ABSTRACT
Autophagy can act either as a tumor suppressor or as a survival mechanism for established tumors. To understand how autophagy plays this dual role in cancer, in vivo models are required. By using a highly heterogeneous C. elegans germline tumor, we show that autophagy-related proteins are expressed in a specific subset of tumor cells, neurons. Inhibition of autophagy impairs neuronal differentiation and increases tumor cell number, resulting in a shorter life span of animals with tumors, while induction of autophagy extends their life span by impairing tumor proliferation. Fasting of animals with fully developed tumors leads to a doubling of their life span, which depends on modular changes in transcription including switches in transcription factor networks and mitochondrial metabolism. Hence, our results suggest that metabolic restructuring, cell-type specific regulation of autophagy and neuronal differentiation constitute central pathways preventing growth of heterogeneous tumors.
Introduction
Germ cell-derived tumors usually result from precocious differentiation of cells in the gonads or the embryo.Citation1 In C. elegans, induction of germline tumors can be achieved by depletion of gld-1, a member of the STAR KH-domain family of RNA binding proteins and a suppressor of translation.Citation2 Depletion results in tumors containing fully differentiated neurons, muscle and gut cells ().Citation2 Their overproliferation leads to gonad swelling and invasion of other tissues by germline tumor cells, resulting in premature death of animals.Citation3,4 Mutations that generally extend C. elegans life span inhibit gld-1 tumor growth and prevent tumor-dependent life-span shortening, although the mechanism is not fully understood.Citation4 Interestingly, metabolic adjustments including upregulation of autophagy have been proposed for these long life-span mutants.Citation5-7
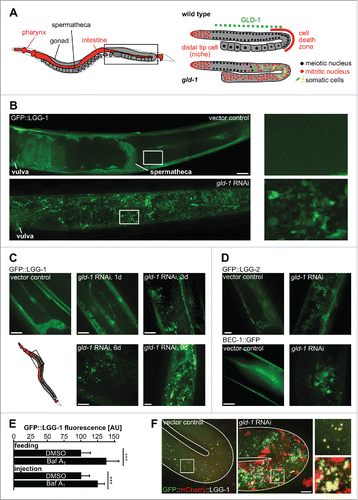
Figure 1. Autophagy-related proteins are expressed in a subset of cells within germline tumors. (A) The C. elegans gonad as a model for tumorigenesis. Anatomy of an adult hermaphrodite. One gonad arm (boxed area) is shown enlarged on the right for wild-type and gld-1 (mutant or RNAi) animal. GLD-1/quaking expression region is indicated by the green dashed line. (B) GFP::LGG-1 expression in the gonad of wild-type and gld-1 RNAi-treated animals. Left panels: Representative gonad arms reconstructed from maximum projections of gonad z-planes. Boxed areas are shown enlarged on the right. The region reconstructed is depicted in the boxed area of the scheme in panel A. (C) GFP::LGG-1 expression in the gonad of a gld-1 RNAi-treated animal from d 1 to d 9 of adulthood. Maximum projections of gonad z-planes. Boxed area in the scheme represents the region depicted in the z-projections of panels (C, D, and F) GFP::LGG-2 and BEC-1::GFP expression in the gonad of wild-type and gld-1 knockdown animals at d 3 of adulthood. Maximum projections of gonad z-planes. Scale bars: 20 μm. (E) Quantification of GFP::LGG-1 fluorescence intensity in gld-1 tumors in DMSO (control) and Baf-treated animals (n = 10 each), ***, P ≤ 0.001. (F) Representative maximum projections of GFP::mCherry::LGG-1 expressing animals. A control animal's gonad turn region (left) and a gld-1 RNAi-treated animal's germline tumor turn region (right) are shown. Boxed areas are shown enlarged on the right. Scale bar: 10 μm
Autophagy is thought to play a dual role in cancer. It can either act as a tumor suppressor mechanism and prevent tumor initiation, or act as an adaptive response to cancer survival-associated stress and sustain cancer metabolism of established tumors.Citation8 Therefore, it is essential to understand the context-specific functions of autophagy in tumorigenesis. These will depend on tumor type, stage, and genetic alterations, among others.Citation9 A recent study showed that intratumor heterogeneity in the form of clonal diversity occurs over clinically relevant timeframes and can lead to clinically important phenotypic properties.Citation10 Hence, the heterogeneity of C. elegans germline tumors suggests that they could constitute an appealing model to study the role of autophagy in tumorigenesis.
Autophagy pathways have been shown to regulate key stages of C. elegans development.Citation11 During embryogenesis, autophagy selectively eliminates paternal mitochondriaCitation12-14 and P granulesCitation15 and contributes to clearance of cell corpses.Citation16 Autophagy also regulates cell size,Citation17 is essential for dauer developmentCitation7,18 and, together with apoptosis, is required for proper embryonic development.Citation19 In the former examples, regulation of autophagy has been mostly studied at the post-transcriptional level. Recently, the nuclear hormone receptor NHR-62,Citation20 the transcription factor HLH-30/TFEBCitation6,21 and the conserved transcription factor PHA-4/FOXACitation5,22 have been identified as transcriptional regulators of autophagy. In addition, pathways such as XBP-1-mediated ER stress and ATFS-1-mediated mitochondrial stress also regulate autophagy at the transcriptional level in C. elegans.Citation23 However, transcriptional and post-transcriptional regulation of autophagy during differentiation remains to be explored.
We reasoned that the heterogeneity of C. elegans germline tumors, with cells expressing a variety of markers otherwise only found in differentiated somatic cells, calls for studying the role of autophagy in tumorigenesis. Thus, we analyzed expression of autophagy markers and performed a comprehensive transcriptome analysis of animals with germline tumors. Genes related to stress response and proteolysis, such as autophagy-related genes, are upregulated in animals with tumors. In agreement, autophagy-related proteins are expressed in germline tumor neurons and inhibition of autophagy impairs neuronal differentiation. Conversely, autophagy inhibition leads to an increase in germ cell number and a rapid invasion of head and vulva by the tumor. In addition, depletion of autophagy-related genes shortens the life span of animals with germline tumors, while induction of autophagy extends their life span. In animals with fully developed tumors, fasting doubles their life span by triggering both an upregulation of autophagy-related genes and a remodeling of the metabolic transcriptome, including an upregulation of oxidative metabolism. Mitochondrial oxidative stress signaling and potentially also modular activation of gene networks governed by nuclear hormone receptors seem to be part of a positive feedback that leads to upregulation of autophagy. This suggests that even in mature tumors a straight-forward dietary intervention can inhibit tumor growth and that autophagy, metabolic restructuring and mitochondrial stress signaling constitute central antitumorigenic pathways in the C. elegans germline tumor model.
Results
Autophagy-related proteins are expressed in gld-1 germline tumors
Since many tumors require drastic changes in metabolism and particularly in catabolism to grow, we asked whether autophagy is induced in germline tumors in C. elegans. To test this, we examined if autophagy-related proteins are expressed in the wild-type gonad and/or in the gld-1 germline tumor. We used transgenic animals that express fluorescence-tagged versions of the following proteins: GFP::LGG-1 (GABARAP ortholog), GFP::LGG-2 (LC3 ortholog), and BEC-1::GFP (BECN1 ortholog). Consistent with previous reports,Citation7,18,24 we observed that autophagy-related proteins are highly expressed in the nervous system, pharynx, intestine, hypodermis, vulva, spermatheca and somatic gonad of wild-type animals, but not in wild-type germline cells (). However, cells within the germline tumor in gld-1 RNAi animals show high expression of the autophagy pathway proteins from the first day of adulthood throughout their life span (). Notably, these transgenes (with the exception of BEC-1::GFP) are not expressed in wild-type germlines and only become activated during zygotic gene activation in embryos () or somatic trans-differentiation in the tumor.
To test whether tumor formation leads to a general induction of autophagy rather than to a block of autophagosome maturation, we treated animals with germline tumors with bafilomycin A1 (Baf) which blocks autophagosome-lysosome fusion. This leads to an increase in GFP::LGG-1 levels in germline tumors, consistent with the idea that autophagy is generally upregulated during tumor formation (). Additionally, we also observed a massive increase of autophagosomes and lysosomes when using a germline-expressed transgene, Ppie-1::GFP::mCherry::lgg-1, that allows to monitor autophagosome maturation due to quenching of GFP fluorescence and maintenance of mCherry fluorescence after acidification following autophagosome-lysosome fusion ().
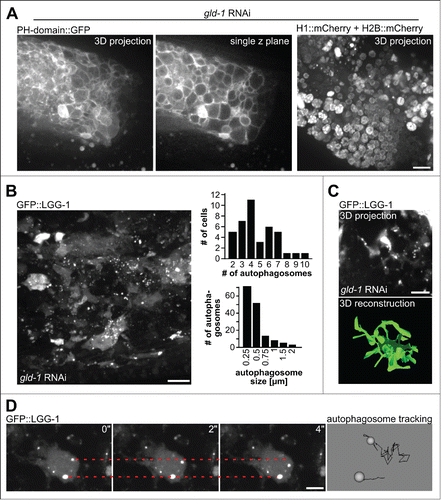
Consistent with germline tumors containing cells expressing different fate markers,Citation2,4 we also observed heterogenetity in nuclear shape, size and nucleolar structure as well as cell shape (). Importantly, germline tumor cells show a heterogenous pattern of GFP::LGG-1 expression, with many cells showing elevated cytoplasmic expression and about 90% of them contain clearly discernible autophagosomes with rather uniform size of around 0.5 μm (). Thus, unlike what has been reported for larval seam cells where only few autophagosomes can be detected, germline tumor cells show a distribution of autophagosome numbers with 4 or 5 autophagosomes per cell on average (). Moreover, many cells expressing GFP::LGG-1 have elongated protrusions which are often connected to neighboring cells, thereby resembling neurons in their shape (). By performing time-lapse microscopy of germline tumors, we also found that while most tumor cells show little intracellular autophagosome movement, in a few tumor cells with high levels of GFP::LGG-1, fast, random-walking autophagosomes can be observed (; Movie S1).
Figure 2. Characterization of tumor cells and autophagosomes. (A) Tumor cells as visualized by their plasma membranes (left, PH domain of PLCd1 fused to GFP) or their nuclei (histone-mCherry fusion proteins). Scale bar: 10 μm. (B) Left: Autophagosomes in tumor cells as visualized by GFP::LGG-1, a representative maximum projection of a central tumor area is shown, scale bar: 10 μm. Top right: Quantification of numbers of autophagosomes per cell. Bottom right: Quantification of autophagosome size. Quantifications were performed by manual counting/measurements. (C) 3D reconstruction of cell shapes of those cells in the germline tumor expressing GFP::LGG-1. Top: Original microscopy 3D projection data, scale bar: 5 μm; bottom: 3D reconstruction performed in imod (see Methods). (D) Movement of autophagosomes inside tumor cells. Left: Stills from time-lapse recordings, the dashed lines mark the initial position of an autophagosome, scale bar: 5 μm. Right: Magnified data from autophagosome tracking using Endrov (see Methods). Autophagosome paths are shown as black lines. Note that the movement of the bottom autophagosome is due to the sample shifting (see Movie S1). All panels in show animals at the d 3 of adulthood.
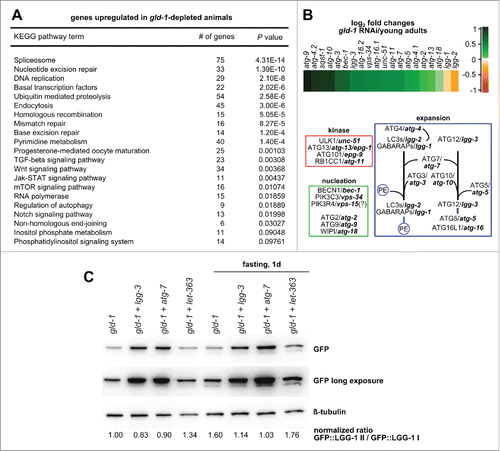
In gld-1 animals, changes in translation seem to be accompanied by transcriptional changes. Several gld-1 targets have been identified;Citation25-27 some of them are transcriptional regulators, like pal-1/Caudal, whose translation is repressed by GLD-1.Citation2 Additionally, GLD-1 can bind and protect its targets from nonsense-mediated mRNA decay (NMD), for instance the glucosamine 6-phosphate N-acetyltransferase gna-2 and the putative transposase Y75B12B.1.Citation28 To confirm our data using integrated reporters () and to gain deeper understanding of the transcriptional regulation in gld-1 tumors, we performed a comprehensive analysis of the transcriptome in wild-type and gld-1 RNAi animals. Genome-wide transcriptome analysis of tumor-containing gld-1 RNAi animals (third d of adulthood) was carried out and compared with wild-type animals of the same age as well as young adults. First, we analyzed the transcripts of the 2 targets protected from NMD. These are strongly downregulated when comparing gld-1 tumor animals with staged gravid adults and young adults (5.2- and 2.6-fold for gna-2, 3.5- and 2.0-fold for Y75B12B.1, respectively). Second, we performed a gene ontology (GO) analysis, which showed that genes related to stress response pathways (e.g., DNA repair, ubiquitin-mediated proteolysis) and autophagy (9 genes) are upregulated in gld-1 RNAi animals compared to young adults (; Table S1), but not compared to wild-type adults of the same age (Table S2) since expression of these genes in the embryos seems to mask the differences between gld-1-depleted and older wild-type animals. All together, these findings are consistent with a transcriptional upregulation of autophagy-related genes followed by protein synthesis in tumor cells.
Figure 3. Stress-response related genes are upregulated in animals with germline tumors. (A) KEGG pathway enrichment analysis of gld-1 RNAi animals. Only upregulated pathways in gld-1 RNAi animals compared to young adults with P values lower than 0.01 are shown. (B) Top: Heatmap depicting expression profiles of autophagy-related genes in gld-1 RNAi animals compared to young adult controls. log2-fold changes are shown. Bottom: The autophagy pathway in C. elegans. Genes were classified in functional modules according to ref. Citation77. (C) Modulation of autophagy by RNAi in the RNAi-mediated germline tumor model. GFP::LGG-1 animals with the respective RNAi were treated as indicated for 24 h and lysed. Protein samples (25 µg) were separated by SDS-PAGE and immunoblotted with the indicated antibodies. Where indicated, RNAi(s) against 2 different genes cloned in the same vector were used only when more than 80% of the animals presented a germline tumor.
Autophagy-related proteins are preferentially expressed in germline tumor neurons
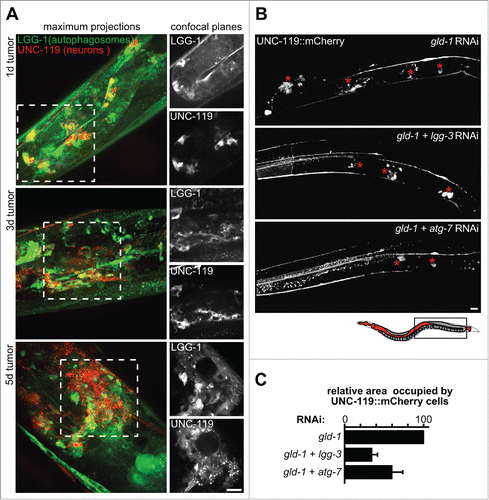
gld-1 germline tumors do not only comprise germ cells but also ectopically differentiated somatic cells, such as muscle, neurons and intestinal cells ( and S1A).Citation2 We observed that the morphology of cells expressing autophagy markers within the germline tumor resembles the morphology of neuronal cells (). Therefore, we used a C. elegans strain expressing a pan-neuronal marker (UNC-119::mCherry) and GFP::LGG-1. Confocal microscopy of germline tumors induced by gld-1 RNAi, showed that LGG-1 is expressed in cells differentiating as neurons (). Concomitantly, we examined differentiation of other somatic cell types in gld-1 germline tumors, by knocking down gld-1 in reporter strains expressing GFP or mCherry under the control of tissue-specific—neuronal (unc-119), muscle (myo-3) or pharyngeal (pha-4)—promoters. Although gld-1-deficient germ cells reprogram mainly as neurons, we also observed—to a lesser extent—differentiation of muscle and pharynx cells (Fig. S1A). Consistently, transcriptome analysis showed that neuronal-specific genes are consistently upregulated in gld-1-depleted animals (Fig. S1B) and, to a lesser extent, muscle- and pharynx-specific genes are also upregulated (Fig. S1B). Hence, the concurrence of neuronal differentiation and high-level expression of autophagy pathway genes suggests that both processes might depend on each other.
Figure 4. Inhibition of autophagy impairs neuronal differentiation within germline tumors. (A) LGG-1-expressing cells in germline tumors are neurons that coexpress UNC-119. Left panels: Maximum projections of merged z-planes of gld-1 RNAi gonads at d 1, 3 and 5 of adulthood. Right panels: Confocal planes showing LGG-1 (top) and UNC-119 (bottom). Scale bar: 20 μm. (B) UNC-119::mCherry expression in the gonad of gld-1, gld-1+lgg-3 and gld-1+atg-7 RNAi-treated animals (d 1.5 of adulthood). Representative gonad arm reconstructed from maximum projections. Asterisks mark neuronal cells in germline tumors. Top: The region reconstructed is depicted in the schema (boxed area). Scale bar: 20 μm. (C) Area occupied by UNC-119 expressing cells per gonad arm normalized by gld-1 RNAi-treated animals. Qualitative scoring of the gonadal area occupied by UNC-119 expressing cells was performed blindly using maximum z-projections covering whole gonad arms (d 1.5 to 2.5 of adulthood. Data represent mean ± SEM of 3 independent experiments, n = 3 to 6 (6 to 12 gonad arms), per experiment.
Inhibition of autophagy impairs neuronal differentiation within germline tumors
A growing body of evidence suggests a crucial role for autophagy during neuronal differentiation.Citation29-33 To investigate whether modulation of autophagy affects neuronal differentiation within germline tumors, we depleted 2 mRNAs simultaneously.Citation34,35 We used dual depletion for gld-1 and an autophagy-related protein to induce both the formation of a tumor and to inhibit autophagy in the same animal. lgg-1 RNAi and lgg-1+gld-1 RNAi lead to similar depletion of lgg-1; likewise, lgg-3 RNAi (Atg12 ortholog) and lgg-3+gld-1 RNAi result in similar accumulation of the unmodified form of LGG-1 and the percentage of animals displaying tumors was similar to single depletion of gld-1 (Fig. S2C and D). To assess neuronal differentiation, the tumor was induced in the pan-neuronal marker (UNC-119::mCherry) strain by gld-1 RNAi and maximum z-projections covering whole gonad arms were used to score the area occupied by UNC-119::mCherry-expressing cells. Concomitant downregulation of either lgg-3 or atg-7 – both required for autophagy () – leads to a reduction of the area occupied by UNC-119::mCherry-expressing cells in the tumor by around 50% and 30%, respectively (). Therefore, downregulation of autophagy results in an impairment of neuronal differentiation within the germline tumor.
Autophagy affects tumor growth and apoptosis
A balance between self-renewal and differentiation of stem cells is critical for proliferation control and maintenance of a stem cell pool in mammalian tissues.Citation36 Hence, we tested whether autophagy downregulation impinges on this balance in gld-1 germline tumors. Contrary to the effect of autophagy depletion on neuronal differentiation, simultaneous knockdown of gld-1 and autophagy-related genes leads to a higher number of germ cells, indicative of increased tumor growth (). In accordance, invasion of the head and vulva region by the tumor occurred earlier in autophagy-deficient animals than in controls (Tables S1 and S2). Moreover, quantitative analysis of gld-1 RNAi-induced tumors shows that nuclei density increases by ∼40% in the central and proximal part of the tumor in atg-7(bp411) animals (), and large and dense accumulations of nuclei can be observed under these conditions (, red arrows; ). Also, a large number of chromosome threads that emerge from apoptotic corpses can be observed. These show a ∼3-fold increase in atg-7(bp411) animals (), particularly in the gonad turn () but also in the proximal part of the tumor ( and data not shown). However, upregulation of apoptosis cannot compensate for the strong upregulation of proliferation since the tumor apoptosis rate is very low (0.7 ± 0.1% apoptotic cells in atg-7(bp411) vs. 0.3 ± 0.1% in wild-type animals with gld-1 induced tumors).
Figure 5. Modulation of autophagy affects tumor growth. (A) N2 L1 or L3 (in the case of let-363 RNAi) larvae treated with the indicated dsRNA(s) till the fourth d of adulthood and stained with the DNA-intercalating dye DAPI (4′,6′-diamidino-2-phenylindole). Left: Representative animals. Right: Enlargement of the head region (boxed areas on the left). Dashed lines indicate the outline of the animal, white lines indicate the gonadal basement membrane. Scale bars: 20 μm. (B) Cell density visualized by fluorescent histone reporters (H1 and H2B-mCherry fusion proteins) in central (left) and proximal (right) parts of gld-1 RNAi-induced germline tumors in wild type and atg-7(bp411) animals. 3D projections are shown. Scale bar: 10 μm. (C) Quantification of cells per area and apoptotic corpses per gonad turn. n = 5 animals each; *** P ≤ 0.001. (D) Gonad turn nuclei and apoptotic corpses in wild-type and atg-7(bp411) animals treated with gld-1 RNAi. 3D projections. Note the chromatin threads that can be observed on goad surfaces in atg-7(bp411) (surface projection, right). Scale bar: 5 μm. (E) Quantification of animals with cell clusters, indicative of high tumor cell density. Animals were treated with the respective RNAis and tumor arms were imaged and scored at d 3 of adulthood for the appearance of bright nuclei clusters (as indicated by red arrows in panel [B]).
![Figure 5. Modulation of autophagy affects tumor growth. (A) N2 L1 or L3 (in the case of let-363 RNAi) larvae treated with the indicated dsRNA(s) till the fourth d of adulthood and stained with the DNA-intercalating dye DAPI (4′,6′-diamidino-2-phenylindole). Left: Representative animals. Right: Enlargement of the head region (boxed areas on the left). Dashed lines indicate the outline of the animal, white lines indicate the gonadal basement membrane. Scale bars: 20 μm. (B) Cell density visualized by fluorescent histone reporters (H1 and H2B-mCherry fusion proteins) in central (left) and proximal (right) parts of gld-1 RNAi-induced germline tumors in wild type and atg-7(bp411) animals. 3D projections are shown. Scale bar: 10 μm. (C) Quantification of cells per area and apoptotic corpses per gonad turn. n = 5 animals each; *** P ≤ 0.001. (D) Gonad turn nuclei and apoptotic corpses in wild-type and atg-7(bp411) animals treated with gld-1 RNAi. 3D projections. Note the chromatin threads that can be observed on goad surfaces in atg-7(bp411) (surface projection, right). Scale bar: 5 μm. (E) Quantification of animals with cell clusters, indicative of high tumor cell density. Animals were treated with the respective RNAis and tumor arms were imaged and scored at d 3 of adulthood for the appearance of bright nuclei clusters (as indicated by red arrows in panel [B]).](/cms/asset/a1739ab4-c573-4dd7-828d-47e507a5d229/kaup_a_1136771_f0005_c.gif)
Next, we asked whether induction of autophagy would have an opposite effect on tumor growth. Indeed, induction of autophagy by silencing the gene encoding the C. elegans TOR ortholog (let-363) () results in a significant reduction of the percentage of animals displaying invasion of the head and vulva region, even at the seventh day of adulthood, both in gld-1 RNAi and mutant animals (; Tables S3 and S4). Invasion of the vulva region often results in the appearance of a tumor prolapse that protrudes from the vulva (Fig. S2C; Movies S2 and S3). Prolapses form by a flow of individual tumor cells and animals with tumor prolapses are motile and often shed their prolapse. In line with modulation of autophagy affecting tumor growth, inhibition of autophagy leads to an increase in the percentage of animals displaying a tumor prolapse both in gld-1 RNAi and mutant animals (Fig. S2D and E). Conversely, let-363 RNAi results in a reduced percentage of gld-1 mutant animals with a tumor prolapse, consistent with impaired tumor growth (Fig. S2). Moreover, the inhibitory effect of let-363 RNAi is partially mediated through autophagy (). Taken together, our data demonstrate that autophagy is crucial for curbing tumor proliferation.
Autophagy impinges on gld-1 animals' life span
As autophagy inhibition resulted in faster tumor growth, we asked whether it affects life span of gld-1 mutants. Overproliferation of gld-1 tumors is lethal and manifests in strongly shortened life spans when compared to wild-type animals without tumors (; as reported previously refs. 3 and 4). Interfering with autophagy by lgg-3 RNAI further shortens the life span of gld-1 RNAi and mutant animals by ∼12% but did not affect wild-type life span ().Citation5,22 The functionality of the RNAi approach is also obvious from the changes in the ratio between lipidated/unlipidated LGG-1 ().
Figure 6. Inhibition of autophagy shortens life span of animals with tumors. (A) N2 L1 larvae were fed with the indicated RNAi(s). A representative cumulative survival curve and Kaplan Meier statistics are depicted. (B) gld-1 mutant L1 larvae were fed with the indicated dsRNAs. A cumulative survival curve and Kaplan Meier statistics are depicted.
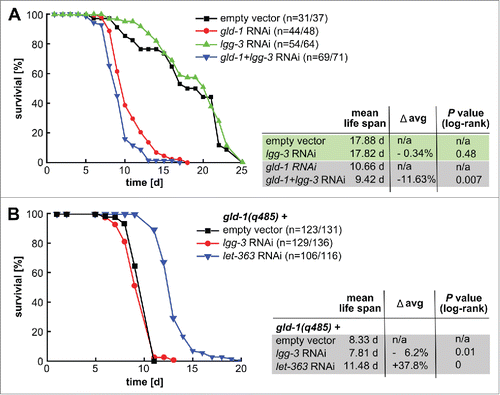
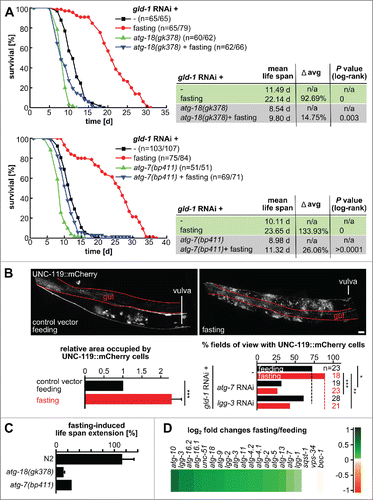
Since we observed that induction of autophagy, by let-363 knockdown, extends the life span of gld-1 mutants by ∼40% (), we wondered whether inducing autophagy after tumors are already established would affect longevity of gld-1 animals. We triggered autophagy by uninterrupted fasting () and found that gld-1 worms fasting from d 2 of adulthood display a striking increase in life span by ∼100% (). Remarkably, fasting inhibited tumor growth most likely by enhancing neuronal differentiation within the tumor (), in line with a function for autophagy on tumor growth or differentiation balance.
Figure 7. Fasting delays death caused by germline tumors in an autophagy-dependent manner and promotes neuronal differentiation. N2, atg-18 and atg-7 mutant L1 larvae were fed with gld-1 RNAi till the d 2 of adulthood and then they were either fed with bacteria expressing empty vector or fasted throughout their life span. (A) Representative cumulative survival curves and Kaplan Meier statistics are presented. (B) Top: UNC-119::mCherry L1 larvae were treated with gld-1 dsRNA till d 2 of adulthood and then treated as indicated for 1 d. Representative gonad arm reconstructed from maximum projections. Scale bar: 20 μm. Bottom left: Area occupied by UNC-119 expressing cells per gonad arm normalized by the animals kept under feeding conditions. Qualitative scoring of the gonadal area occupied by UNC-119 expressing cells was performed blindly using maximum z-projections covering whole gonad arms (d 3 of adulthood). Data represent mean ± SEM of 2 independent experiments, n = 5 (10 gonad arms), per experiment; *** P ≤ 0.001. Bottom right: Quantification of tumors with UNC-119-expressing cells. Animals were treated with the RNAis as indicated and either continued feeding or were starved for 1d before imaging and scoring. (C) Fasting-induced life span of gld-1 RNAi-treated N2, atg-18 and atg-7 mutants. For N2 and atg-18 mutant, data represent mean ± SEM of 2 independent experiments. (D) Expression profiles of transcripts in the autophagy pathway of fasted gld-1 RNAi animals compared to fed gld-1 RNAi animals. log2-fold changes are shown.
Autophagy is required for fasting-induced life-span extension
Next, we analyzed whether autophagy is required for the fasting-induced life-span extension observed in gld-1 deficient animals. To this end, atg-18(gk378) and atg-7(bp411) autophagy mutants were fed with gld-1 RNAi till d 2 of adulthood and then either fasted or kept under feeding conditions throughout their life span. Fasting extends the life span of gld-1 RNAi-treated wild-type animals by ∼100% (). This is a substantially longer life-span extension than previously reported for fasting of wild-type animals without tumors.Citation37,38 On the other hand, fasting prolongs the life span of atg-18(gk378) and atg-7(bp411) mutants by only ∼15% and ∼26%, respectively (), showing that autophagy is a central pathway for fasting-induced life-span extension.
Fasting induces modular metabolic switches
Fasting does not exclusively induce autophagy, but it triggers changes at both transcriptional and post-transcriptional levels.Citation39 To acquire a broader picture, we performed a transcriptome analysis of gld-1 RNAi animals fasted for 48 h, from d 2 of adulthood, and compared their transcriptome with fed gld-1 RNAi animals. Consistent with autophagy being required for fasting-induced life-span extension, we observed that autophagy-related genes, are upregulated in fasted gld-1 RNAi animals (). In accordance with previous reportsCitation40,41 fasting also induced upregulation of components of SCF-E3 ubiquitin ligase complexs, members of the P450 family of cytochromes, C-type lectins, secreted cysteine-rich, and saposin-like proteins (Table S5).
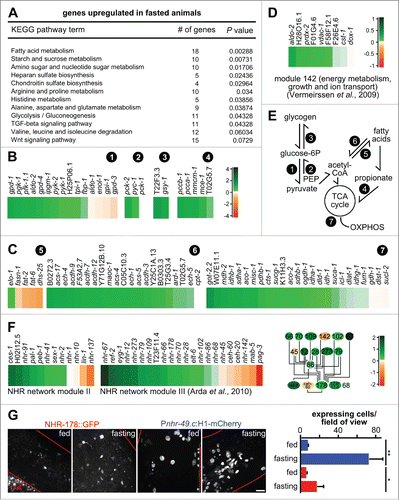
Functional clustering showed a modular reshuffle of metabolism in fasted, gld-1-depleted animals (). A consistent upregulation of genes involved in fatty acid β−oxidation, citric acid cycle, amino acid and propionate catabolism (; Table S5) was detected in fasted gld-1 RNAi animals. Conversely, fatty acid synthesis genes are downregulated in fasted animals (). Regarding carbohydrate metabolism, upregulation of genes involved in glycolysis, gluconeogenesis and glycogenesis was detected (). In line with a metabolic reorganization in fasted gld-1 animals, module 142 ‘Generation of precursor metabolites and energy, positive regulation of growth, ion transport’ described by Vermeirssen and colleaguesCitation42 is coregulated in fasted animals (; all components except cst-1 and dox-1, which are also found inversely regulated in other experimentsCitation42). This module was identified using transcription profiles from various experiments by applying a reverse-engineering algorithm to extract ensembles of coexpressed genes. Module 142 is related to mitochondrial metabolism and contains transcripts of the mitochondrial phosphate carrier F01G4.6, the complex IV subunit F26E4.6, the ATP synthase subunits F58F12.1 and H28O16.1, the outer membrane voltage-gated anion channel vdac-1, the fructose bisphosphate aldolase aldo-2, and the peroxidase prdx-2. Taken together, the coregulation of several metabolic pathways and of module 142 during fasting leads to an upregulation of the major catabolic pathways and a downregulation of fatty acid synthesis, suggesting that mitochondrial respiration will increase under these conditions ().
Figure 8. Fasting induces modular changes in the metabolic gene regulatory network. (A) KEGG pathway enrichment analysis of gld-1 RNAi animals that were fasted. Only upregulated pathways in fasted compared to fed gld-1 RNAi animals with P values lower than 0.01 are shown. (B) Changes in transcript levels in selected carbohydrate and branched-chain amino acid catabolism pathways. log2-fold changes between fasting and feeding are shown. (circled numbers: 1 = glycolysis; 2 = gluconeogenesis; 3 = glycogen metabolism; 4 = propionate metabolism). (C) Changes in transcript levels in fatty acid and mitochondrial metabolic pathways. log2-fold changes between fasting and feeding are shown. log2-scale as in panel (B). (circled numbers: 5 = fatty acid synthesis; 6 = ß-oxidation; 7 = tricarboxylic acid (TCA) cycle, mitochondrial catabolism) (D) Changes in transcript levels in module 142 from ref. Citation42. log2-fold changes between fasting and feeding are shown. (E) Core metabolic processes in C. elegans and the pathways identified to be regulated after fasting (circled numbers, see above). PEP = phosphoenol-pyruvate, OXPHOS = oxidative phosphorylation. (F) Left: Changes in transcript levels in modules II and III from ref. Citation45. log2-fold changes between fasting and feeding are shown. Right: Connection between the metabolic regulators in module III. Colors represent log2-fold changes. (G) NHR-178 and NHR-49 expression in germline tumors and regulation by fasting. Left: Representative z-projections of central germline tumor regions of the indicated transgenic animals. Scale bar: 10 μm. Right: Quantification of tumor cells expressing NHR-49 (blue) or NHR-178 (red) at d 3 of adulthood with or without starvation for 1 d; n = 3 animals each; *P ≤ 0.05; **P ≤ 0.01.
To corroborate whether changes on the transcript level correspond to metabolic alterations, we analyzed fat content of gld-1 RNAi animals by vital staining of lipid droplets with BODIPY 493/503.Citation43 In agreement with the observed transcriptional upregulation of genes involved in β-oxidation and downregulation of fatty acid synthesis, the number of lipid droplets in fasted animals is lower than in fed animals (Fig. S3; lipid droplets are also observed in tumor prolapses). This observation sustains the hypothesis that metabolic adjustments reflect the observed transcriptional changes. We additionally examined N-acetyl-D-glucosamine (GlcNAc) levels, as we observed an upregulation of the hexosamine pathway in the tumor and a downregulation when the animals containing tumors were fasted (Fig. S4A). This pathway has been associated with life-span extension.Citation44 Again in agreement with the transcriptome data, we observe that O-linked GlcNAc-modified proteins are increased when tumors are formed and are not further increased under starvation since the transferase OGA-1 is downregulated (Fig. S4B).
Additionally, we analyzed the expression of genes comprised in a central C. elegans metabolic gene regulatory network.Citation45 This network was generated from experimental interactions between genes involved in lipid metabolismCitation46 and fasting responsive genes.Citation47 A substantial number of genes in this network are nuclear hormone receptors (NHRs), that can function as metabolic sensors and represent an expanded gene family (284 members in C. elegans vs. 48 in H. sapiens). Using topological overlap analysis, 5 modules could be identified in this network.Citation45 Two of these 5 modules, II and III, are coregulated in fasted gld-1-depleted animals (), both containing several NHRs. Importantly, all components of module III that are directly linked to lipid catabolism are consistently upregulated during fasting, including nhr-28, -66, -70, -79, -109, -178, and -273, while transcription factors involved in developmental regulation are inversely regulated, ceh-20, -40, mab-5, and pag-3 (, see also in ref. Citation45).
To establish whether these NHRs indeed play roles in tumor metabolism, we focused on NHR-178. For this NHR, it has been shown that depletion by RNAi results in increased lipid levels and that its expression is upregulated in the epidermis on food withdrawal.Citation45 Consistent with these results and a function of NHR-178 in tumor metabolism, we observed weak expression in germline tumors that increases ∼3-fold during starvation (, red). This demonstrates that NHR-178 and most likely also additional NHRs of module III involved in metabolism function in adapting tumor metabolism to environmental conditions.
Although only moderately upregulated during tumorigenesis (1.1-fold) and further upregulated during fasting (1.1-fold), we also analyzed expression of the transcription factor NHR-49 in germline tumors by using an integrated reporter construct that drives histone H1 expression from an nhr-49 promoter. NHR-49 is a major regulator of metabolism and autophagy.Citation22,47 Notably, we detect expression of the nhr-49 reporter in germline tumors which increases ∼10-fold during fasting (, blue). Therefore, the global changes that we detect by RNAseq seem to underestimate the expression changes inside the tumor. Thus, far-ranging, coordinated changes in transcriptional networks seem to govern the response to fasting, including known (NHR-49, NHR-178, mitochondrial energy metabolism) and previously unknown regulators (). These findings also support the notion that NHRs seem to act redundantly during metabolic adaptations.Citation45
Two other regulators, PHA-4 and HLH-30, have been previously proposed as regulators of metabolic changes and autophagy. To gain an even broader picture, we also analyzed their contribution to autophagy-related gene expression in the tumor. Expression of PHA-4 within the tumor is found only in a very low number of tumor cells (Fig. S1) and we could not detect coregulation of PHA-4 targets in the transcriptome data (Table S6). Hence, PHA-4 does not seem to be involved in autophagy regulation in the tumor. Regarding HLH-30, we examined whether HLH-30 depletion in the tumor affects LGG-1 expression or autophagosome formation. We did not observe major changes in GFP::LGG-1 levels in the tumor upon hlh-30 RNAi, although we observed a slight decrease in the number of autophagosomes and an increase in number of large autophagosomes, the latter statistically not being significant (Fig. S5A to C). To examine the expression of HLH-30 in the tumor, we generated a strain expressing both HLH-30::GFP and UNC-119::mCherry transgenes. Very few cells express HLH-30 compared to the number of cells expressing UNC-119 and those expressing HLH-30 are not tumor neurons (Fig. S5D). This suggests that HLH-30 is not a key transcription factor in autophagy regulation either. In agreement, transcriptome analysis showed that HLH-30 targets are not coregulated in the tumor (Table S7) and lysosomal genes, previously shown to be influenced by HLH-30 levels,Citation6 are also not coregulated (Table S8).
ROS stress signaling affects tumor autophagy and differentiation
Our analysis reveals that all major pathways leading to mitochondrial catabolism are upregulated in fasted animals, including pathways that lead to anaplerotic reactions for the tricarboxylic acid cycle (). Activation of these pathways under low glucose supply could lead to activation of AMP-activated protein kinase (aak-2), the main activator of general catabolism, and result in increased reactive oxygen species (ROS) production. Under these conditions, increased ROS levels have been suggested to act as a hormetic stressor, which induces defense mechanisms like phase II detoxification, transduced by a peroxiredoxin (prdx-2), thereby prolonging life span.Citation48,49 Remarkably, activators, transducers and downstream factors of mitochondrial stress signaling are upregulated in germline tumors animals under starvation, including prdx-2 (being part of module 142, ), aak-2, and many components of phase II detoxification ().Citation49,50
Figure 9. Mitochondrial stress signaling is induced during fasting and is required for fasting-induced tumor survival and autophagy. (A) Top left: Changes in transcript levels in regulators implicated in mitochondrial ROS signaling. log2-fold changes between fasting and feeding are shown. Bottom left: Model of mitochondrial ROS signaling. Right: Changes in transcript levels of class II detoxification genes. log2-fold changes between fasting and feeding are shown. (B) Life-span analysis under feeding conditions. Number of animals and Mantel-Cox test P values are shown. (C) Life-span analysis under fasting conditions. Number of animals and Mantel-Cox test P values are shown. (D) Quantification of neuronal differentiation in gld-1 germline tumors with and without NAC. (E) Changes in autophagosomes in control and NAC-treated gld-1 RNAi animals. Left: Representative z-projections of central germline tumor regions. Line scans of greyscale values along the dashed red lines in the projections are shown on the right side of each projection. Scale bar: 10 μm. (F) Quantification of autophagosome numbers; n = 3 animals each; **P ≤ 0.01. (G) Model showing the pathways contributing to tumor cell growth and differentiation. See discussion for details. Scale bar: 10 μm.
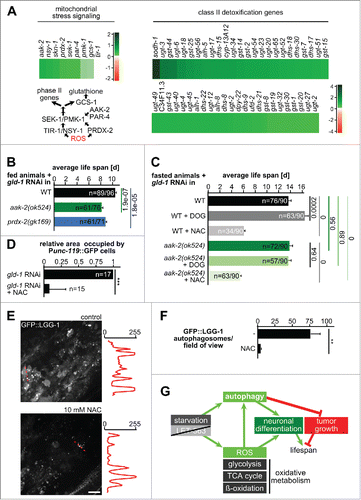
To test a function of mitochondrial stress signaling in tumor survival during starvation, we used mutants in this signaling pathway, prdx-2(gk169) and aak-2(ok425). After induction of tumor formation by gld-1 RNAi, we performed life span analysis either under feeding (aak-2 and prdx-2; and S6A) or starvation conditions (aak-2 only; and S6B) with animals being additionally exposed either to DOG (2-deoxy-D-glucose, to increase ROS levels), or an antioxidant (N-acetyl-cysteine, NAC). Notably, life span under feeding in animals with tumors depends on both aak-2 and prdx-2 function (), whereas starvation-induced life-span extension is not affected in the aak-2 mutant (), consistent with recent findings.Citation51 Moreover, glucose restriction by DOG only extends life span in wild-type animals but not in aak-2 mutants () and life span under fasting conditions was severely reduced in animals exposed to NAC, demonstrating that ROS production is crucial for starvation-induced life-span extension in animals with tumors (). To understand whether ROS scavenging also influences tumor cell autophagy and differentiation, we first quantified the extent of neuronal differentiation in the tumor of fasted animals treated with NAC. NAC led to a severe reduction in the tumor area occupied by neurons () and exposure to NAC also leads to a severe reduction in the number of autophagosomes (). Collectively, these findings suggest that a reciprocal dependence exists between induction of autophagy and mitochondrial oxidative metabolism during germline tumorigenesis.
Discussion
A metabolic model for tumor growth control in C. elegans
We show here that catabolic pathways including autophagy influence cell differentiation and proliferation in germline tumors. Regulation of tumor growth by autophagy, a pathway of central importance when animals rely on internal energy stores, has important implications for the life span of animals with tumors. Autophagy is required for life-span extension by various longevity pathways in C. elegans.Citation5,7,52-55 Here, however, we show that inhibition of autophagy further decreases the life span of severely short-lived, tumor-bearing animals (). Strikingly, induction of autophagy by fasting animals with completely developed tumors from d 2 of adulthood extends their life span by ∼100% (), fully rescuing the short life-span phenotype of these animals. Fasting from d 2 of adulthood was also reported to extend the life span of wild-type animals, but only by 40 to 50%.Citation37,38
Furthermore, our genome-wide transcriptome analysis uncovered a reshuffle of the metabolic transcriptome (). Our observations, together with previous reports, suggest a model () in which induction of autophagy seems to collaborate with other pathways to affect tumor proliferation through induction of cell-type specific effects: (1) stress-response genes are upregulated in animals with germline tumors (); (2) long life-span mutations (eat-2, isp-1, clk-1, daf-2), known to induce autophagy, trigger tumor proliferation arrest;Citation4-7,22,38 and (3) autophagy promotes neuronal differentiation vs. proliferation within germline tumors while inhibition of autophagy leads to the opposite (). Additionally, ROS produced by metabolic activity were also reported to be early activators of fasting-induced autophagy,Citation56 by oxidizing both AMPK,Citation57 leading to its activation, and Atg4, resulting in inhibition of its delipidating activity toward the LC3 and GABARAP subfamilies at the autophagosome formation site.Citation58 Therefore, it is possible that increasing oxidative metabolism and inducing autophagy collaboratively establish cytoprotection, being crucial for positive regulation of life span during fasting.
Strengthening the link between neurogenesis and autophagy
To date, a role for autophagy in cell differentiation during development in C. elegans is restricted to a microRNA-regulated case of bilaterally asymmetric neuron specification.Citation59 In mammals, accumulating evidence supports a role for autophagy in cell differentiation, particularly neuronal differentiation.Citation60 We suggest that autophagy might be similarly required for differentiation of neurons or the maintenance of the metabolic needs of this cell type in C. elegans gld-1 tumors since systemic inhibition of autophagy reduces neuronal differentiation (). In addition, autophagy proteins are strongly reexpressed in transdifferentiating neurons and not much in any other cell type within gld-1 tumors (). Their regulation appears to be mainly transcriptional () with the exception of LGG-1/GABARAP and LGG-2/MAP1LC3 (LC3) whose mRNAs are direct GLD-1 targetsCitation26,27 and their expression is probably regulated post-transcriptionally. Interestingly, in line with our observations, a role for autophagy in neuroendocrine differentiation of human prostate cancer cells has been recently reported,Citation61 suggesting that a conserved interdependency exists in C. elegans and mammalian tumors.
Moreover, we observed that fasting not only inhibits tumor growth but also induces neuronal differentiation within the tumor (), in line with observations showing that dietary restriction in mice enhances neurogenesis.Citation62 Fasting and dietary restriction have shown positive effects in both cancer prevention and treatment in diverse mice models.Citation39 However, Pinkston and colleagues did not observe changes in the frequency of neuronal differentiation within the tumor in the daf-2 mutant,Citation4 pointing to the possibility that specific pathways inducing tumor growth arrest, likely also through autophagy,Citation7 might affect differentiation differently. It is therefore essential to better understand the molecular mechanisms that underlie tumor growth arrest upon activation of different catabolic pathways.
Triggering modular changes in metabolic networks
Our data show that fasted animals with germline tumors upregulate major catabolic pathways and downregulate fatty acid synthesis (), reminiscent of DAF-16-dependent metabolic restructuring. Citation63 However, DAF-16 targetsCitation64 are not consistently regulated in fasted animals, suggesting that other regulatory mechanisms are responsible for the metabolic remodeling of fasted animals with germline tumors. Nonetheless, gene expression profiles of fasted animals with germline tumors partially overlap with those of daf-2 mutantsCitation64 and fasted wild-type animals,Citation41 indicating that related pathways underlie longevity extension regardless of the presence of germline tumors. For instance, it has been suggested that the coregulated module 142 ‘Generation of precursor metabolites and energy, positive regulation of growth, ion transport’ may be part of insulin-like signaling coordinating oxidative stress tolerance, metabolic programs and life span since all promoters of module 142 genes contain the DAF-16 consensus binding element.Citation42
At the organismal level, we observed coregulation of bioinformatically predicted gene modules ().Citation42,45 Remarkably, 2 modules that are part of a metabolic gene regulatory networkCitation45 are coregulated in fasted animals (). These modules are enriched for specific NHRs, the majority of which are consistently upregulated in fasted animals (). NHRs are transcription factors that integrate intrinsic and environmental cues to coordinate transcriptional cascades that control reproduction, development, metabolism and homeostasis.Citation65 In particular, NHR-49 and NHR-62 have been shown to mediate the response to nutrient availabilityCitation20,66 and NHR-62 is required for dietary restriction- induced autophagy and longevity.Citation20 Recent studiesCitation52 show that autophagy is also regulated by the NHRs FXR and PPARA (the ortholog of NHR-49) in mammals and that these factors might have antagonistic effects on transcription of autophagy genes. Additionally, NHR-49 has been shown to be involved in the control of fat consumption and the upregulation of mitochondrial β-oxidationCitation47,67, which we find is also upregulated in our RNAseq analysis (). Our findings also implicate NHR-49 in this regulation (). Furthermore, additional NHRs (e.g., NHR-67, NHR-91 - the latter a likely ortholog of FXR, and the fasting-responsive NHR-178; ) seem to be involved in the response to fasting most likely as part of a transcription factor network. Further studies will be required to identify the NHRs directly affecting autophagy gene expression and which additional regulatory roles these NHRs have.
Altogether, our data suggest that spontaneous neuronal differentiation in germline tumors seems to rely on the induction of autophagy, which in turn depends on metabolic restructuring. Differentiation in this context apparently has a positive effect on animal survival. Thus, manipulation of metabolic restructuring seems to constitute a powerful way to drive cell differentiation and to interfere with tumor growth.
Materials and methods
C. elegans strains
Worms were maintained using standard procedures.Citation68 The following strains were obtained from the Caenorhabditis Genetics Center (CGC): AZ212 (ruIs32[pie-1::GFP::H2B + unc-119(+)] III); DA2123 (adIs2122[lgg-1p::GFP::lgg-1 + rol-6(su1006)]); EG4887 (oxIs322[myo-2p::mCherry::H2B + myo-3p::mCherry::H2B + Cbr-unc-119(+)]); JK1466 (gld-1(q485)/dpy-5(e61) unc-13(e51) I); HZ1686 (atg-7(bp411) IV; him-5(e1490) V. bnIs1 [pie-1p::GFP::pgl-1 + unc-119(+)] I); MS1180 (irIs83 [pMM824 (unc-119::mCherry) + pMM768 (end-3(+))]); wild isolate N2, Bristol variety; OP37 (wgIs37 [pha-4::TY1::EGFP::3xFLAG + unc-119(+)]); OP433 (wgIs433 [hlh-30::TY1::EGFP::3xFLAG + unc-119(+)]); OP454 (wgIs454[nhr-178::TY1::EGFP::3xFLAG + unc-119(+)]); VC893 (atg-18(gk378) V); RB754 (aak-2(ok524) X); RW10226 (itIs37[pie-1P::mCherry::H2B::pie-1 3′UTR + unc-119(+)]. stIs10226[his-72 pomoter HIS-24::mCherry translational fusion with let-858 3′ UTR + unc-119(+)]); RW11084 (zuIs178[his-72(1kb 5′ UTR)::his-72::SRPVAT::GFP::his-72 (1KB 3′ UTR) + 5.7 kb XbaI - HindIII unc-119(+)]. stIs10024[pie-1P::H2B::GFP::pie-1 3′ UTR + unc-119(+)]. stIs10932[nhr-49.c::H1-wCherry + unc-119(+)]); VC289 (prdx-2(gk169) II). CHP8 was generated by crossing DA2123 and MS1180. For the analyses shown in , atg-7(bp411) was outcrossed from strain HZ1686 into RW10226. CHP17 [bec-1::TY1::EGFP::3xFLAG(92C12)+unc-119(+)] was generated by biolistic transformation of DP38 worms (unc‐119(ed3)III)) with a fosmid obtained from the C. elegans TransgeneOme resource (https://transgeneome.mpi-cbg.de/) according to published protocols and progeny that showed integration of the vector were selected after 3 or 4 wk.Citation69 Transgenes Ex[gfp::lgg-2; rol-6(su1006)] and Ex[unc-119(+); Ppie-1::GFP::mCherry::lgg-1] were kindly provided by Renaud Legouis.
Preparation of L1 larvae
To obtain synchronized L1 larvae, gravid adults were bleached. The eggs recovered were left to hatch in M9 (3.0 g KH2PO4, 6.0 g Na2HPO4, 5 g NaCl, 1 ml 1 M MgSO4, H2O to 1 L), overnight, at room temperature.
RNAi treatment
RNAi was performed by feeding according to standard protocols.Citation70 Clones used were prepared from cDNA or obtained from commercially available libraries (http://www.lifesciences.sourcebioscience.com/clone-products/mammalian/rnai/mirna-rnai-resources.aspx). To knock down 2 target genes simultaneously, a gld-1 cDNA fragment was amplified, digested with XhoI and inserted into a vector (pL4440) containing already a cDNA fragment of the other gene, as described elsewhere.Citation34,35 Vectors were transformed into the HT115 E. coli strain. Synchronized L1 or L3 (for let-363 clones) larvae were used for RNAi feeding. The following gene‐specific primers were used to generate RNAi clones: let-363 (forward: CATCGTAGATCTTCGGAATTCTTGGAGCAATC; reverse: GATACTAGATCTTCATCAACCACTGCAACCAT).
RNA sequencing
Synchronized L1 larvae were placed on L4440 control or gld-1 RNAi plates and kept at 20°C. Animals were either collected as young adults or picked as synchronized adults at the first d of adulthood into fresh RNAi plates. When performing gld-1 RNAi, only animals displaying a tumor at the first d of adulthood were picked. gld-1 RNAi animals were kept in gld-1 RNAi plates until d 3 of adulthood, when harvested. Control animals, kept in L4440 plates, were washed off the plates twice a day with M9 to remove the progeny and harvested at d 3 of adulthood. Fasted gld-1 RNAi animals were moved to unseeded plates at d 2 of adulthood for 48 h, before harvesting. Fed gld-1 RNAi control animals were moved to L4440 seeded plates at d 2 of adulthood for 48 h, before harvesting. A total of approximately 3000 animals pulled from 2 independent experiments were collected per condition. The animals were washed off the plates with M9, washed 4 additional times with M9 to remove remaining bacteria and the pellet of worms was frozen in liquid nitrogen. RNA isolation, cDNA libraries construction and sequencing were conducted by GATC Biotech AG.
RNAseq data analysis
3′-adaptor sequences were trimmed from the RNA sequencing reads using custom-built Perl script. Resulting reads were then aligned to Ensembl WS220 assembly using the TopHat.Citation71 Uniquely mapped reads were quantified by counting the overlap of reads with genes using the HtSeq python package (http://www-huber.embl.de/users/anders/HTSeq). Differential expression analysis was performed using the DESeq software.Citation72 Count data were variance-stabilized to overcome the influence of bias due to difference in library size. DAVIDCitation73 was used for gene ontology analysis of genes with 1.2 or higher fold-change between the experimental conditions. Heat maps of log2 fold-changes in gene expression were produced using the heatmap2 package of R.Citation74
BODIPY 493/503 staining
For vital lipid droplet staining, live animals were incubated in M9/BODIPY 493/503 (6.7 μg/ml; ThermoFisher Scientific, D-3922) solution for 20 min.Citation43 Animals were then washed 3 times with M9 and used immediately for microscopy.
DAPI staining
As described previously,Citation3 intact worms were fixed in cold methanol for 5 min at −20°C and incubated for 30 min in 100 ng/mL DAPI (Sigma-Aldrich, D9542) in modified M9 (no Mg2+).
Bafilomycin A1 (Baf) treatment
Animals were either injected with 50 μM Baf (LC Laboratories, B-1080) or DMSO (Carl Roth, A994.1) and imaged 3 h later or incubated in an M9 solution with 25 μM of Baf or DMSO for 6 h.
Microscopy
Animals were mounted in 3 μl of 15 mM tetramisole (Sigma-Aldrich, L9756) in M9 buffer suspension containing 45-μm polystyrene microspheres (Polysciences, 07314-5), and sealed between 2 coverslips (Corning, 2845-18 and 2975-246) with vaseline. Alternatively, for time-lapse recordings (Movies S1, S2 and S3), a recently described procedure was used.Citation75 Microscopy was performed with a VisiScope spinning disk confocal microscope system (Visitron Systems, Puchheim, Germany) based on a Leica DMI6000B inverted microscope (Leica Microsystems, Wetzlar, Germany), a Yokogawa CSU X1 scan head (Yokogawa Electric Corporation, Tokyo, Japan), and a Hamamatsu ImagEM EM-CCD (Hamamatsu Photonics, Hamamatsu, Japan). All acquisitions were performed at 21°C to 23°C using a Leica HC PL APO 40x/1.3 oil or a Leica HC PL APO 63x/1.4–0.6 oil objective.
Animals stained with DAPI were imaged using a Leica DM IL epifluorescence microscope (Leica Microsystems, Wetzlar, Germany) equipped with a Leica HC PL FL 10×/0.3 objective and a Motic Moticam 1SP camera (Motic Deutschland GmbH, Wetzlar, Germany) with Motic acquisition software.
3D reconstructions were performed using imod (http://bio3d.colorado.edu/imod/) for cell outlines and Endrov (http://www.endrov.net/) for autophagosome movement.
Quantifications
To quantify the area occupied by UNC-119 expressing cells per gonad arm, a qualitative scoring was performed blindly from maximum z-projections covering the whole gonad arms (0% to 25%, 25% to 50%, 50% to 75% or 75% to 100% of area occupied by the tumor scored 0, 1, 2 or 3 respectively) and the average score per gonad arm was calculated. The average score per condition was normalized by the control. Mean and standard error are shown including P values from Student t test statistics or from N-1 2 proportion statistics. The number of animals displaying head or vulva invasion (with clear disruption of the gonadal basement membrane) was counted in DAPI stained animals. Nuclei density and apoptotic nuclei number were counted from maximum projections of the superficial cell layer of the tumor. Autophagosomes were counted using fluorescence intensity cut-offs and defined circular windows in ImageJ.
Life-span analysis
Life-span experiments were carried at 20°C. Synchronized L1 larvae were placed on RNAi plates. Animals were picked as synchronized adults at the first day of adulthood into fresh RNAi plates. When gld-1 RNAi was used, only animals displaying a tumor at the first d of adulthood were picked. Worms were transferred to new plates every other day until the end of the reproductive period and every 4 or 5 d in the case of gld-1 animals or after the end of the reproductive period. Animals were fasted by transferring them to empty feeding plates at d 2 of adulthood; the respective control animals, kept under feeding conditions, were transferred to RNAi feeding plates with bacteria expressing the empty pL4440 vector. Animals were examined every day for touch-provoked movement, until death. Animals that crawled off the plate, displayed extruded internal organs during the first 3 d of adulthood, or died from internally hatched progeny were censored. Kaplan-Meier method and Log-Rank (Mantel-Cox) test were performed using OriginPro 8 or OASIS (Online Application for Survival Analysis).Citation76
In parallel with life-span analysis, the number of animals displaying tumor-extruded tissue through the vulva from d 3 of adulthood throughout their life span was quantified and the percentage of animals showing tumor prolapses calculated.
Protein extraction and western blot analysis
To prepare total worm extracts, worm pellets were resuspended in lysis buffer (62.5 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS (Carl Roth, 0183.3) with glass beads (Carl Roth, N030.1), vortexed for 2 min and heated with shaking at 95°C for 5 min. The samples were centrifuged at 20,000 g for 5 min and supernatant fractions were collected. Protein concentration was determined by BCA assay (ThermoFisher Scientific, 23225).
The indicated amounts of protein were separated by AnyKD polyacrylamide gels (Bio-Rad, 4569033) and transferred onto polyvinylidene difluoride (Bio-Rad, 1620174) membranes. The following antibodies were used: anti-GFP (1:1000; Santa Cruz Biotechnology, sc-9996), anti-β-tubulin (1:1000; Developmental Studies Hybridoma Bank, E7), anti-O-linked N-acetyl-D-glucosamine antibody (1:1000; antibody RL2, abcam, ab2739).
Abbreviations
| aak/AAK | = | AMP-activated kinase |
| atg/Atg | = | autophagy (yeast Atg homolog) |
| bec-1 | = | Beclin 1 (human autophagy) homolog |
| C. elegans | = | Caenorhabditis elegans |
| daf | = | abnormal dauer formation |
| DOG | = | 2-deoxy-D-glucose |
| dsRNA | = | double-stranded ribonucleic acid |
| GFP | = | green florescent protein |
| GlcNAc | = | N-acetyl-D-glucosamine |
| gld/GLD | = | defective in germline development |
| GO | = | gene ontology |
| hlh/HLH | = | helix loop helix |
| MAP1LC3/LC3 | = | microtubule-associated protein 1 light chain 3 |
| let/LET | = | lethal |
| lgg/LGG | = | LC3 GABARAP and GATE-16 family |
| mCherry | = | monomeric Cherry |
| mRNA | = | messenger ribonucleic acid |
| myo | = | myosin heavy chain structural genes |
| NAC | = | N-acety-l-cysteine |
| nhr/NHR | = | nuclear hormone receptor |
| pha/PHA | = | defective pharynx development |
| prdx | = | peroxiredoxin |
| RNAi | = | ribonucleic acid interference |
| RNAseq | = | ribonucleic acid sequencing |
| ROS | = | reactive oxygen species. |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary_files.zip
Download Zip (17.4 MB)Acknowledgments
The authors want to thank members of the Pohl and Dikic labs for helpful suggestions and comments as well as Renaud Legouis for reagents.
Funding
Research in the lab of ID and CP is funded by the Deutsche Forschungsgemeinschaft (EXC 115) and the LOEWE Research Cluster Ubiquitin Networks. CP is further supported by the European Union Framework Program 7 (Marie Curie Actions Project 326632) and ID by the LOEWE Center for Cell and Gene Therapy, Frankfurt, and a European Research Council Advanced Grant. LCG is supported by a long-term FEBS fellowship.
References
- Cabral FC, Krajewski KM, Rosenthal MH, Hirsch MS, Howard SA. Teratoma with malignant transformation: report of three cases and review of the literature. Clin Imaging 2014; 38:589-93; PMID:24908364; http://dx.doi.org/10.1016/j.clinimag.2014.04.011
- Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 2006; 311:851-3; PMID:16469927; http://dx.doi.org/10.1126/science.1122491
- Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 1995; 139:579-606; PMID:7713419
- Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science 2006; 313:971-5; PMID:16917064; http://dx.doi.org/10.1126/science.1121908
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 2008; 4:e24; PMID:18282106; http://dx.doi.org/10.1371/journal.pgen.0040024
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 2013; 4:2267; PMID:23925298
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003; 301:1387-91; PMID:12958363; http://dx.doi.org/10.1126/science.1087782
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 2012; 12:401-10; PMID:22534666; http://dx.doi.org/10.1038/nrc3262
- Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev 2011; 25:1999-2010; PMID:21979913; http://dx.doi.org/10.1101/gad.17558811
- Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 2014; 514:54-8; PMID:25079331; http://dx.doi.org/10.1038/nature13556
- Yang P, Zhang H. You are what you eat: multifaceted functions of autophagy during C. elegans development. Cell Res 2014; 24:80-91; PMID:24296782; http://dx.doi.org/10.1038/cr.2013.154
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011; 334:1144-7; PMID:22033522; http://dx.doi.org/10.1126/science.1211878
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 2011; 334:1141-4; PMID:21998252; http://dx.doi.org/10.1126/science.1210333
- Zhou Q, Li H, Xue D. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res 2011; 21:1662-9; PMID:22105480; http://dx.doi.org/10.1038/cr.2011.182
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, Miao L, Zhang H. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 2009; 136:308-21; PMID:19167332; http://dx.doi.org/10.1016/j.cell.2008.12.022
- Huang S, Jia K, Wang Y, Zhou Z, Levine B. Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy 2013; 9:138-49; PMID:23108454; http://dx.doi.org/10.4161/auto.22352
- Aladzsity I, Toth ML, Sigmond T, Szabo E, Bicsak B, Barna J, Regos A, Orosz L, Kovacs AL, Vellai T. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegans. Genetics 2007; 177:655-60; PMID:17890369; http://dx.doi.org/10.1534/genetics.107.075762
- Alberti A, Michelet X, Djeddi A, Legouis R. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 2010; 6:622-33; PMID:20523114; http://dx.doi.org/10.4161/auto.6.5.12252
- Erdélyi P, Borsos E, Takács-Vellai K, Kovács T, Kovács AL, Sigmond T, Hargitai B, Pásztor L, Sengupta T, Dengg M, et al. Shared developmental roles and transcriptional control of autophagy and apoptosis in Caenorhabditis elegans. J Cell Sci 2011; 124:1510-8; PMID:21502138; http://dx.doi.org/10.1242/jcs.080192
- Heestand BN, Shen Y, Liu W, Magner DB, Storm N, Meharg C, Habermann B, Antebi A. Dietary restriction induced longevity is mediated by nuclear receptor NHR-62 in Caenorhabditis elegans. PLoS Genet 2013; 9:e1003651; PMID:23935515; http://dx.doi.org/10.1371/journal.pgen.1003651
- O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol 2013; 15:668-76; PMID:23604316; http://dx.doi.org/10.1038/ncb2741
- Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 2015; 11:867-80; PMID:25836756; http://dx.doi.org/10.1080/15548627.2015.1034410
- Guo B, Huang X, Zhang P, Qi L, Liang Q, Zhang X, Huang J, Fang B, Hou W, Han J, Zhang H. Genome-wide screen identifies signaling pathways that regulate autophagy during Caenorhabditis elegans development. EMBO Rep 2014; 15:705-13; PMID:24764321
- Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, Kovacs AL, Muller F. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol 2005; 15:1513-7; PMID:16111945; http://dx.doi.org/10.1016/j.cub.2005.07.035
- Doh JH, Jung Y, Reinke V, Lee MH. C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation. Worm 2013; 2:e26548; PMID:24744981; http://dx.doi.org/10.4161/worm.26548
- Jungkamp AC, Stoeckius M, Mecenas D, Grun D, Mastrobuoni G, Kempa S, Rajewsky N. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell 2011; 44:828-40; PMID:22152485; http://dx.doi.org/10.1016/j.molcel.2011.11.009
- Wright JE, Gaidatzis D, Senften M, Farley BM, Westhof E, Ryder SP, Ciosk R. A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1. EMBO J 2011; 30:533-45; PMID:21169991; http://dx.doi.org/10.1038/emboj.2010.334
- Lee MH, Schedl T. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev 2004; 18:1047-59; PMID:15105376; http://dx.doi.org/10.1101/gad.1188404
- Aburto MR, Sanchez-Calderon H, Hurle JM, Varela-Nieto I, Magarinos M. Early otic development depends on autophagy for apoptotic cell clearance and neural differentiation. Cell Death Dis 2012; 3:e394; PMID:23034329; http://dx.doi.org/10.1038/cddis.2012.132
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447:1121-5; PMID:17589504
- Morgado AL, Xavier JM, Dionisio PA, Ribeiro MF, Dias RB, Sebastiao AM, Sola S, Rodrigues CM. MicroRNA-34a Modulates Neural Stem Cell Differentiation by Regulating Expression of Synaptic and Autophagic Proteins. Mol Neurobiol 2014; 51:1168-83; PMID:24973144; http://dx.doi.org/10.1007/s12035-014-8794-6
- Vazquez P, Arroba AI, Cecconi F, de la Rosa EJ, Boya P, de Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy 2012; 8:187-99; PMID:22240590; http://dx.doi.org/10.4161/auto.8.2.18535
- Zeng M, Zhou JN. Roles of autophagy and mTOR signaling in neuronal differentiation of mouse neuroblastoma cells. Cell Signal 2008; 20:659-665; PMID:18207367; http://dx.doi.org/10.1016/j.cellsig.2007.11.015
- Gouda K, Matsunaga Y, Iwasaki T, Kawano T. An altered method of feeding RNAi that knocks down multiple genes simultaneously in the nematode Caenorhabditis elegans. Biosci Biotechnol Biochem 2010; 74:2361-5; PMID:21071832; http://dx.doi.org/10.1271/bbb.100579
- Min K, Kang J, Lee J. A modified feeding RNAi method for simultaneous knock-down of more than one gene in Caenorhabditis elegans. Biotechniques 2010; 48:229-32; PMID:20359305; http://dx.doi.org/10.2144/000113365
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 2006; 441:1068-74; PMID:16810241; http://dx.doi.org/10.1038/nature04956
- Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, Fields S, Kennedy BK, Kaeberlein M. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 2006; 5:487-94; PMID:17081160; http://dx.doi.org/10.1111/j.1474-9726.2006.00238.x
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell 2006; 5:515-24; PMID:17096674; http://dx.doi.org/10.1111/j.1474-9726.2006.00241.x
- Longo VD, Mattson MP. Fasting: molecular mechanisms and clinical applications. Cell Metab 2014; 19:181-192; PMID:24440038; http://dx.doi.org/10.1016/j.cmet.2013.12.008
- Eckley DM, Rahimi S, Mantilla S, Orlov NV, Coletta CE, Wilson MA, Iser WB, Delaney JD, Zhang Y, Wood W 3rd, et al. Molecular characterization of the transition to mid-life in Caenorhabditis elegans. Age 2013; 35:689-703; PMID:22610697; http://dx.doi.org/10.1007/s11357-012-9401-2
- Uno M, Honjoh S, Matsuda M, Hoshikawa H, Kishimoto S, Yamamoto T, Ebisuya M, Matsumoto K, Nishida E. A fasting-responsive signaling pathway that extends life span in C. elegans. Cell Rep 2013; 3:79-91; PMID:23352664; http://dx.doi.org/10.1016/j.celrep.2012.12.018
- Vermeirssen V, Joshi A, Michoel T, Bonnet E, Casneuf T, Van de Peer Y. Transcription regulatory networks in Caenorhabditis elegans inferred through reverse-engineering of gene expression profiles constitute biological hypotheses for metazoan development. Mol Biosyst 2009; 5:1817-30; PMID:19763340; http://dx.doi.org/10.1039/b908108a
- Klapper M, Ehmke M, Palgunow D, Böhme M, Matthäus C, Bergner G, Dietzek B, Popp J, Döring F.Fluorescence-based fixative and vital staining of lipid droplets in Caenorhabditis elegans reveal fat stores using microscopy and flow cytometry approaches. J Lipid Res 2011; 52:1281-93; PMID:21421847; http://dx.doi.org/10.1194/jlr.D011940
- Denzel MS, Storm NJ, Gutschmidt A, Baddi R, Hinze Y, Jarosch E, Sommer T, Hoppe T, Antebi A. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell 2014; 156:1167-78; PMID:24630720; http://dx.doi.org/10.1016/j.cell.2014.01.061
- Arda HE, Taubert S, MacNeil LT, Conine CC, Tsuda B, Van Gilst M, Sequerra R, Doucette-Stamm L, Yamamoto KR, Walhout AJ. Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol Syst Biol 2010; 6:367; PMID:20461074; http://dx.doi.org/10.1038/msb.2010.23
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 2003; 421:268-72; PMID:12529643; http://dx.doi.org/10.1038/nature01279
- Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A 2005; 102:13496-501; PMID:16157872; http://dx.doi.org/10.1073/pnas.0506234102
- Ristow M, Schmeisser S. Extending life span by increasing oxidative stress. Free Radic Biol Med 2011; 51:327-36; PMID:21619928; http://dx.doi.org/10.1016/j.freeradbiomed.2011.05.010
- Crook-McMahon HM, Olahova M, Button EL, Winter JJ, Veal EA. Genome-wide screening identifies new genes required for stress-induced phase 2 detoxification gene expression in animals. BMC Biol 2014; 12:64; PMID:25204677; http://dx.doi.org/10.1186/s12915-014-0064-6
- De Haes W, Frooninckx L, Van Assche R, Smolders A, Depuydt G, Billen J, Braeckman BP, Schoofs L, Temmerman L. Metformin promotes lifespan through mitohormesis via the peroxiredoxin PRDX-2. Proc Natl Acad Sci U S A 2014; 111:E2501-9; PMID:24889636; http://dx.doi.org/10.1073/pnas.1321776111
- Schmeisser S, Priebe S, Groth M, Monajembashi S, Hemmerich P, Guthke R, Platzer M, Ristow M. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Mol Metab 2013; 2:92-102; PMID:24199155; http://dx.doi.org/10.1016/j.molmet.2013.02.002
- Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. Autophagy regulates ageing in C. elegans. Autophagy 2007; 3:93-5; PMID:17204841; http://dx.doi.org/10.4161/auto.3636
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 2007; 3:597-9; PMID:17912023; http://dx.doi.org/10.4161/auto.4989
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 2008; 4:330-8; PMID:18219227; http://dx.doi.org/10.4161/auto.5618
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 2003; 426:620; PMID:14668850; http://dx.doi.org/10.1038/426620a
- Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ 2014; 22:377-88; PMID:25257172; http://dx.doi.org/10.1038/cdd.2014.150
- Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. Exposure to hydrogen peroxide induces oxidation and activation of AMP-activated protein kinase. J Biol Chem 2010; 285:33154-64; PMID:20729205; http://dx.doi.org/10.1074/jbc.M110.143685
- Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 2007; 17:422-7; PMID:17804237; http://dx.doi.org/10.1016/j.tcb.2007.07.009
- Zhang P, Zhang H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep 2013; 14:568-76; PMID:23619095; http://dx.doi.org/10.1038/embor.2013.53
- Wu X, Won H, Rubinsztein DC. Autophagy and mammalian development. Biochem Soc Trans 2013; 41:1489-94; PMID:24256242; http://dx.doi.org/10.1042/BST20130185
- Chang PC, Wang TY, Chang YT, Chu CY, Lee CL, Hsu HW, Zhou TA, Wu Z, Kim RH, Desai SJ, et al. Autophagy pathway is required for IL-6 induced neuroendocrine differentiation and chemoresistance of prostate cancer LNCaP cells. PLoS One 2014; 9: e88556; PMID:24551118; http://dx.doi.org/10.1371/journal.pone.0088556
- Lee J, Seroogy KB, Mattson MP. Dietary restriction enhances neurotrophin expression and neurogenesis in the hippocampus of adult mice. J Neurochem 2002; 80:539-47; PMID:11905999; http://dx.doi.org/10.1046/j.0022-3042.2001.00747.x
- Depuydt G, Xie F, Petyuk VA, Smolders A, Brewer HM, Camp DG 2nd, Smith RD, Braeckman BP. LC-MS proteomics analysis of the insulin/IGF-1-deficient Caenorhabditis elegans daf-2(e1370) mutant reveals extensive restructuring of intermediary metabolism. J Proteome Res 2014; 13:1938-56; PMID:24555535; http://dx.doi.org/10.1021/pr401081b
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 2003; 424:277-83; PMID:12845331; http://dx.doi.org/10.1038/nature01789
- Magner DB, Antebi A. Caenorhabditis elegans nuclear receptors: insights into life traits. Trends Endocrinol Metab 2008; 19:153-60; PMID:18406164; http://dx.doi.org/10.1016/j.tem.2008.02.005
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014; 516:112-5; PMID:25383539
- Ratnappan R, Amrit FR, Chen SW, Gill H, Holden K, Ward J, Yamamoto KR, Olsen CP, Ghazi A. Germline signals deploy NHR-49 to modulate fatty-acid β-oxidation and desaturation in somatic tissues of C. elegans. PLoS Genet 2014; 10:e1004829; PMID:25474470; http://dx.doi.org/10.1371/journal.pgen.1004829
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77:71-94; PMID:4366476
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 2001; 157:1217-26; PMID:11238406
- Ahringer, J., ed. Reverse genetics (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, http://dx.doi.org/10.1895/wormbook.1.47.1, http://www.wormbook.org.
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25:1105-11; PMID:19289445; http://dx.doi.org/10.1093/bioinformatics/btp120
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/10.1038/nprot.2008.211
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:R80; PMID:15461798; http://dx.doi.org/10.1186/gb-2004-5-10-r80
- Kim E, Sun L, Gabel CV, Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One 2013; 8:e53419; PMID:23301069; http://dx.doi.org/10.1371/journal.pone.0053419
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 2011; 6:e23525; PMID:21858155; http://dx.doi.org/10.1371/journal.pone.0023525
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/10.1038/nature09204
