ABSTRACT
BECN1/Beclin 1 has a critical role in the early stages of autophagosome formation. Recently, structures of its central and C-terminal domains were reported, however, little structural information is available on the N-terminal domain, comprising a third of the protein. This lack of structural information largely stems from the inability to produce this region in a purified form. Here, we describe the expression and purification of the N-terminal domain of BECN1 (residues 1 to 150) and detailed biophysical characterization, including NMR spectroscopy. Combined, our studies demonstrated at the atomic level that the BECN1 N-terminal domain is intrinsically disordered, and apart from the BH3 subdomain, remains disordered following interaction with a binding partner, BCL2L1/BCL-XL. In addition, the BH3 domain α-helix induced upon interaction with BCL2L1 reverts to a disordered state when the complex is dissociated by exposure to a competitive inhibitor. No significant interactions between N- and C-terminal domains were detected.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a highly conserved catabolic process involving the formation of autophagosomes.Citation1,2 These vesicles encapsulate cellular components (such as macromolecules and organelles) and pathogens, which are delivered to lysosomes for degradation. BECN1, a haploinsufficient tumor suppressor, is the first identified mammalian autophagy gene.Citation3-5 It plays a central role in the autophagy initiation process involving nucleation of the autophagic vesicle through interactions with other proteins including PIK3R4/Vps15, ATG14/Barkor, and the class III phosphatidylinositol 3-kinase (PtdIns3K; PIK3C3/Vps34)Citation6 which all assemble into a PtdIns3K complex.
Structurally, BECN1 is a 450 amino-acid protein with 3 well-characterized domains (). Residues 112 to 123 comprise a BH3 domain, which is a short conserved sequence motif important for mediating interactions between members of the BCL2 family in the intrinsic apoptotic pathway. BECN1 was actually discovered as a BCL2-interacting proteinCitation3,7 and interactions between the BECN1(BH3) domain and BCL2 and BCL2L1 are well-characterized both biochemically and structurally,Citation8–16 though their physiological relevance has been recently challenged.Citation17,18 A potential helical domain has been proposed (based on unpublished data) between residues 141 to 170,Citation8 which is followed by a coiled-coil region (residues 174 to 266) that mediates BECN1 homodimerization as well as heterodimerization with UVRAG/Vps38 and ATG14.Citation16 A crystal structure of this domain shows it forms an antiparallel dimer characterized by an imperfect interface.Citation19 The C-terminal region of BECN1 after the coiled-coiled domain is called the β-α repeat autophagic-specific (BARA) domain,Citation20,21 which contains a highly conserved region known as the evolutionarily conserved domain (ECD) (residues 244 to 337). The BARA domain is critical for BECN1 activity as it mediates interactions with membrane lipids. A crystal structure of the BARA domain of yeast and human BECN1 orthologs has been solved and showed it to represent a novel fold and a new class of membrane-binding domains.Citation20,22
Figure 1. Expression, purification and characterization of the BECN1 N-terminal domain. (A) Schematic of the structural domains of BECN1 and BCL2L1 used in this study. BH, BCL2-homology domain; TM, transmembrane domain; ECD, evolutionarily conserved domain; BARA, β-α repeated, autophagy-specific. (B) Chromatogram following gel-filtration chromatography of the BECN1 N terminus (BECN1(1–150)[4CS]Y construct). The protein eluted at an elution volume consistent with a protein larger than the calculated molecular weight, Inset: SDS-PAGE analysis of peak fractions following gel-filtration chromatography. Protein fractions used for NMR studies were >90% pure. (C) Top panel: Continuous sedimentation coefficient [c(s)] distribution plotted as a function of standardized sedimentation coefficient (s20,w) for BECN1(1–150)[4CS]Y at an initial concentration of 50 µM. Inset: residuals resulting from the c(s) distribution best fit plotted as a function of radial position (cm). Bottom panel: Continuous mass [c(M)] distribution (Da−1) plotted as a function of molar mass (Da) for BECN1(1–150)[4CS]Y at an initial concentration of 50 µM. Inset: Residuals resulting from the c(M) distribution best fit plotted as a function of radial position (cm). (D) Solution competition assays comparing binding of BECN1 N terminus (BECN1(1–150)[4CS]Y) (red curve) with BECN1(BH3) peptide (black curve) to BCL2L1. Both bind with a similar IC50 value (determined from n=5 to 8 independent experiments).
![Figure 1. Expression, purification and characterization of the BECN1 N-terminal domain. (A) Schematic of the structural domains of BECN1 and BCL2L1 used in this study. BH, BCL2-homology domain; TM, transmembrane domain; ECD, evolutionarily conserved domain; BARA, β-α repeated, autophagy-specific. (B) Chromatogram following gel-filtration chromatography of the BECN1 N terminus (BECN1(1–150)[4CS]Y construct). The protein eluted at an elution volume consistent with a protein larger than the calculated molecular weight, Inset: SDS-PAGE analysis of peak fractions following gel-filtration chromatography. Protein fractions used for NMR studies were >90% pure. (C) Top panel: Continuous sedimentation coefficient [c(s)] distribution plotted as a function of standardized sedimentation coefficient (s20,w) for BECN1(1–150)[4CS]Y at an initial concentration of 50 µM. Inset: residuals resulting from the c(s) distribution best fit plotted as a function of radial position (cm). Bottom panel: Continuous mass [c(M)] distribution (Da−1) plotted as a function of molar mass (Da) for BECN1(1–150)[4CS]Y at an initial concentration of 50 µM. Inset: Residuals resulting from the c(M) distribution best fit plotted as a function of radial position (cm). (D) Solution competition assays comparing binding of BECN1 N terminus (BECN1(1–150)[4CS]Y) (red curve) with BECN1(BH3) peptide (black curve) to BCL2L1. Both bind with a similar IC50 value (determined from n=5 to 8 independent experiments).](/cms/asset/0ea77b88-fb3a-4bec-b829-a30ea02be9a3/kaup_a_1140292_f0001_c.gif)
A low resolution (28 Å) structure, determined by single particle electron microscopy, of one of the entire human PtdIns3K complexes (there are at least 3 such complexes that contain common as well as unique components) has been reported.Citation23 This showed BECN1 has an extended structure that makes contact with ATG14 and PIK3R4. The model also suggested the BECN1 and ATG14 coiled-coil domains are parallel unlike the antiparallel arrangement in homodimeric BECN1.Citation19 Although the BECN1 N terminus was mapped to the pivot point of the V-shaped PtdIns3K complex in the EM structure, no density was visible for this region. Very recently, a crystal structure of the yeast PtdIns3K complex II vacuolar sorting complex (PtdIns3K complex I functions in macroautophagy), that also involves the BECN1 ortholog (Vps30/Atg6), has been determined.Citation24 This complex has a similar overall architecture to the mammalian PtdIns3K autophagy complex, although Atg14 is replaced with Vps38/UVRAG. The Vps30/Atg6 structure shows 2 adjacent coiled-coil domains (a short CC1 and longer CC2) with several contacts between both of these with PIK3R4 and PIK3C3/Vps34. Interestingly, while the Vps30/Atg6 N-terminal domain was mostly undefined in this structure, there was some evidence that this region could potentially contain 3 disconnected helices that interact with Vps38/UVRAG.
Prior to this crystal structure of Vps30/Atg6 in the context of the PtdIns3K complex II, there was no detailed structural information on the entire N-terminal region of BECN1 beyond studies on the ∼20 residue BH3 domain, which is unstructured but becomes helical upon engagement of BCL2 prosurvival proteins.Citation8,10-13 As this region comprises approximately one-third of the protein (150 amino acid residues), it is of fundamental interest to characterize it structurally. Recently, extensive primary sequence analysis using multiple algorithms to search for intrinsically disordered regions in autophagy regulators indicated that a large proportion of the BECN1 N terminus (residues 42 to 115) could be intrinsically disordered (consistent with the absence of density in the EM and crystallography studies), although subsequent analyses using NMR spectroscopy could only focus on the BH3 region due to difficulties associated with producing the entire domain.Citation8 An earlier study using NMR to examine a construct that encompasses residues 101 to 154 concluded it was structured,Citation25 although the fragment they purified and analyzed was associated with a “solubility tag” (the 8-kDa streptococcal protein G B1 domain) which itself is a fully folded protein.Citation26,27 Hence, the interpretation of these data must be carefully considered. Intrinsically disordered regions are functionally very important in proteins because they are often the sites of protein-protein interactions and/or post-translational modifications.Citation28
In this paper we report the expression, purification and structural characterization of the entire N-terminal domain of BECN1. We show that it is essentially entirely disordered, and with the exception of the BH3 subdomain, remains unstructured following complex formation with the BCL2 family member BCL2L1. Our results also showed, for the first time, the helical conformation adopted by the BH3 domain of BECN1 upon binding BCL2L1 is reversed to an intrinsically disordered state in solution when the complex is dissociated.
Results
Expression, purification and activity of the BECN1 N terminus
Initial attempts to produce the BECN1 N-terminal domain (residues 1 to 150) were hindered by its sensitivity to bacterial proteases. This was largely overcome by incorporation of a cocktail of protease inhibitors in the purification protocol, and ensuring all procedures were performed at low temperature where possible. As a result of these changes, we were generally able to purify 2 to 4 mg of the various BECN1 constructs per liter of culture media (both minimal and complex). Upon gel-filtration chromatography, the major protein-containing peak eluted at an earlier retention time than expected for a protein of the calculated molecular mass based on the amino acid sequence (). This could indicate that the protein eluted as some type of oligomer (e.g., a dimer or trimer based on the molecular weight standards). Alternatively, aberrant behavior on gel-filtration chromatography can indicate that the protein is intrinsically disordered, as these proteins have a large hydrodynamic radius compared to globular proteins.Citation29 To ascertain the exact oligomeric state of the BECN1(1–150)[4CS]Y protein (see and Materials and Methods for details of this and other constructs) we examined its sedimentation velocity by analytical ultracentrifugation. These data demonstrated that the protein exists as a 1.2S monomer (), hence the behavior on gel-filtration is indicative of the protein being intrinsically disordered rather than oligomeric. The highly elongated shape of the molecule, as determined by its high frictional ratio in the sedimentation velocity experiment (f/f0: 2.2), is also a characteristic of an intrinsically disordered protein.Citation30 Characterization by Coomassie-stained SDS-PAGE indicated the protein in the major gel filtration peak fractions was highly pure (>90%) with minimal proteolysis (, inset). The slightly anomalous migration on SDS-PAGE is likely due to the high positive charge content of the molecule, another characteristic of an intrinsically disordered protein.Citation31
It is important to note that the purified protein tended to form visible aggregates during attempts to concentrate it following gel-filtration chromatography, therefore subsequent NMR and other analyses were generally performed on the unmanipulated peak fraction (which was typically at ∼150 μM) obtained in this purification step. Mutation of the 4 cysteine residues to serine reduced the amount of aggregated material we observed in the gel-filtration step (present in the minor peaks that eluted between 50 to 80 mls) and improved long-term storage without significantly influencing the NMR spectrum (data not shown).
To check the “activity” of the purified BECN1 N-terminal domain (BECN1(1–150)[4CS]Y) we determined its relative affinity for BCL2L1 (). In solution competition assays, the IC50 (2.0 μM) value of BECN1(1–150)[4CS]Y was similar to that of a synthetic peptide whose sequence overlaps with the BECN1(BH3) domain (IC50 2.9 μM) and consistent with previously published binding data on BECN1(BH3) domain peptides.Citation10,12,16 The similarity in the values between the BECN1(BH3) peptide and the entire N-terminal domain suggests the protein produced is fully functional and that regions outside of the BH3 domain are not involved in interactions with BCL2 family members.
Circular dichroism studies demonstrate the BECN1 N terminus does not possess any significant secondary structure
Although various characteristics of the BECN1 N terminus during purification suggested it was intrinsically disordered (sensitivity to proteases, anomalous behavior on gel-filtration and SDS-PAGE), we next performed circular dichroism (CD) analysis to establish whether there was any significant secondary structure present. The CD spectrum showed a single minimum at approximately 200 nm () that is indicative of random or disordered secondary structure. Calculation of α-helix content from these data indicated none was present, demonstrating that the BH3 domain in its native context (rather than in short peptides used in previous studies) must only assume the helical conformation observed in complexes with BCL2L1Citation11,12 upon binding to the protein.
Figure 2. The BECN1 N-terminal domain is intrinsically disordered. (A) Circular dichroism spectra of BECN1(1–150)[4CS]Y. The mean residue ellipticity ([θ]) is plotted as a function of wavelength. (B) Overlay of 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y (black) and BECN1(1-76)[2CS]Y (green) with backbone assignments of BECN1(1–150)[4CS]Y annotated. Resonances corresponding to side-chain NH2 amides of Asn and Gln are connected by horizontal lines. Resonances arising from the short linker sequence at the N terminus and the extra tyrosine residue added to the C terminus are labeled in red. (C) Secondary structure of BECN1(1–150)[4CS]Y as predicted by CSI 2.0Citation34 based on experimentally observed backbone chemical shifts (13Cα, 13Cβ, 13C, 15N, 1HN, and 1Hα). C: random coil, H: helix. Serine substitutions for native cysteines are indicated in red.
![Figure 2. The BECN1 N-terminal domain is intrinsically disordered. (A) Circular dichroism spectra of BECN1(1–150)[4CS]Y. The mean residue ellipticity ([θ]) is plotted as a function of wavelength. (B) Overlay of 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y (black) and BECN1(1-76)[2CS]Y (green) with backbone assignments of BECN1(1–150)[4CS]Y annotated. Resonances corresponding to side-chain NH2 amides of Asn and Gln are connected by horizontal lines. Resonances arising from the short linker sequence at the N terminus and the extra tyrosine residue added to the C terminus are labeled in red. (C) Secondary structure of BECN1(1–150)[4CS]Y as predicted by CSI 2.0Citation34 based on experimentally observed backbone chemical shifts (13Cα, 13Cβ, 13C, 15N, 1HN, and 1Hα). C: random coil, H: helix. Serine substitutions for native cysteines are indicated in red.](/cms/asset/2d019292-ae61-4dc2-b527-1549c023b41c/kaup_a_1140292_f0002_c.gif)
Backbone chemical shift assignment of BECN1(1–150)[4CS]Y demonstrates the BECN1 N terminus is disordered
As all of the data described above provided strong evidence for the BECN1 N terminus being intrinsically disordered, we next performed NMR studies to gain residue-specific structural information. The 1H-15N HSQC spectrum of BECN1(1–150)[4CS]Y () displayed very narrow chemical shift dispersion (between 7.7 and 8.7 ppm) on the 1H dimension that is a characteristic feature of intrinsically disordered proteins.Citation32,33 Analyses of triple resonance 3-dimensional spectra of BECN1(1–150)[4CS]Y together with spectra of BECN1(1-76)[2CS]Y (), which helped to resolve a number of ambiguous resonances due to narrow spectral dispersion of BECN1(1–150)[4CS]Y, led to complete assignments of backbone 13Cα, 13Cβ, 13C, 15N, 1HN, and 1Hα resonances for all non-proline residues, and 13C (13Cα, 13Cβ and 13C) resonances for the prolines except P84. These assignments have been deposited in BioMagResBank (http://www.bmrb.wisc.edu, Accession code: 25384). Experimentally observed backbone chemical shifts of BECN1(1–150)[4CS]Y were then used to evaluate its secondary structure with the improved version of Chemical Shift Index software (CSI 2.0)Citation34 which makes use of all 6 backbone chemical shifts (13Cα, 13Cβ, 13C, 15N, 1HN, and 1Hα), along with sequence-derived features. This analysis confirmed that BECN1 is disordered throughout its entire N terminus (1 to 150) except for a potential half-helical turn involving residues A44 and P45 (). Furthermore, the nearly identical chemical shifts observed for resonances arising from BECN1(1–76)[2CS]Y and BECN1(1–150)[4CS]Y (apart from those arising from the C-terminal truncation site of BECN1(1–76)[2CS]Y) indicates that it is unlikely there are any direct contacts between BECN1 residues 1 to 76 and BECN1 residues 77 to 150.
BCL2L1 engages only the BH3 domain and all other residues remain disordered
Although there was some evidence of electron density associated with the N-terminal domain of the yeast BECN1 ortholog, Vps30/Atg6, in the crystal structure of the yeast PtdIns3K complex II, structural studies on the mammalian BECN1 N terminus have been restricted to the BH3 domain in complex with BCL2L1. However, as we had obtained a fully assigned 1H-15N HSQC spectrum of the N-terminal 150 residues, we were now able to assess whether there were any global structural changes along the whole N-terminal domain upon interaction with BCL2L1. When BCL2L1ΔC25 was titrated into the BECN1(1–150)[4CS]Y sample, only resonances located within or adjacent to the BH3 domain (residues 98 to 132) were significantly perturbed () with only a small number of new resonances detected in the BCL2L1ΔC25-bound form (highlighted in cyan circles in ). This indicates the exchange between the free and bound conformations of BECN1(1–150)[4CS]Y is intermediate (in the millisecond range) on the NMR chemical shift timescale. Indeed, protein-protein interactions involving intrinsically disordered proteins in solution, as studied by NMR, are frequently hampered by excessive line broadening due to the exchange between free and receptor-bound conformations occurring on this timescale which precludes direct detection of the bound-state resonances.Citation35 Interestingly, the large majority of resonances of BECN1(1–150)[4CS]Y were completely unaffected (both peak intensity and linewidth) by the binding of its BH3 domain with BCL2L1ΔC25 (except for a small loss in intensity in residues adjacent to Q40, ), indicating that nearly all residues outside of the BH3 domain are tumbling as freely when bound to BCL2L1 as in the unbound form, and do not adopt any secondary or tertiary structure elements that are not present in the free form.
Figure 3. Interaction of BECN1(1–150)[4CS]Y and BCL2L1ΔC25. (A) Superimposed full 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y in the absence (black) and presence of BCL2L1ΔC25 (red). Molar ratio of BECN1(1–150)[4CS]Y:BCL2L1ΔC25 is 1.00:1.32. Cyan circles indicate resonances that shift upon binding. (B) Ratio of peak intensities of BECN1(1–150)[4CS]Y in the presence of BCL2L1ΔC25 (molar ratio 1.00:1.32) to those in the absence of BCL2L1ΔC25 plotted against the sequence of BECN1(1–150)[4CS]Y. The most significant loss of backbone amide intensities is located within or adjacent to the BH3 domain indicating those residues are most important in the binding with BCL2L1ΔC25. Sequence positions of proline residues are indicated with circles on the X-axis. (C) Partial 1H-15N HSQC spectrum of BECN1(1–150)[4CS]Y showing just the region of glycine residues (corresponding to the boxed region in [A]). (D to H) Superimposition of spectrum in region B (black) with the same region of the spectrum for BECN1(1–150)[4CS]Y in the presence of BCL2L1ΔC25 (colored) at a BECN1(1–150)[4CS]Y:BCL2L1ΔC25 molar ratio of (D) 1.00:0.26, (E) 1.00:0.53, (F) 1.00:0.79, (G) 1.00:1.06, (H) 1.00:1.32. (I to K) Similar to (D to H) but after BCL2L11(BH3) was added to the (BECN1(1–150)[4CS]Y-BCL2L1ΔC25 complex mixture shown in (H). Molar ratios of BECN1(1–150)[4CS]Y:BCL2L1ΔC25:BCL2L11(BH3) are (I) 1.00:1.32:0.75, (J) 1.00:1.32:1.50, (K) 1.00:1.32:3.00. (L) One-dimensional traces from 1H-15N HSQC spectra recorded over the entire course of the titration highlights changes of peak intensities for G55 (not affected by binding with BCL2L1ΔC25) and G101 (which is significantly affected) along with an adjacent peak (annotated as G101') that appeared/disappeared in the presence of BCL2L1ΔC25 and/or BCL2L11(BH3). (M) Superimposed full 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y in the absence (black) and presence of BCL2L1ΔC25 and BCL2L11(BH3) peptide (green). Molar ratio of BECN1(1–150)[4CS]Y:BCL2L1ΔC25:BCL2L11(BH3) is 1.00:1.32:3.00. (N) Ratio of peak intensities of BECN1(1–150)[4CS]Y in the presence (molar ratio 1.00:1.32:3.00) and absence of BCL2L1ΔC25 and BCL2L11(BH3). Peaks of BECN1(1–150)[4CS]Y that were lost due to binding to BCL2L1ΔC25 (see panel B) were completely restored when BCL2L11(BH3) is added and in excess. The overall reduction of peak intensity is due to dilution. Sequence positions of proline residues are indicated with circles on the X-axis.
![Figure 3. Interaction of BECN1(1–150)[4CS]Y and BCL2L1ΔC25. (A) Superimposed full 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y in the absence (black) and presence of BCL2L1ΔC25 (red). Molar ratio of BECN1(1–150)[4CS]Y:BCL2L1ΔC25 is 1.00:1.32. Cyan circles indicate resonances that shift upon binding. (B) Ratio of peak intensities of BECN1(1–150)[4CS]Y in the presence of BCL2L1ΔC25 (molar ratio 1.00:1.32) to those in the absence of BCL2L1ΔC25 plotted against the sequence of BECN1(1–150)[4CS]Y. The most significant loss of backbone amide intensities is located within or adjacent to the BH3 domain indicating those residues are most important in the binding with BCL2L1ΔC25. Sequence positions of proline residues are indicated with circles on the X-axis. (C) Partial 1H-15N HSQC spectrum of BECN1(1–150)[4CS]Y showing just the region of glycine residues (corresponding to the boxed region in [A]). (D to H) Superimposition of spectrum in region B (black) with the same region of the spectrum for BECN1(1–150)[4CS]Y in the presence of BCL2L1ΔC25 (colored) at a BECN1(1–150)[4CS]Y:BCL2L1ΔC25 molar ratio of (D) 1.00:0.26, (E) 1.00:0.53, (F) 1.00:0.79, (G) 1.00:1.06, (H) 1.00:1.32. (I to K) Similar to (D to H) but after BCL2L11(BH3) was added to the (BECN1(1–150)[4CS]Y-BCL2L1ΔC25 complex mixture shown in (H). Molar ratios of BECN1(1–150)[4CS]Y:BCL2L1ΔC25:BCL2L11(BH3) are (I) 1.00:1.32:0.75, (J) 1.00:1.32:1.50, (K) 1.00:1.32:3.00. (L) One-dimensional traces from 1H-15N HSQC spectra recorded over the entire course of the titration highlights changes of peak intensities for G55 (not affected by binding with BCL2L1ΔC25) and G101 (which is significantly affected) along with an adjacent peak (annotated as G101') that appeared/disappeared in the presence of BCL2L1ΔC25 and/or BCL2L11(BH3). (M) Superimposed full 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y in the absence (black) and presence of BCL2L1ΔC25 and BCL2L11(BH3) peptide (green). Molar ratio of BECN1(1–150)[4CS]Y:BCL2L1ΔC25:BCL2L11(BH3) is 1.00:1.32:3.00. (N) Ratio of peak intensities of BECN1(1–150)[4CS]Y in the presence (molar ratio 1.00:1.32:3.00) and absence of BCL2L1ΔC25 and BCL2L11(BH3). Peaks of BECN1(1–150)[4CS]Y that were lost due to binding to BCL2L1ΔC25 (see panel B) were completely restored when BCL2L11(BH3) is added and in excess. The overall reduction of peak intensity is due to dilution. Sequence positions of proline residues are indicated with circles on the X-axis.](/cms/asset/556cbb5f-619a-4b74-9630-eea65734e8a6/kaup_a_1140292_f0003_c.gif)
BECN1 N terminus reverts to a disordered structure following dissociation from BCL2L1
Although the conformation of BECN1(1–150)[4CS]Y in complex with BCL2L1 under the current conditions cannot be quantified due to the absence of resonances in the BH3 region, our NMR data combined with published structures of BCL2L1-BECN1(BH3) complexes,Citation11,12 enable us to conclude that residues between 98 to 132 that engage BCL2L1 likely adopt a helical conformation, while the rest of BECN1(1–150)[4CS]Y remains unstructured. To establish whether this secondary structure transition is reversible, we next acquired 1H-15N HSQC spectra following titration of a BCL2L11(BH3)/BIM(BH3) synthetic peptide into the BCL2L1ΔC25-BECN1(1–150)[4CS]Y complex. BCL2L11(BH3) binds BCL2L1 with ∼1000-fold higher affinity than BECN1(BH3), hence would be expected to outcompete the associated BECN1 N-terminal domain. Indeed, after addition of the BCL2L11(BH3) peptide, all of the resonances of BECN1(1–150)[4CS]Y residues that were disturbed by binding to BCL2L1ΔC25 (i.e., those within or adjacent to the BH3 domain) began to re-emerge at their original locations (). This is most obvious by examining just the characteristic glycine region of the 1H-15N HSQC spectra (). Moreover, when BCL2L11(BH3) was in molar excess to BCL2L1, the 1H-15N HSQC spectrum of BECN1(1–150)[4CS]Y completely reverted to that seen in the absence of BCL2L1 suggesting that any structural change that occurred was fully reversible ().
BECN1 N terminus interactions with the C-terminal BARA/ECD domain
Our fully assigned spectrum for the BECN1 N terminus also enabled us to assess whether part(s) of it engages in any significant interactions with other regions on the BECN1 protein, at least when present in cis. Accordingly, a 1H-15N HSQC spectrum for 15N-labeled BECN1(1–150)[4CS]Y was recorded in the presence of an equimolar concentration of purified BECN1(BARA/ECD) domain (residues 248 to 450) produced recombinantly in E. coli (). When compared to a spectrum of BECN1(1–150)[4CS]Y recorded in the presence of an equal volume of buffer (to account for dilution effects), we observed only very minor changes in peak intensities along the N terminus of BECN1 (e.g., residues 96 to 101, 115, 116, 121, 122, 128, 145) (), unlike the very large changes observed for residues within or adjacent to the BH3 domain upon titration of BCL2L1 (). Hence, while these data suggest there is some potential for regions within the BECN1 N-terminal region to interact with its C-terminal domain, these interactions are likely very weak and/or transient, and probably not critical for the overall structure of full-length BECN1.
Figure 4. Interaction of BECN1(1–150)[4CS]Y and the BECN1(BARA/ECD) domain. (A) Coomassie-stained SDS-PAGE gel showing peak fractions following gel-filtration chromatography of BECN1(BARA/ECD) domain demonstrating it is very pure. (B) Superimposed 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y in the absence (black) and presence of BECN1(BARA/ECD) (red). Molar ratio of BECN1(1–150)[4CS]Y:BECN1(BARA/ECD) domain is 1:1. The reference spectrum (black) was acquired by mixing the BECN1(1–150)[4CS]Y with the equivalent volume of the buffer solution used for preparation of the BARA/ECD domain. (C) Ratio of peak intensities of BECN1(1–150)[4CS]Y in the presence of BECN1(BARA/ECD) to those in its absence plotted against the sequence of BECN1(1–150)[4CS]Y.
![Figure 4. Interaction of BECN1(1–150)[4CS]Y and the BECN1(BARA/ECD) domain. (A) Coomassie-stained SDS-PAGE gel showing peak fractions following gel-filtration chromatography of BECN1(BARA/ECD) domain demonstrating it is very pure. (B) Superimposed 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y in the absence (black) and presence of BECN1(BARA/ECD) (red). Molar ratio of BECN1(1–150)[4CS]Y:BECN1(BARA/ECD) domain is 1:1. The reference spectrum (black) was acquired by mixing the BECN1(1–150)[4CS]Y with the equivalent volume of the buffer solution used for preparation of the BARA/ECD domain. (C) Ratio of peak intensities of BECN1(1–150)[4CS]Y in the presence of BECN1(BARA/ECD) to those in its absence plotted against the sequence of BECN1(1–150)[4CS]Y.](/cms/asset/7e20f9a3-398d-45c6-ae30-3170a0ff6250/kaup_a_1140292_f0004_c.gif)
Apoptotic potential of the BECN1(BH3) domain
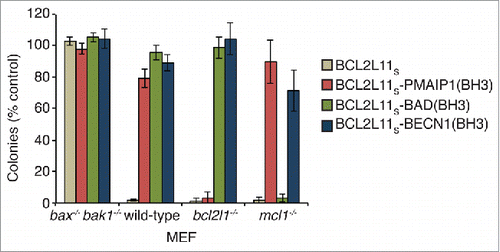
Several reports have demonstrated that deletion of a large portion (residues 88 to 144) of the BECN1 N-terminal domain or indeed the entire N terminus of yeast Vps30/Atg6, does not affect its autophagic activity.Citation7,20,36 Similar deletion studies have also shown the N-terminal domain does not have autophagic activity on its own.Citation37 However, as it contains a BH3 domain, which is the critical functional domain in proapoptotic BH3-only proteins, it was of interest to establish whether the BECN1(BH3) sequence possesses any apoptotic activity. As such we generated a chimeric construct in which the BH3 domain of the proapoptotic BH3-only protein BCL2L11 (BCL2L11Sisoform) was replaced with the corresponding BECN1(BH3) sequence, or those from other proapoptotic BH3-only proteins (BAD/BCL2L8 and PMAIP1/NOXA) as specificity controls. This is a well-established approach for comparing the killing activity of different BH3 domains in vitro and in vivo.Citation38–40 Killing activity was assessed by long-term (7 d) colony formation assays in mouse embryonic fibroblasts (MEFs) of different genotypes. MEFs are dependent on BCL2L1 and MCL1/BCL2L3 for their survival,Citation39 hence both of these proteins must be neutralized for cell-killing activity. BCL2L11S, which can neutralize all BCL2 prosurvival proteins,Citation40 killed all cell lines except those deficient in the critical mediators of apoptosis, BAX and BAK1, indicating its mechanism-based activity. In contrast, the BCL2L11S-BAD(BH3) chimera which targets BCL2L1, BCL2 and BCL2L2/BCLW only killed mcl1 knockout MEFs, while the BCL2L11S-PMAIP1(BH3) chimera (which only targets MCL1) only killed bcl2l1/bclx knockout MEFs, consistent with the specificity profiles of both of these constructs.Citation40 Unlike all of these other constructs, no killing activity was observed with the BCL2L11S-BECN1(BH3) chimera in any cell line tested suggesting its affinity for BCL2 proteins, especially MCL1 and BCL2L1, is too low to functionally neutralize them in cells.
Discussion
In this paper we provide the first report of the expression and purification of the BECN1 N-terminal domain (residues 1 to 150). This was the critical first step toward the structural evaluation of this previously mostly uncharacterized region that comprises a third of the entire BECN1 protein. Our results from CD and NMR analysis showed that this entire region (residues 1 to 150) is intrinsically disordered, lacking any secondary or tertiary structure. This conclusion was supported by a range of other findings including its susceptibility to proteolysis, aberrant behavior on SDS-PAGE and gel-filtration chromatography, and the high frictional ratio (f/f0) upon analytical centrifugation. This finding is consistent with recent sequence analysis studies that used various algorithms to come to a similar conclusion, though the “consensus” disordered region derived from the results of 4 different prediction programs suggested only residues 42 to 115 were unstructured. It is also consistent with the data on the recent EM structure reported for the PtdIns3K complex where no density was apparent for the BECN1 N terminus (indicating it is disordered),Citation23 and provides an explanation for the difficulties in obtaining crystals of full-length BECN1 in the absence of stabilizing binding partners.
The N-terminal domain of Vps30/Atg6 in the crystal structure of the yeast PtdIns3K vacuolar sorting complex (complex II) was also not well defined, consistent with it being disordered, though there was some electron density that was assigned to this region which appears to form 3 disconnected helices involved in interactions with Vps38 (UVRAG).Citation24 Unfortunately, the identity of these potentially structured residues was not discernable due to the low resolution of the structure. Nevertheless, if these residues can be definitively assigned to the N terminus, then this result is interesting as it suggests that regions outside of the BH3 domain can also undergo disordered-to-ordered structural transitions upon interactions with other proteins. Such structural transitions are often observed in intrinsically disordered proteins or domains.Citation41 It should also be noted, however, that the yeast N-terminal domain has a long insertion (∼55 residues) relative to the corresponding domain in mammalian BECN1 so there could be some difference(s) in how this region behaves in different species. It is also unclear from the available data whether the same structural transition occurs in the context of the PtdIns3K autophagy complex.
We were not able to detect any significant interactions between the BECN1 N terminus and C-terminal BARA/ECD domains when examined in cis (), suggesting these domains do not interact in the full-length BECN1 protein. This agrees with the PtdIns3K complex EM structure and PtdIns3K complex II crystal structure in which BECN1/Vps30 appears highly elongated with the N and C termini being located distal to each other, separated by the coiled-coil domains. Similarly, our NMR studies on the truncated BECN1 residues 1 to 76 N-terminal fragment indicated there are no significant intradomain interactions between the N- and C-terminal halves of the N-terminal domain. More importantly, our NMR analysis showed that upon binding to BCL2L1, the whole of the N-terminal region except the BH3 domain remains unstructured, suggesting that this interaction does not “nucleate” additional structural transitions beyond the formation of the α-helical BH3 domain.Citation8,10–12 In addition, we showed that the BH3 domain reassumes its disordered conformation once it dissociates from its binding partner. Although the physiological relevance of interactions between BECN1 and mammalian prosurvival members of the BCL2 family has recently been questioned,Citation17,18 interactions between BECN1 and viral BCL2 orthologs have also been shown to be mediated by the BECN1(BH3) domain.Citation3,7,13,42 Hence, such interactions could be important for virus survival through inhibition of host cell autophagy following infection. As such, structural studies on BECN1 following such interactions, such as described here, will be important for fully understanding BECN1 biology.
In the past 2 decades the extent and importance of structural disorder in proteins has become increasingly apparent and is now recognized as being widespread and a key factor driving protein-protein interactions.Citation31,43-46 In the BCL2 family, the majority of the BH3-only subclass of proteins, critical for triggering the apoptotic cascade, are intrinsically unstructured with only the BH3 domain becoming α-helical after binding the prosurvival family members.Citation47,48 In this context, our data suggests the BECN1 N terminus fits the BH3-only protein structure archetype perfectly. However, in contrast to all BCL2 family BH3-only proteins, the affinity of the BECN1(BH3) domain for prosurvival protein targets is relatively weak, providing an explanation for the lack of cell-killing we showed here in mammalian cell survival assays (). Other studies using overexpressed BECN1 have also shown it to be lacking apoptosis-inducing activity despite possessing a BH3 domain.Citation15 This makes sense because, although a BH3 domain with high affinity for BCL2 prosurvival proteins would better enable autophagy-inhibitory interactions, it would also be more capable of inducing cell death, which would not be desirable. Interestingly the affinity of the BECN1(BH3) domain is significantly higher for some viral BCL2 proteins compared to its mammalian orthologs, suggesting a critical role for the inhibition of autophagy in some viral infections.Citation14,42
Figure 5. Cell-killing activity of the BECN1(BH3) domain. Colony formation assay showing mouse embryonic fibroblast (MEF) cell-killing activity of BCL2L11S chimeras with different BCL2 prosurvival protein specificities. BCL2L11S-BECN1(BH3) did not have any significant effect on any cell line tested.
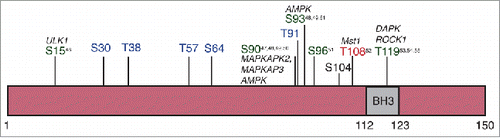
Greater than 50% of autophagy effector proteins are predicted to possess an intrinsically disordered region.Citation8 The role of these unstructured regions in autophagy however is still poorly understood, including the function of the N-terminal domain of BECN1 (beyond the BH3 domain). However, intrinsically disordered regions in proteins can be functionally very important as their lack of structure often enables them to engage in protein-protein interactions with multiple binding partners, including those that can create post-translational modifications such as kinases and ubiquitin ligases.Citation28,49 Indeed, the BECN1 N terminus contains a ubiquitination siteCitation50 and numerous phosphorylation sitesCitation51–60 () including several within the BH3 domain that can both positively and negatively regulate autophagy. Hence, although the BECN1 N-terminal domain might be dispensable for its autophagic activity,Citation7,20,36 our finding that the BECN1 N-terminal domain is intrinsically disordered is entirely consistent with its critical role as a regulatory hub that influences autophagy through phosphorylation, ubiquitination and other protein:protein interactions.
Figure 6. Phosphorylation sites in the BECN1 N-terminal domain. Summary of phosphorylation sites in the BECN1 N-terminal domain. Sites in green promote autophagy while those in red inhibit it. Those sites indicated in blue were identified from proteomics-based experiments however their functional consequences are unknown.
Materials and methods
E. coli expression constructs
The human BECN1 DNA encoding residues 1 to 150 was codon-optimized for expression in E. coli and cloned into the pET DUET-1 vector (Site I) (Novagen, 71146–3) at the BamHI/NotI sites, which fuses it to a hexahistdidine purification tag followed by a short linker sequence (MGSSHHHHHHSQDP). This served as a base construct for a subsequent construct in which all cysteines (i.e., residues 18, 21, 137, 140) were mutated to serine plus a C-terminal tyrosine was added to facilitate quantification by UV spectroscopy (called “BECN1(1–150)[4CS]Y”). An additional truncated version of this construct (residues 1 to 76, BECN1(1–76)[2CS]Y), also with a C-terminal tyrosine, was made to facilitate NMR spectrum assignment. The BECN1(BARA/ECD) domain (encoding residues 248 to 450) construct was cloned identically into the pET DUET-1 vector.
Protein expression
To produce BCL2L1 we used a C-terminally truncated construct (BCL2L1ΔC25), which removes the hydrophobic C-terminal transmembrane region that causes the protein to aggregate. This construct was expressed as described previously.Citation40 BECN1 constructs were transformed and expressed in the BL21 DE3 E. coli strain. Cultures were grown at 37°C in Super Broth or minimal mediumCitation61 for 15N or 15N/13C labeling to OD600nm 0.5–1.0 before expression was induced by addition of IPTG to a final concentration of 500 µM. Expression was induced for 3 h at 37°C. Cell pellets were stored at −80°C until proteins were purified.
Protein purification
BCL2L1ΔC25 was purified as described previously.Citation40 All BECN1 protein constructs including N-terminal and BARA/ECD domains were histidine-tagged and purified using the same purification steps which were all conducted at 4°C with prechilled solutions. The cell pellet was resuspended in Tris-buffered saline (TBS; 20 mM Tris, pH 8.0, 150 mM NaCl) using ∼40 mL for the pellet from a 2 L culture, containing 250 units of DNase 1 (Roche, 04536282001) and 3 tablets of protease inhibitor cocktail (Roche, 05056489001). Cells were lysed by 2 passages through a pressure cell homogenizer (Stansted, model SPCH-10). After centrifugation for 30 min at 48,000g the supernatant fraction was filtered through a 0.2-μm filter prior to loading samples onto a 1 mL HiTrap Chelating HP column (GE Healthcare, 17–0408–01) precharged with nickel sulfate solution. The column was then washed with 20 column volumes TBS containing 20 mM imidazole and the bound protein eluted with 5 column volumes TBS containing 250 mM imidazole. The eluted protein was then injected directly onto a Superdex S75 16/60 column equilibrated with 50 mM phosphate buffer, pH 6.8 containing 50 mM NaCl, at 1 mL/min. Peak fractions (based on absorbance 280 nm for tyrosine-containing constructs or 214 nm for native construct) were analyzed on NuPAGE 4–12% Bis-Tris gels (Invitrogen, NPO335BOX) stained with Coomassie Brilliant Blue R-250. The concentration of the eluted peak protein fractions was determined using the Beer-Lambert equation from the absorption at 280 nm determined using a Nano drop spectrophotometer. The purest fraction(s) were then used directly for subsequent analysis without further manipulation. For long-term storage of NMR samples, NaN3 was added to a final concentration of 0.02 % (w/v).
Peptides
Peptides were synthesized by Mimotopes (Clayton, Victoria, Australia) and purified by reversed-phase HPLC to >90 % purity. The sequences were: BECN1(BH3) - GEASDGGTMENLSRRLKVTGDLFDIMSGQTDVDH; BCL2L11(BH3)/BIM(BH3) –DMRPEIWIAQELRRIGDEFNAYYARR, used previously for NMR studies.Citation62
Surface plasmon resonance solution competition assay
Solution competition assays were performed using a Biacore 3000 instrument (GE Healthcare, Little Chalfont, UK) as described previously.Citation63 Briefly, recombinant BCL2L1ΔC25 (5 nM) was incubated with the BECN1(BH3) synthetic peptide or BECN1(1–150)[4CS]Y for at least 2 h prior to injection onto a CM5 sensor chip on which either wild-type BCL2L11(BH3) peptide or an inert BCL2L11(BH3) mutant peptide (BCL2L114E) was immobilized. Specific binding of BCL2L1 to the surface in the presence and absence of BECN1 competitors was quantified by subtracting the signal obtained on the BCL2L11(BH3)-mutant channel from that obtained on the wild-type-BCL2L11(BH3) channel. The ability of the BECN1(BH3) synthetic peptide or BECN1(1–150)[4CS]Y to inhibit BCL2L1 binding to immobilized BCL2L11(BH3) was expressed as IC50, calculated by nonlinear curve fitting of the data by using GraphPad Prism (GraphPad Software)
Analytical ultracentrifugation
Sedimentation velocity experiments were conducted in a Beckman model XL-A analytical ultracentrifuge at a temperature of 20°C. BECN1(1–150)[4CS]Y (50 µM in 50 mM sodium phosphate, 50 mM sodium chloride, pH 6.8) was loaded into a conventional double sector quartz cell and mounted in a Beckman 4-hole An-60 Ti rotor (Beckman Coulter Indianapolis, IN, USA). Both the sample (380 µL) and buffer solution (400 µL) were centrifuged at a rotor speed of 116,297 x g, and the data were collected at a single wavelength (236 nm) in continuous mode, using a time interval of 0 s and a step-size of 0.003 cm without averaging. Solvent density (1.0061 g/mL at 20°C) and viscosity (1.0287 cp), and an estimate of the partial specific volume (0.7153 mL/g) were computed using the amino-acid composition of the BECN1(1–150[4CS]Y peptide and the program SEDNTERP.Citation64 Sedimentation velocity data at multiple time points were fitted to continuous size-distribution modelsCitation65–67 using the program SEDFIT, which is available at www.analyticalultracentrifugation.com.
Circular dichroism spectroscopy
Circular dichroism spectra were recorded at 20°C using an AVIV Model 420 CD spectrometer (Aviv Biomedical, NJ, USA). Wavelength scans were performed between 195 and 250 nm in aqueous buffer (50 mM sodium phosphate, 50 mM NaCl, pH 6.8) using 0.15 mg/mL of protein in a 1 mm quartz cuvette. The resulting raw data were converted to mean residue elipticity ([θ]) and the α-helical content estimated from ([θ]222nm) as described previously.Citation65
Nuclear magnetic resonance spectroscopy
15N-labeled (∼0.18 mM) and 13C/15N labeled (∼0.12 mM) BECN1(1–150)[4CS]Y were prepared in 50 mM sodium phosphate containing 50 mM sodium chloride and 0.02 % (w/v) sodium azide at pH 6.8. A 15N-labeled N-terminal half of BECN1(1–150)[4CS]Y, BECN1–76[2CS]Y (∼0.30 mM), was also prepared in the same buffer solution for the present study. Two-dimensional 1H-15N HSQC spectra of BECN1(1–150)[4CS]Y over the temperature range of 5 to 30°C were first acquired and evaluated. Spectra at 15°C provided a good compromise between molecular rotational tumbling and relatively rapid backbone amide exchange of BECN1(1–150)[4CS]Y. All spectra used for backbone sequential assignments were subsequently recorded at 15°C on a Bruker Avance800 spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) equipped with a cryoprobe. Backbone assignments were achieved from standard triple resonance HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HNCO and HN(CA)CO spectraCitation68 whereas backbone Hα chemical shifts were obtained from HNHA and 15N-HSQC-TOCSY experiments. NMR data were processed in Topspin (Version 3.2, Bruker) with the 1H chemical shifts referenced indirectly to DSS at 0 ppm via the H2O signal (4.885 ppm at 15°C), and the 13C and 15N chemical shifts were referenced indirectly using absolute frequency ratios.Citation69 Spectra analyses were carried out in XEASY Version 1.3.Citation70 Experimentally determined backbone chemical shifts (13Cα, 13Cβ, 13C, 15N, 1HN, and 1Hα) of BECN1(1–150)[4CS]Y were then used for the identification of secondary structure using the program CSI 2.0.Citation34
To validate the interaction between BECN1(1–150)[4CS]Y and BCL2L1ΔC25, BCL2L1ΔC25 (0.46 mM) was titrated into the 13C/15N BECN1(1–150)[4CS]Y solution (0.145 mM) with the course of interaction monitored by 1H-15N HSQC spectra. After a molar excess of BCL2L1ΔC25 in solution was reached (i.e., BECN1(1–150)[4CS]Y became fully bound), the BCL2L11(BH3) peptide (2 mM) was subsequently titrated into the solution containing the (BECN1(1–150)[4CS]Y)-BCL2L1ΔC25 complex until a molar excess of BCL2L11(BH3) (versus BCL2L1ΔC25) was reached (no bound BECN1(1–150)[4CS]Y present in solution). For studies looking at the interactions between the N-terminal and BARA/ECD domains, 15N BECN1(1–150)[4CS]Y solution (0.150 mM) was mixed in a 1:1 molar ratio with unlabelled BARA/ECD domain or equivalent volume of buffer and the interaction monitored by the 1H-15N HSQC spectra.
Cell-killing assay
Long-term clonogenic assays were performed exactly as described recently.Citation71 Briefly MEFs (wild-type, bax−/− bak1−/−, bcl2l1−/−, mcl1−/−) were infected with retroviruses to express BCL2L11S, BCL2L11S-BECN1(BH3), BCL2L11S-BAD(BH3) or BCL2L11S-PMAIP1(BH3) where their expression was linked to that of green fluorescent protein via an internal ribosome entry site. Infected (green fluorescent protein+) cells were sorted and plated, then colonies counted 7 d later. The BCL2L11S-BECN1(BH3) construct has the BCL2L11(BH3) domain (DMRPEIWIAQELRRIGDEFNAYYARR) replaced with the analogous sequence (DGGTMENLSRRLKVTGDLFDIMSGQT) in BECN1. BCL2L11S-BAD(BH3) and BCL2L11S-PMAIP1(BH3) have been described previously.Citation40
Abbreviations
| BARA | = | β-α repeat autophagic-specific |
| BAK1 | = | BCL2-antagonist/killer 1 |
| BAX | = | BCL2-associated X protein |
| BCL2 | = | B-cell CLL/lymphoma 2 |
| BECN1 | = | Beclin 1, autophagy related |
| BH3 | = | BCL2 Homology 3 |
| CD | = | circular dichroism |
| ECD | = | evolutionarily conserved domain |
| MEF | = | mouse embryonic fibroblast |
| PtdIns3K | = | class III phosphatidylinositol 3-kinase |
| MCL1 | = | myeloid cell leukemia 1 |
| NMR | = | nuclear magnetic resonance |
| TBS | = | Tris-buffered saline |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Dr Jeff Babon (WEHI) for helpful discussions.
Funding
This work was supported by grants and fellowships from the NHMRC of Australia (Project Grant 1049949 and Career Development Fellowship 1024620 to E.F.L). Infrastructure support from NHMRC IRIISS grant #361646 and the Victorian State Government OIS grant is gratefully acknowledged.
References
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 2013; 368:651–62; PMID:23406030; http://dx.doi.org/10.1056/NEJMra1205406
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280–93; PMID:20965422; http://dx.doi.org/10.1016/j.molcel.2010.09.023
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 1998; 72:8586–96; PMID:9765397
- Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999; 59:59–65; PMID:10395800; http://dx.doi.org/10.1006/geno.1999.5851
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672–6; PMID:10604474; http://dx.doi.org/10.1038/45257
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22:140–9; PMID:20097051; http://dx.doi.org/10.1016/j.ceb.2010.01.001
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927–39; PMID:16179260; http://dx.doi.org/10.1016/j.cell.2005.07.002
- Mei Y, Su M, Soni G, Salem S, Colbert CL, Sinha SC. Intrinsically disordered regions in autophagy proteins. Proteins 2014; 82:565–78; PMID:24115198; http://dx.doi.org/10.1002/prot.24424
- Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy 2007; 3:374–6; PMID:17438366; http://dx.doi.org/10.4161/auto.4237
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 2007; 26:2527–39; PMID:17446862; http://dx.doi.org/10.1038/sj.emboj.7601689
- Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol 2007; 372:223–35; PMID:17659302; http://dx.doi.org/10.1016/j.jmb.2007.06.069
- Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem 2007; 282:13123–32; PMID:17337444; http://dx.doi.org/10.1074/jbc.M700492200
- Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy 2008; 4:989–97; PMID:18797192; http://dx.doi.org/10.4161/auto.6803
- Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene 2008; 27 Suppl 1:S137–48; PMID:19641499; http://dx.doi.org/10.1038/onc.2009.51
- Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene 2009; 28:2128–41; PMID:19347031; http://dx.doi.org/10.1038/onc.2009.60
- Noble CG, Dong JM, Manser E, Song H. Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1. J Biol Chem 2008; 283:26274–82; PMID:18641390; http://dx.doi.org/10.1074/jbc.M804723200
- Lindqvist LM, Vaux DL. BCL2 and related prosurvival proteins require BAK1 and BAX to affect autophagy. Autophagy 2014; 10:1474–5; PMID:24991825; http://dx.doi.org/10.4161/auto.29639
- Lindqvist LM, Heinlein M, Huang DC, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci U S A 2014; 111:8512–7; PMID:24912196; http://dx.doi.org/10.1073/pnas.1406425111
- Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, Zhang M, Yue Z, Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun 2012; 3:662; PMID:22314358; http://dx.doi.org/10.1038/ncomms1648
- Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J Biol Chem 2012; 287:16256–66; PMID:22437838; http://dx.doi.org/10.1074/jbc.M112.348250
- Hurley JH, Schulman BA. Atomistic autophagy: the structures of cellular self-digestion. Cell 2014; 157:300–11; PMID:24725401; http://dx.doi.org/10.1016/j.cell.2014.01.070
- Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res 2012; 22:473–89; PMID:22310240; http://dx.doi.org/10.1038/cr.2012.24
- Baskaran S, Carlson LA, Stjepanovic G, Young LN, Kim do J, Grob P, Stanley RE, Nogales E, Hurley JH. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. eLife 2014; 3:e05115; PMID:25490155; http://dx.doi.org/10.7554/eLife.05115
- Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 2015; 350:aac7365; PMID:26450213; http://dx.doi.org/10.1126/science.aac7365
- Priyadarshi A, Roy A, Kim KS, Kim EE, Hwang KY. Structural insights into mouse anti-apoptotic Bcl-xl reveal affinity for Beclin 1 and gossypol. Biochem Biophys Res Commun 2010; 394:515–21; PMID:20206602; http://dx.doi.org/10.1016/j.bbrc.2010.03.002
- Gronenborn AM, Clore GM. Identification of the contact surface of a streptococcal protein G domain complexed with a human Fc fragment. J Mol Biol 1993; 233:331–5; PMID:8411147; http://dx.doi.org/10.1006/jmbi.1993.1514
- Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 1991; 253:657–61; PMID:1871600; http://dx.doi.org/10.1126/science.1871600
- Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem 2014; 83:553–84; PMID:24606139; http://dx.doi.org/10.1146/annurev-biochem-072711-164947
- Uversky VN. Size-Exclusion Chromatography in Structural Analysis of Intrinsically Disordered Proteins. In: Uversky VN, Dunker AK, eds. Intrinsically Disordered Protein Analysis. UK: Humana Press, 2012:179–94
- Salvay AG, Communie G, Ebel C. Sedimentation Velocity Analytical Ultracentrifugation for Intrinsically Disordered Proteins. In: Uversky VN, Dunker AK, eds. Intrinsically Disordered Protein Analysis. UK: Humana Press, 2012:91–105
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6:197–208; PMID:15738986; http://dx.doi.org/10.1038/nrm1589
- Kosol S, Contreras-Martos S, Cedeno C, Tompa P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules 2013; 18:10802–28; PMID:24008243; http://dx.doi.org/10.3390/molecules180910802
- Konrat R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J Magn Reson 2014; 241:74–85; PMID:24656082; http://dx.doi.org/10.1016/j.jmr.2013.11.011
- Hafsa NE, Wishart DS. CSI 2.0: a significantly improved version of the Chemical Shift Index. J Biomol NMR 2014; 60:131–46; PMID:25273503; http://dx.doi.org/10.1007/s10858-014-9863-x
- Schneider R, Maurin D, Communie G, Kragelj J, Hansen DF, Ruigrok RW, Jensen MR, Blackledge M. Visualizing the molecular recognition trajectory of an intrinsically disordered protein using multinuclear relaxation dispersion NMR. J Am Chem Soc 2015; 137:1220–9; PMID:25551399; http://dx.doi.org/10.1021/ja511066q
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem 2006; 281:14474–85; PMID:16522639; http://dx.doi.org/10.1074/jbc.M600364200
- Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 2005; 1:46–52; PMID:16874027; http://dx.doi.org/10.4161/auto.1.1.1542
- Lee EF, Takiguchi M, Pettikiriarachchi A, Evangelista M, Huang DC, Dickins RA, Fairlie WD. A transgenic mouse model to inducibly target prosurvival Bcl2 proteins with selective BH3 peptides in vivo. Cell Death Dis 2015; 6:e1679; PMID:25766318; http://dx.doi.org/10.1038/cddis.2015.54
- Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. Proapoptotic Bak is sequestered by Mcl−1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev 2005; 19:1294–305; PMID:15901672; http://dx.doi.org/10.1101/gad.1304105
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 2005; 17:393–403; PMID:15694340; http://dx.doi.org/10.1016/j.molcel.2004.12.030
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol 2009; 19:31–8; PMID:19157855; http://dx.doi.org/10.1016/j.sbi.2008.12.003
- Ku B, Woo JS, Liang C, Lee KH, Jung JU, Oh BH. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy 2008; 4:519–20; PMID:18334862; http://dx.doi.org/10.4161/auto.5846
- Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn 2012; 30:137–49; PMID:22702725; http://dx.doi.org/10.1080/07391102.2012.675145
- Pancsa R, Tompa P. Structural disorder in eukaryotes. PLoS One 2012; 7:e34687; PMID:22496841; http://dx.doi.org/10.1371/journal.pone.0034687
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 2007; 447:1021–5; PMID:17522630; http://dx.doi.org/10.1038/nature05858
- Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 2008; 9 Suppl 2:S1; http://dx.doi.org/10.1186/1471-2164-9-S2-S1
- Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC, Day CL. Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ 2007; 14:128–36; PMID:16645638; http://dx.doi.org/10.1038/sj.cdd.4401934
- Rogers JM, Oleinikovas V, Shammas SL, Wong CT, De Sancho D, Baker CM, Clarke J. Interplay between partner and ligand facilitates the folding and binding of an intrinsically disordered protein. Proc Natl Acad Sci U S A 2014; 111:15420-5.
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 2004; 32:1037–49; PMID:14960716; http://dx.doi.org/10.1093/nar/gkh253
- Abrahamsen H, Stenmark H, Platta HW. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: Tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett 2012; 586:1584–91; PMID:22673570; http://dx.doi.org/10.1016/j.febslet.2012.04.046
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741–50; PMID:23685627; http://dx.doi.org/10.1038/ncb2757
- Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, Levine B. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife 2015; 4:e05289.
- Fogel AI, Dlouhy BJ, Wang C, Ryu SW, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol 2013; 33:3675–88; PMID:23878393; http://dx.doi.org/10.1128/MCB.00079-13
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013; 152:290–303; PMID:23332761; http://dx.doi.org/10.1016/j.cell.2012.12.016
- Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal 2010; 3:ra64; PMID:20736484; http://dx.doi.org/10.1126/scisignal.2000998
- Houck SA, Ren HY, Madden VJ, Bonner JN, Conlin MP, Janovick JA, Conn PM, Cyr DM. Quality control autophagy degrades soluble ERAD-resistant conformers of the misfolded membrane protein GnRHR. Mol Cell 2014; 54:166–79; PMID:24685158; http://dx.doi.org/10.1016/j.molcel.2014.02.025
- Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med 2013; 19:1478–88; PMID:24141421; http://dx.doi.org/10.1038/nm.3322
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 2009; 10:285–92; PMID:19180116; http://dx.doi.org/10.1038/embor.2008.246
- Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy 2009; 5:720–2; PMID:19395874; http://dx.doi.org/10.4161/auto.5.5.8625
- Gurkar AU, Chu K, Raj L, Bouley R, Lee SH, Kim YB, Dunn SE, Mandinova A, Lee SW. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun 2013; 4:2189; PMID:23877263; http://dx.doi.org/10.1038/ncomms3189
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol 1974; 119:736–47; PMID:4604283
- Yao S, Westphal D, Babon JJ, Thompson GV, Robin AY, Adams JM, Colman PM, Czabotar PE. NMR studies of interactions between Bax and BH3 domain-containing peptides in the absence and presence of CHAPS. Arch Biochem Biophys 2014; 545:33–43; PMID:24434006; http://dx.doi.org/10.1016/j.abb.2014.01.003
- Smith BJ, Lee EF, Checco JW, Evangelista M, Gellman SH, Fairlie WD. Structure-guided rational design of α/β-peptide foldamers with high affinity for BCL−2 family prosurvival proteins. Chembiochem 2013; 14:1564–72; PMID:23929624; http://dx.doi.org/10.1002/cbic.201300351
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. Analytical Ultracentrifugation in Biochemistry and Polymer Science: The Royal Society of Chemistry, Cambridge., 1992:90–125
- Perugini MA, Schuck P, Howlett GJ. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J Biol Chem 2000; 275:36758–65; PMID:10970893; http://dx.doi.org/10.1074/jbc.M005565200
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 2000; 78:1606–19; PMID:10692345; http://dx.doi.org/10.1016/S0006-3495(00)76713-0
- Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J 2002; 82:1096–111; PMID:11806949; http://dx.doi.org/10.1016/S0006-3495(02)75469-6
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Progress in Nuclear Magnetic Resonance Spectroscopy 1999; 34:93–158; http://dx.doi.org/10.1016/S0079-6565(98)00025-9
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 1995; 6:135–40; PMID:8589602
- Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J Biomol NMR 1995; 6:1–10; PMID:22911575; http://dx.doi.org/10.1007/BF00417486
- Lee EF, Dewson G, Evangelista M, Pettikiriarachchi A, Gold GJ, Zhu H, Colman PM, Fairlie WD. The functional differences between pro-survival and pro-apoptotic B cell lymphoma 2 (Bcl-2) proteins depend on structural differences in their Bcl-2 homology 3 (BH3) domains. J Biol Chem 2014; 289:36001–17; PMID:25371206; http://dx.doi.org/10.1074/jbc.M114.610758
