abstract
Macroautophagy (hereafter referred to as autophagy) is controlled by a number of core proteins that are critical for all autophagy responses. In addition, a number of autophagy regulators have been found that are not critical for macroautophagy per se, but which play roles in regulating autophagy in either selective situations or in response to specific stimuli. In a recent study, we reported the initial characterization of a new autophagy regulator encoded by TMEM150B that is related to the Damage-Regulated Autophagy Modulator, DRAM1. We have termed this factor DRAM3 for DRAM-Related/Associated Member 3. Interestingly, like DRAM1, DRAM3 regulates both autophagy and cell death, but we found these two functions of the protein are not intrinsically connected.
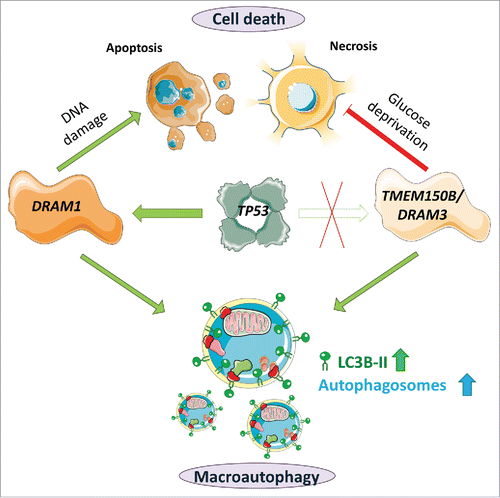
Some 10 years ago, we identified DRAM1 as the first direct molecular link between the tumor suppressor TP53/p53 and autophagy. Since then, various roles for DRAM1 have been described in several processes, including autophagy, cell death, immunity and cellular differentiation. We and others have also become interested in the function of DRAM-related proteins, which have been identified by in silico analysis. The DRAM-family of proteins contains 5 members in humans and other vertebrates whereas only one ortholog is present in Drosophila melanogaster, which has a function similar to that of DRAM1. In addition to DRAM1, human DRAM2 has been researched previously, but much less is known about the remaining DRAM-related proteins, consisting of the 3 TMEM150 proteins (TMEM150A/B/C).
TMEM150B/DRAM3 is the most closely related protein to DRAM1 after DRAM2, with an amino acid sequence overlap of 30% and a sequence similarity of 43%. Structure predictions indicate that TMEM150B/DRAM3 contains 6 hydrophobic transmembrane domains—a structure similar to that predicted for DRAM1. Due to its conservation in structure and amino acid sequence, we hypothesized that TMEM150B/DRAM3 may also be functionally related to DRAM1, and we therefore aimed to investigate whether it plays a role in autophagy and in mechanisms of cell death.
Interestingly, we found that TMEM150B/DRAM3, in contrast to DRAM1, does not seem to be regulated by the tumor suppressor TP53 (). We could neither detect a consensus TP53 binding site in TMEM150B/DRAM3 or its promoter, nor could we detect increased expression of TMEM150B/DRAM3 mRNA after TP53 induction. In addition, none of the potential inducers tested, including stimuli that are described to induce DRAM1, such as chemotherapeutic drugs and inflammatory agents, and other treatments such as different starvation conditions significantly upregulated TMEM150B/DRAM3 mRNA expression. However, as varied expression of TMEM150B/DRAM3 was observed in different cell lines and tissues, we assume that TMEM150B/DRAM3 expression may be differentially regulated. Moreover, it may also be the case that TMEM150B/DRAM3 is induced by agents and processes we have not yet tested.
Despite its unresponsiveness to TP53, we considered that TMEM150B/DRAM3 may regulate cell death and autophagy in a similar fashion to DRAM1 either at unstimulated levels or in response to other, as yet unknown, regulators. We tested this possibility by both overexpression and CRISPR/Cas9-mediated TMEM150B/DRAM3 gene disruption. Collectively, our findings indicate that TMEM150B/DRAM3 modulates autophagy, at least in unstarved (fed) conditions (). We observed, however, that in cells starved of amino acids the effect of TMEM150B/DRAM3 on autophagy was mostly abolished. This could be due to the strong activation of autophagy following complete depletion of amino acids, which may induce maximal autophagic flux in the cell lines tested, irrespective of TMEM150B/DRAM3's role in controlling baseline levels of autophagy.
As another assay of autophagy, we used the enhanced mitophagy assay and found that TMEM150B/DRAM3 overexpression promotes mitophagy, while TMEM150B/DRAM3 disruption has no apparent effect. Since members of the DRAM family are divergent and yet share significant homology, it is conceivable that one or more members of the family can compensate for TMEM150B/DRAM3's loss in this specific form of stress-induced autophagy, whereas they are unable to compensate for the protein's function in the homeostatic autophagy of unstressed cells.
In line with this hypothesis, similar nonreciprocal effects could also be seen with survival after glucose starvation. Whereas TMEM150B/DRAM3 overexpression significantly increases clonogenic survival and decreases overall cell death after glucose deprivation, TMEM150B/DRAM3 disruption in this situation does not conversely seem to increase cell death. Interestingly, we observed that the cell death following glucose starvation is nonapoptotic, as it could not be blocked by caspase inhibition and is not accompanied by DNA fragmentation. Therefore, we concluded that while DRAM1 has a prominent role in TP53-mediated apoptosis, TMEM150B/DRAM3's prosurvival effect largely affects a type of necrotic cell death following glucose starvation (). Future investigations are required to determine whether TMEM150B/DRAM3 affects cell death through other necrotic stimuli and/or starvation from different nutrients, and which cellular pathways promote survival in TMEM150B/DRAM3 overexpressing cells subjected to glucose deprivation.
Our results indicate that TMEM150B/DRAM3 expression has effects on both autophagy and cell death following starvation, and this led us to question whether these 2 observations may be linked. Starvation is a well-described autophagy inducer, and prolonged starvation often leads to cell death. We therefore wanted to test whether TMEM150B/DRAM3 modulates glucose deprivation-induced death by increasing autophagic flux. To investigate this possibility, we added the late-stage autophagy inhibitor chloroquine to the starvation medium and tested whether this abolished the difference in cell survival conferred by TMEM150B/DRAM3 overexpression. To our surprise, even though the chloroquine concentrations used effectively blocked autophagic flux, as assessed by LC3B-II accumulation, this had no effect on cell survival in control or glucose-starved conditions. We therefore concluded that TMEM150B/DRAM3 may regulate cell survival and autophagy through independent processes.
Our studies of TMEM150B/DRAM3 function were performed in cancer cell line systems, and how our findings translate to normal cells and whole organisms is a matter for further investigation. The effect of TMEM150B/DRAM3 on survival after glucose deprivation is particularly intriguing, and warrants investigation in metabolic diseases—such as hypoglycemia or diabetes—and in situations where tissues are deprived of glucose plus other nutrients, such as occurs in strokes and during the development of solid tumors. A transgenic mouse model with Tmem150b/Dram3 knockout would certainly facilitate these studies.
In summary, our study provides a first insight into the function of TMEM150B/DRAM3, a new autophagy and cell death modulator. Since TMEM150B/DRAM3 belongs to a family of proteins, this naturally begs the question of whether these proteins have similar functions, but in different contexts, or if they work together in some way in a concerted manner. Future studies are therefore required to look at the effects of combined expression or loss of 2 or more members of this intriguing protein family in cell death, autophagy and general physiology.
Figure 1. Functional comparison of TMEM150B/DRAM3 and DRAM1. The transcription factor TP53 can bind to the promoter of DRAM1 and induce DRAM1 protein expression, but it does not modulate TMEM150B/DRAM3 levels. Increased levels of DRAM1 and TMEM150B/DRAM3 can increase autophagic flux, but these proteins have differing roles in cell death—DRAM1 is required for maximal apoptosis after TP53 activation, for example after chemotherapy-induced DNA damage, whereas TMEM150B/DRAM3 protects against glucose deprivation-induced necrotic cell death. Image contains components from Servier Medical Art (by Servier), licensed under CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
