only. The graph shows the relative increased expression in the treated versus the corresponding untreated samples. White bars show the fold change of the mRNA levels of target genes in treated vs. untreated control larvae. Black bars show the fold change of mRNA levels in treated vs. untreated Mitf-silenced larvae. Gene expression was normalized relative to Act5C gene. Data are mean of replicates (n=3) ± SEM. (D) Confocal microscopy images of mammalian Daoy cells stably expressing mRFP-ATXN1-82Q and transfected with MYC-Mitf construct. ˜65% of total cells showed aggregates. Dashed lines indicate transfected cells. Graph shows percentage of cells with aggregates in the cytosol, or in the nucleus, or in both. Error bars represent SEM of 3 different experiments, at least 100 cells/experiment were counted. *, P < 0.05; **, P < 0.005 by Student t test.
Mitf mediates clearance of expanded ataxin-1
Since mammalian TFEB can clear cytoplasmic protein aggregates,Citation4,6,45-49 we investigated the potential of Drosophila Mitf to carry out a similar function. Spinocerebellar ataxia type 1 (SCA1) is a neurodegenerative disorder caused by expansion of a polyglutamine tract in ATXN1 (ataxin 1) protein.Citation50 We used a stable line of human medulloblastoma-derived cell line (Daoy cells) expressing glutamine-expanded ATXN1 fused with monomeric red fluorescent protein (mRFP-ATXN1[82Q])Citation51 in which expanded ATXN1 is found to aggregate in both nuclear and cytoplasmic compartments. A vector containing MYC-tagged Mitf was transfected into mRFP-ATXN1(82Q) Daoy cells. We counted cells displaying mRFP-ATXN1(82Q) and MYC-Mitf-positive cells discriminating between cytoplasmic and nuclear aggregates. Cells accumulating expanded ATXN1 in the cytoplasm or in the nucleus (or in both) showed significantly less ATXN1 when they were also Mitf-positive ( iii, dashed lines highlight transfected cells). These data suggest Mitf-mediated clearance of both cytoplasmic and nuclear ATXN1 aggregates.
Discussion
In this study we show that the Drosophila Mitf transcription factor regulates lysosomal biogenesis, autophagy and lipid metabolism in addition to its previously known role in eye development.Citation23 We also show that Mitf is required for the cellular response to starvation and its downregulation impairs the normal cellular response to nutrient deprivation. These data indicate that Mitf, the only Drosophila MiTF-TFE family member, performs multiple functions that in mammals are carried out by different family members. Thus, duplication during evolution led to functional specialization although mammalian members of the MiTF-TFE family still retain overlapping functions.Citation18,19 The absence of MiTF-TFE family paralogs in Drosophila facilitates the study of Mitf/TFEB-like functions since there are no compensatory effects from other MIT genes. Taking advantage of this exclusive feature we investigated the effects on lysosomes, autophagosomes and lipid droplets in conditions in which the sole member of the family was knocked down. Interestingly, Mitf-knockdown animals display cellular phenotypes that are reminiscent of lysosomal storage disorders. These phenotypes include the accumulation of autophagy substrates such as of polyubiquitinated proteins and ref(2)P-positive aggregates, as well as enlarged lysosomes.
The genes encoding the subunits of V-ATPase stand out as a subset of functionally related genes whose expression is modulated by Mitf. They contain multiple CLEAR sites near their transcription start sites and are particularly responsive to Mitf levels in qRT-PCR experiments. This is consistent with the idea that the proton pump is an ancient protein complex with a key, evolutionarily conserved, role in multiple physiological contexts including lysosomal acidification and nutrient sensing.Citation14,53 These observations support the use of Drosophila to further investigate Mitf role in the cellular adaptation to environmental cues, especially in light of the increasing interest in the nutrient-sensing machinery.
Knockdown of Mitf leads to the formation of LAMP-GFP-positive vesicles that do not stain with the LysoTracker Green dye. We interpret these vesicles as immature lysosomes with impaired acidification, which is consistent with the idea that proton pump subunit genes are particularly sensitive to Mitf regulation. We also found that Mitf is required for starvation-induced fusion of lysosomes and autophagosomes as revealed by Atg8a and LAMP1 colocalization experiments. In this context a recent publication reports that individual V-ATPase subunits (i.e., VhaSFD, VhaAC39-1 and Vha36-1) are not required for autophagosome-lysosome fusion in Drosophila.Citation52 Mitf, however, controls several stages of the autophagy process from the biogenesis of new autophagosomes to their fusion with lysosomes for cargo degradation, and it also regulates many other autophagy and lysosomal genes besides VhaSFD, VhaAC39-1 and Vha36-1 ATPase subunits including the entire proton pump complex; see and S1B.
We investigated the potential of Mitf to clear ATXN1 toxic protein aggregates, which accumulate in the cytosol as well as in the nuclear compartment. Interestingly, we found that Mitf not only promotes the clearance of cytoplasmic aggregates, but also prevents the formation of nuclear aggregates, possibly through decreasing the abundance of ATXN1 in both compartments. These observations suggest that the cytoplasmic machinery can be exploited to ameliorate neurodegenerative diseases in which nuclear inclusions are found. Importantly, the discovery of Mitf as a key regulator of lysosome biogenesis and autophagy in Drosophila provides the framework for using genetic approaches to investigate Mitf functions and modulate its activities in vivo. These unknown modulators may provide therapeutic opportunities to treat proteinopathies or other disorders in which activation of autophagy may reduce cellular burden.
Materials and methods
Maintenance of Drosophila strains
Overexpression and transgenic RNAi studies were performed using the GAL4-UAS system.Citation29 Flies were raised at low density on standard cornmeal/molasses/agar fly food at 25°C. The following lines were obtained from the Bloomington Drosophila Stock Center: the standard laboratory strain w1118; the pan-neuronal driver elav-GAL4, the mushroom body driver ok107-GAL4, and the fat body driver lsp2-GAL4. UAS-Mitf RNAi (ID: 108519) was obtained from the Vienna Drosophila RNAi Center (VDRC). UAS-Mitf-9B1 (Mitf-overexpressing line) and UAS-Mitf-EA2 (Mitf dominant negative, DN) were a gift from Francesca Pignoni (Upstate Medical University, NY). Crosses were performed by standard procedures, and progeny was raised at 28°C, unless otherwise specified. Drosophila genotypes are listed in Table S1.
Bioinformatic analysis
Multiple sequence alignments and the phylogenetic tree were generated with ClustalW (www.ebi.ac.uk/Tools/msa/clustalw2) and the alignment was edited with Jalview editor. Human orthologs of Drosophila genes were identified using Flybase (http://www.flybase.org) and HGNC (http://www.genenames.org) databases, except in the cases of ATP6AP1 and VPS11, which we identified as the single closest homologs to VhaAC45 and Vps11 Drosophila genes, respectively.
Expression analysis
Gene network expression analysis was performed as described previously, with minor modifications. Briefly, lysosomal genes were analyzed by using the g:Profiler tool.Citation54 For a selected gene, the g:Profiler subprogram, g:Sorter, returns genes with most similar coexpressed profiles within a specified Gene Expression Omnibus data set.Citation55 The analysis was carried out using 58 heterogeneous expression microarray datasets, available at the time of the analysis, based on the Affymetrix DrosGenome1 platform. g:Sorter was queried with the gene probes for the lysosomal genes. For each analyzed probe, lysosomal probes were scored based on their cumulative occurrence in the top 3% correlated gene probes in the 58 data sets. The MeV toolCitation56 was subsequently used to create a heat map of lysosomal genes based on coexpression scores, using default parameters. Probes used for Drosophila lysosomal genes are listed in Table S2.
Genome analysis
The logo of the resulting dCLEAR motif was elaborated by using WebLogo.Citation57 Drosophila melanogaster promoters were retrieved from Flybase database (http://www.flybase.org) and analyzed with the CLEAR position weight matrix by using DNA pattern from the Regulatory Sequence Analysis Tool packageCitation58 with default parameters.
RNA extraction
Third-instar larvae were pinned and immobilized on silicone plates and bathed in ice-cold phosphate-buffered saline (PBS; Life Technologies, 14190250). Total RNA was extracted from fat bodies using the RNeasy Mini Kit (Qiagen, 74104) and quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE).
Real-time quantitative RT-PCR
Real-time quantitative reverse transcription-PCR (real-time qRT-PCR) was performed to measure the mRNA levels of the studied genes using the CFX Connect real-time PCR detection system (Bio-Rad, Hercules, CA, USA). mRNA from 1 μg of total RNA was reverse-transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen, 205310). qRT-PCR amplification was performed using the PerfeCTa® SYBR® Green FastMix® (Quanta Biosciences, 95072), and the parameters used were according to the recommendations of Quanta Biosciences. For expression studies the qRT-PCR results were normalized against an internal control (Act5C). The primers were designed using the Primer 3 Plus softwareCitation59 with the following parameters: product size of 70 to 150 base pairs, melting temperature (Tm) of 58 to 62°C, length of 18 to 23 nucleotides and GC content of 30 to 80%. Sequences of primers used in qRT-PCR analysis were purchased from Integrated DNA Technologies (IDT) and are listed in Table S3.
Treatments
For starvation treatments third-instar larvae were dissected (fed) or placed in vials containing PBS 20% sucrose (Sigma, S0389) for 4 h prior to dissection (starved). Adult flies were dissected (fed) or placed in tubes containing filter paper soaked with PBS 20% sucrose for 24 h prior to dissection (starved). Rapamycin (Sigma, R0395) was administered to animals by dissolving the DMSO stock solutions in the feeding medium. Control animals were fed with medium containing 1% DMSO (Sigma, D-5879) only. Briefly, eggs were collected and transferred to food containing either DMSO or 1 μM rapamycin. RNA was extracted from fat body of third-instar larvae. For in vitro treatments, 24 h after transfection cells were treated for 1 h with Torin-1 (250 nM, Tocris, 4247) or for 72 h with rapamycin (1 μM), and then processed for immunofluorescence.
Immunofluorescence
For the detection of polyubiquitination and ref(2)P-positive aggregates, adult fly brains and third-instar larvae were used, respectively. Tissues were dissected in PBS, fixed in 3.7% formaldehyde (Sigma, F8775) in PBS for 30 min, permeabilized with 1% PBST (PBS containing 0.25% Triton X-100 [Fisher Scientific, BP151]) for 30 min, blocked with 5% normal goat serum (Sigma, G9023) in PBS for 30 min, incubated with mouse monoclonal anti-ubiquitin (1:20; Thermo Fisher Scientific, 13-1600) or rabbit anti-ref(2)P (1:500)Citation60 antibodies overnight at 4°C, followed by incubating with Alexa Fluor 555-conjugated goat anti-mouse or Alexa Fluor 488-conjugated goat anti-rabbit secondary antibodies (1:400; Invitrogen, A-21422 and A-11070) for 1 h at room temperature. The samples were washed with PBS in between and after antibody incubations. Polyubiquitin- and ref(2)P-positive dots were counted manually using ImageJ software analysis. For the detection of Atg8a-positive autophagosomes, third-instar larvae were incubated with rabbit monoclonal anti-Atg8 (1:100; Abcam, ab109364). For in vitro immunofluorescence, cells were fixed in 4% paraformaldehyde (Sigma, 158127) in PBS for 15 min, permeabilized in 0.25% Triton X-100 in PBS for 5 min, blocked with 10% BSA (Fisher BioReagents, BP1605) in PBS for 30 min and incubated with primary antibodies (mouse anti-FLAG [1:500; Sigma, F1804], rabbit anti-LAMP1 [1:500; Abcam, ab30687], mouse anti-MYC [1:200; Life Technologies, 13-2500]) overnight at 4°C. Alexa Fluor 488-conjugated goat anti-mouse or Alexa Fluor 594-conjugated goat anti-rabbit (1:400; Invitrogen, A-11001 and A-11037) were used as secondary antibodies. Samples were mounted in VECTASHIELD Antifade Mounting Medium containing DAPI (Vector Laboratories, H-1200) on glass slides and photographed live on a confocal fluorescence microscope (Leica TCS SP5, Leica Microsystems Inc., Buffalo Grove, IL, USA).
Confocal microscopy and image analysis
The fluorescent images were documented in a confocal fluorescence microscope (Leica TCS SP5, Leica Microsystems Inc., Buffalo Grove, IL, USA) equipped with 63x oil immersion objective. Confocal images were obtained as a single layer or as orthogonal projections throughout the thickness of the analyzed tissue (at least 10 sections per tissue with 1-μm interval), and then the signal was stacked and summed. Image processing was done with Adobe Photoshop C2 (Adobe System Inc., San Jose, USA). To carry out image quantification and analysis, at least 10 randomly chosen fields were scanned, using the same setting parameters (i.e., pinhole, laser power, and offset gain) below pixel saturation. Counts were performed manually using ImageJ software analysis. Colocalization of LAMP1-GFP (green) and Atg8a (red) vesicles was analyzed in ImageJ using the Intensity Correlation Analysis plugin to calculate Pearson correlation coefficient of single red and green channels.
Western blots
Fly heads (8 per sample) were homogenized in sample buffer (Thermo Fisher Scientific, NP0008), proteins fractionated on precast polyacrylamide gels (4-12% gradient; Thermo Fisher Scientific, NP0336), transferred overnight to nitrocellulose membranes (Bio-Rad, 1620145), blocked in TBST (TBS [Bio-Rad, 1706435] 0.1% Tween-20 [Amresco, M147]) 5% nonfat dry milk (Bio-Rad, 1706404) and probed with primary antibodies (anti-ubiquitin [1:100] and anti-ref(2)P [1:8000]Citation60) overnight at 4°C. Membranes were washed 3 times in TBST and incubated with fluorescently-labeled goat anti-mouse secondary antibody (1:15000; Li-Cor, 926-32210). Western blot images were obtained on an Odyssey® imager (Li-Cor Biosciences, Nebraska, USA). Equal loadings were confirmed by reprobing with mouse anti-LamC antibody (1:10000; Hybridoma Bank, LC28.26). Protein levels were quantified by using ImageJ software analysis.
Transmission electron microscopy
Drosophila brain ultrastructure was imaged following standard Electron Microscopy procedures using a Ted Pella Bio Wave processing microwave with vacuum attachments. Briefly, whole heads were dissected in accordance to preserve the brain tissue. The tissue was covered in 2% paraformaldehyde, 2.5% glutaraldehyde, in 0.1 M sodium cacodylate buffer at pH 7.2. After dissection the heads were incubated overnight in the fixative on a rotator. The prefixed heads were then fixed again in the vacuum microwave (640W, 10 s ON - 20 s OFF - 10 s ON), followed by 3x millipore water rinses (3x, 150W, 40 s each), postfixed with 1% aqueous osmium tetroxide (1x, 80W, 2 min ON - 2 min OFF - 2 min ON - cooled on ice in between repeats). Samples were then placed in vacuum microwave for 1 h (80W) and then rinsed again with millipore water (3x, 150W, 40 s each). Concentrations from 30 to 100% of ethanol were used for the initial dehydration series (2x, 250 W, 40s each), followed with propylene oxide as the final dehydrant (3x, 250 W, 40 s each). Samples were gradually infiltrated with 3 ratios of propylene oxide and Embed 812 (Electron Microscopy Sciences, RT14120) (2x, 250W, 15 min each), finally going into 3 changes of pure resin under vacuum (3x, 250W, 3 min each). Samples were allowed to infiltrate in pure resin overnight on a rotator. The samples were embedded into flat silicone molds and cured in the oven at 62°C for 3 d. The polymerized samples were thin-sectioned at 48 to 50 nm and stained with 1% uranyl acetate for 10 min followed by lead citrate for 1 min before TEM examination. Grids were viewed in a JEOL JEM 1010 transmission electron microscope (JEOL USA, Inc., USA) at 80kV. Images were captured using an AMT XR-16 midmount 16 mega-pixel digital camera. Mitochondrial size (area) was measured manually using ImageJ software analysis. At least 100 mitochondria/group were analyzed from 3 different experiments.
LysoTracker Green staining
Fat bodies were dissected in cold PBS from the pinned third-instar larvae (fed or starved) and incubated in PBS containing LysoTracker Green DND-26 (1:500; Molecular Probes, L-7526) at room temperature for 45 min. The samples were then washed 3 times with PBS, transferred to VECTASHIELD Antifade Mounting Medium with DAPI on glass slides, covered, and immediately photographed live on a Leica confocal fluorescent microscope. Acidic vesicles were counted manually using ImageJ software analysis.
Nile red staining
For lipid droplet staining, fed or starved third-instar larvae were dissected in PBS and fixed in 3.7% formaldehyde in PBS for 30 min at room temperature. Tissues were then rinsed twice with PBS, incubated for 30 min in a 1:2500 dilution with PBS of 0.5 mg/ml Nile red (Sigma, N3013), and then rinsed twice with PBS. Stained samples were mounted in VECTASHIELD Antifade Mounting Medium with DAPI on glass slides, covered, and photographed on a Leica confocal fluorescent microscope. Lipid droplet size (area) was measured manually using ImageJ software analysis.
Cell culture and transfection
For cell transfection studies, Mitf cDNA was cloned into the pUAST-FLAG and pCMV-MYC expression vectors. Constructs used for transfection were pUAST-Mitf-FLAG, pACT-GAL4 and pCMV-MYC-Mitf. Drosophila S2 cells were maintained in Schneider medium (Thermo Fisher Scientific, 21720) with 10% FBS (Thermo Fisher Scientific, 10082) at 28°C. Cells were seeded at 1 × 105 cells per well onto sterile concanavalin A (Sigma, L7647)-coated coverslips in 24-well dishes and grown overnight. Cells were transfected using calcium phosphate (Thermo Fisher Scientific, K2780), as indicated by the manufacturer. Daoy cells (gift from Dr. Huda Y. Zoghbi) were cultured in DMEM with 10% FBS at 37°C and transfected using Lipofectamine 2000 (Thermo Fisher Scientific, 11668019), according to the manufacturer's protocol. Two d after transfection immunofluorescence was performed as described in the above paragraph.
Statistical analysis
The 2-tailed Student t test was used to assess the statistical value of differences between analyzed groups. Results are presented as means ± SEM and a P value < 0.05 was considered significant.
Abbreviations
| ATXN1/SCA1 | = | ataxin 1 |
| bHLH-Zip | = | basic helix-loop-helix leucine zipper |
| bp | = | base pair |
| CLEAR | = | coordinated lysosomal expression and regulation |
| LSD | = | lysosomal storage disorder |
| MITF | = | microphthalmia-associated transcription factor |
| MTORC1 | = | mechanistic target of rapamycin (serine/threonine kinase) complex 1 |
| S2 | = | Schneider 2 cells |
| SCA1 | = | spinocerebellar ataxia type 1 |
| TEM | = | transmission electron microscopy |
| TFE3 | = | transcription factor binding to IGHM enhancer 3 |
| TFEB | = | transcription factor EB |
| TFEC | = | transcription factor EC |
| TSS | = | transcription start site |
| V-ATPase | = | vacuolar-type H+-ATPase. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1134081_Supplemental_Material.zip
Download Zip (2.7 MB)Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We thank Drs. Francesca Pignoni for Drosophila Mitf transgenic lines, Vivian Pogenberg for help with the bioinformatic analysis, Lita Duraine for technical support in transmission electron microscopy, Kai Li for technical assistance and Roman Polishchuk for helpful discussion. We are also grateful to Drs. Gabor Juhasz, Thomas Neufeld, Hugo J. Bellen and Huda Y. Zoghbi for kindly providing reagents. We also thank Drs. Rodney C. Samaco, Diego L. Medina, Ismael Al-Ramahi, Andrea Chai, Carmine Settembre and Alessandro Luciani for critical discussion and insightful comments on the manuscript.
Funding
This work was supported by the Robert A. and Renée E. Belfer Family Foundation, the Huffington Foundation and NIH grant NS42179. The project was supported in part by IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. Cores: Confocal Microscopy and Human Cell Lines.
References
- Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 2009; 1793: 664–73; PMID:18706940; http://dx.doi.org/10.1016/j.bbamcr.2008.07.014
- Hale AN, Ledbetter DJ, Gawriluk TR, Rucker EB 3rd. Autophagy: regulation and role in development. Autophagy 2013; 9:951–72; PMID:24121596; http://dx.doi.org/10.4161/auto.24273
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717–21; PMID:11099404; http://dx.doi.org/10.1126/science.290.5497.1717
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science 2009; 325:473–77; PMID:19556463
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429–33; PMID:21617040; http://dx.doi.org/10.1126/science.1204592
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 2011; 21:421–30; PMID:21889421; http://dx.doi.org/10.1016/j.devcel.2011.07.016
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013 15:647–58; PMID:23604321; http://dx.doi.org/10.1038/ncb2718
- Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 2011; 20:3852–66; PMID:21752829; http://dx.doi.org/10.1093/hmg/ddr306
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AM, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 2014; 40:896–909; PMID:24882217; http://dx.doi.org/10.1016/j.immuni.2014.05.002
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, at el. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095–108; PMID:22343943; http://dx.doi.org/10.1038/emboj.2012.32
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903–14; PMID:22576015; http://dx.doi.org/10.4161/auto.19653
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/10.1126/scisignal.2002790
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966; 28:435–92; PMID:5322983; http://dx.doi.org/10.1146/annurev.ph.28.030166.002251
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678–83; PMID:22053050; http://dx.doi.org/10.1126/science.1207056
- Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 2013; 14:133–9; PMID:23361334; http://dx.doi.org/10.1038/nrm3522
- Hemesath TJ, Steingrímsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994; 8:2770–80; PMID:7958932; http://dx.doi.org/10.1101/gad.8.22.2770
- Pogenberg V, Ogmundsdóttir MH, Bergsteinsdóttir K, Schepsky A, Phung B, Deineko V, Milewski M, Steingrímsson E, Wilmanns M. Restricted leucine zipper dimerization and specificity of DNA recognition of the melanocyte master regulator MITF. Genes Dev 2012; 26:2647–58; PMID:23207919; http://dx.doi.org/10.1101/gad.198192.112
- Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014; 7:ra9; PMID:24448649; http://dx.doi.org/10.1126/scisignal.2004754
- Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, Graeber TG, von Euw E, Ribas A, De Robertis EM. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A 2015; 112:E420–9; PMID:25605940; http://dx.doi.org/10.1073/pnas.1424576112
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 2013; 4:2267; PMID:23925298
- O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 2013; 15:668–76; PMID:23604316; http://dx.doi.org/10.1038/ncb2741
- Campello S, Cecconi F. Ho(a)xing autophagy to regulate development. Dev Cell 2014; 28:3–4; PMID:24434134; http://dx.doi.org/10.1016/j.devcel.2013.12.018
- Hallsson JH, Haflidadóttir BS, Stivers C, Odenwald W, Arnheiter H, Pignoni F, Steingrímsson E. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 2004; 167:233–41; PMID:15166150; http://dx.doi.org/10.1534/genetics.167.1.233
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrímsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 1993; 74:395–404; PMID:8343963; http://dx.doi.org/10.1016/0092-8674(93)90429-T
- Hughes AE, Newton VE, Liu XZ, Read AP. A gene for Waardenburg syndrome type 2 maps close to the human homologue of the microphthalmia gene at chromosome 3p12-p14.1. Nat Genet 1994; 7:509–12; PMID:7951321; http://dx.doi.org/10.1038/ng0894-509
- Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res 1999; 9:1106–15; PMID:10568750; http://dx.doi.org/10.1101/gr.9.11.1106
- Gennarino VA, Sardiello M, Mutarelli M, Dharmalingam G, Maselli V, Lago G, Banfi S. HOCTAR database: a unique resource for microRNA target prediction. Gene 2011; 480:51–8; PMID:21435384; http://dx.doi.org/10.1016/j.gene.2011.03.005
- Sardiello M, Tripoli G, Romito A, Minervini C, Viggiano L, Caggese C, Pesole G. Energy biogenesis: one key for coordinating two genomes. Trends Genet 2005; 21:12–6; PMID:15680507; http://dx.doi.org/10.1016/j.tig.2004.11.009
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993; 118:401–15; PMID:8223268
- Liang C, Sir D, Lee S, Ou JH, Jung JU. Beyond autophagy: the role of UVRAG in membrane trafficking. Autophagy 2008; 4:817–20; PMID:18612260; http://dx.doi.org/10.4161/auto.6496
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004; 117:2805–12; PMID:15169837; http://dx.doi.org/10.1242/jcs.01131
- Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene 2004; 23:9314–25; PMID:15602573; http://dx.doi.org/10.1038/sj.onc.1208331
- Young AR, Chan EY, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888–900; PMID:16940348; http://dx.doi.org/10.1242/jcs.03172
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603–14; PMID:16286508; http://dx.doi.org/10.1083/jcb.200507002
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131–45; PMID:17580304; http://dx.doi.org/10.1074/jbc.M702824200
- Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A. A block of autophagy in lysosomal storage disorders. Hum Mol Genet 2008; 17:119–29; PMID:17913701; http://dx.doi.org/10.1093/hmg/ddm289
- Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy 2012; 8:719–30; PMID:22647656; http://dx.doi.org/10.4161/auto.19469
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci 2010; 13:567–76; PMID:20383138; http://dx.doi.org/10.1038/nn.2528
- Janda E, Isidoro C, Carresi C, Mollace V. Defective autophagy in Parkinson's disease: role of oxidative stress. Mol Neurobiol 2012; 46:639–61; PMID:22899187; http://dx.doi.org/10.1007/s12035-012-8318-1
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 2005; 64:113–22; PMID:15751225
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 2008; 118:2190–9; PMID:18497889
- Bartlett BJ, Isakson P, Lewerenz J, Sanchez H, Kotzebue RW, Cumming RC, Harris GL, Nezis IP, Schubert DR, Simonsen A, Finley KD. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 2011; 7:572–83; PMID:21325881; http://dx.doi.org/10.4161/auto.7.6.14943
- Fraldi A, Annunziata F, Lombardi A, Kaiser HJ, Medina DL, Spampanato C, Fedele AO, Polishchuk R, Sorrentino NC, Simons K, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J 2010; 29:3607–20; PMID:20871593; http://dx.doi.org/10.1038/emboj.2010.237
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol 2014; 24:400–6; PMID:24698685; http://dx.doi.org/10.1016/j.tcb.2014.03.003
- Spampanato C, Feeney E, Li L, Cardone M, Lim JA, Annunziata F, Zare H, Polishchuk R, Puertollano R, Parenti G, et al. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol Med 2013; 5:691–706; PMID:23606558; http://dx.doi.org/10.1002/emmm.201202176
- Pastore N, Blomenkamp K, Annunziata F, Piccolo P, Mithbaokar P, Maria Sepe R, Vetrini F, Palmer D, Ng P, Polishchuk E, et al. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med 2013; 5:397–412; PMID:23381957; http://dx.doi.org/10.1002/emmm.201202046
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Björklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci U S A 2013; 110:E1817–26; PMID:23610405; http://dx.doi.org/10.1073/pnas.1305623110
- Xiao Q, Yan P, Ma X, Liu H, Perez R, Zhu A, Gonzales E, Burchett JM, Schuler DR, Cirrito JR, et al. Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J Neurosci 2014; 34:9607–20; PMID:25031402; http://dx.doi.org/10.1523/JNEUROSCI.3788-13.2014
- Polito VA, Li H, Martini-Stoica H, Wang B, Yang L, Xu Y, Swartzlander DB, Palmieri M, di Ronza A, Lee VM, et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med 2014; 6:1142–60; PMID:25069841; http://dx.doi.org/10.15252/emmm.201303671
- Banfi S, Servadio A, Chung MY, Kwiatkowski TJ Jr, McCall AE, Duvick LA, Shen Y, Roth EJ, Orr HT, Zoghbi HY. Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet 1994; 7:513–20; PMID:7951322; http://dx.doi.org/10.1038/ng0894-513
- Park J, Al-Ramahi I, Tan Q, Mollema N, Diaz-Garcia JR, Gallego-Flores T, Lu HC, Lagalwar S, Duvick L, Kang H, et al. RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature 2013; 498:325–31; PMID:23719381; http://dx.doi.org/10.1038/nature12204
- Mauvezin C, Nagy P, Juhász G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun 2015; 6:7007; PMID:25959678; http://dx.doi.org/10.1038/ncomms8007
- Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-sensing mechanisms across evolution. Cell 2015; 161(1):67–83; PMID:25815986; http://dx.doi.org/10.1016/j.cell.2015.02.041
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 2007; 35:W193–200; PMID:17478515; http://dx.doi.org/10.1093/nar/gkm226
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2013; 41:D991–5; PMID:23193258; http://dx.doi.org/10.1093/nar/gks1193
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34:374–8; PMID:12613259
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res 2004; 14:1188–90; PMID:15173120; http://dx.doi.org/10.1101/gr.849004
- Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohée S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res 2008; 36:W119–27; PMID:18495751; http://dx.doi.org/10.1093/nar/gkn304
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 2007; 35:W71–4; PMID:17485472; http://dx.doi.org/10.1093/nar/gkm306
- Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One 2012; 7:e44214; PMID:22952930; http://dx.doi.org/10.1371/journal.pone.0044214
