ABSTRACT
Selective macroautophagy/autophagy targets specific cargo by autophagy receptors through interaction with ATG8 (autophagy-related protein 8)/MAP1LC3 (microtubule associated protein 1 light chain 3) for degradation in the vacuole. Here, we report the identification and characterization of 3 related ATG8-interacting proteins (AT1G17780/ATI3A, AT2G16575/ATI3B and AT1G73130/ATI3C) from Arabidopsis. ATI3 proteins contain a WxxL LC3-interacting region (LIR) motif at the C terminus required for interaction with ATG8. ATI3 homologs are found only in dicots but not in other organisms including monocots. Disruption of ATI3A does not alter plant growth or development but compromises both plant heat tolerance and resistance to the necrotrophic fungal pathogen Botrytis cinerea. The critical role of ATI3A in plant stress tolerance and disease resistance is dependent on its interaction with ATG8. Disruption of ATI3B and ATI3C also significantly compromises plant heat tolerance. ATI3A interacts with AT3G56740/UBAC2A and AT2G41160/UBAC2B (Ubiquitin-associated [UBA] protein 2a/b), 2 conserved proteins implicated in endoplasmic reticulum (ER)-associated degradation. Disruption of UBAC2A and UBAC2B also compromised heat tolerance and resistance to B. cinerea. Overexpression of UBAC2 induces formation of ATG8- and ATI3-labeled punctate structures under normal conditions, likely reflecting increased formation of phagophores or autophagosomes. The ati3 and ubac2 mutants are significantly compromised in sensitivity to tunicamycin, an ER stress-inducing agent, but are fully competent in autophagy-dependent ER degradation under conditions of ER stress when using an ER lumenal marker for detection. We propose that ATI3 and UBAC2 play an important role in plant stress responses by mediating selective autophagy of specific unknown ER components.
Abbreviations
| ATI3 | = | ATG8-interacting 3 |
| ATG | = | autophagy related |
| BiFC | = | bimolecular fluorescence complementation |
| concA | = | concanamycin A |
| ER | = | endoplasmic reticulum |
| ERAD | = | ER-associated degradation |
| LIR | = | LC3-interacting region |
| TM | = | tunicamycin |
| UBAC2 | = | ubiquitin-associated (UBA) domain-containing protein 2 |
| WT | = | wild type |
Introduction
Macroautophagy/autophagy is an evolutionarily conserved pathway for degradation of cytoplasmic components in eukaryotes [Citation1]. Unlike the ubiquitin-proteasome system, which usually targets short-lived proteins, autophagy has the capacity to degrade both short- and long-lived proteins and organellesCitation[1,Citation2]. Autophagic degradation is required to maintain cell homeostasis in a wide range of physiological processes ranging from stress adaptation, cell differentiation and development to host immunity, cell survival and death [Citation2–4]. The molecular components of autophagy were initially discovered in the budding yeast, Saccharomyces cerevisiae, where more than 40 autophagy-related (ATG) proteins have been identified [Citation1]. These ATG proteins are involved in the induction of autophagy, phagophore nucleation, elongation, maturation of the autophagosome and fusion with the vacuole [Citation1]. Most of these ATG proteins have also been identified and analyzed in other eukaryotes including plants, indicating the conserved nature of the core autophagy process.
Autophagy in plants has also been extensively analyzed. Genes encoding most of the conserved ATG proteins have been identified in Arabidopsis and other plants and subjected to functional analysis through genetic and molecular approaches [Citation5–10]. These studies have shown that autophagy is involved in almost all aspects of plant life. Autophagy plays an important role in nutrient recycling and utilization in plants. Under nitrogen- or carbon-limiting conditions, both the formation of the autophagosome and expression of ATG genes are induced [Citation11,Citation12]. Furthermore, Arabidopsis autophagy mutants are hypersensitive to nitrogen- or carbon-limiting conditions [Citation11–13]. Autophagy also plays a role in the regulation of plant senescence, and plant mutants defective in autophagy undergo early senescence [Citation13–15]. Autophagy is induced by abiotic stresses such as oxidative, high salt and osmotic stress conditions and autophagy mutants are hypersensitive to these abiotic stresses [Citation16–18]. We have also reported that autophagy is induced under high temperature and Arabidopsis autophagy mutants are highly sensitive to heat stress [Citation19–21]. Therefore, autophagy plays a critical role in plant responses to a wide range of abiotic stresses.
Autophagy also affects plant responses to microbial pathogens. In Tobacco mosaic virus-inoculated Nicotinana benthamiana expressing the N resistance gene, virus-induced silencing of tobacco ATG6 and ATG7 genes results in expansion of N gene-mediated hypersensitive cell death to uninfected tissue in inoculated leaves and uninfected distant leaves [Citation7]. When infected by the avirulent Pseudomonas syringae strain expressing the avirulent gene AvrRpm1, transgenic ATG6 antisense Arabidopsis plants containing the AT3G07040/RPM1 resistance gene develop expanded hypersensitive cell death beyond the infiltrated areas [Citation22]. Thus, autophagy limits the spread of hypersensitive cell death beyond infected cells. Intriguingly, others have found that autophagy promotes defense-associated hypersensitive cell death induced by avirulent or related pathogens [Citation23]. The discrepancy in the effect of autophagy on pathogen-induced hypersensitive cell death could be related to such factors as the ages of plants and the time of cell death monitoring [Citation24,Citation25]. Unlike the conflicting results with biotrophic pathogens, there is consistent and compelling evidence that autophagy plays a critical and positive role in plant resistance to necrotrophic pathogens, which kill plant cells very early during infection. Autophagy is induced by infection of the necrotrophic fungal pathogen Botrytis cinerea and Arabidopsis autophagy mutants exhibit enhanced susceptibility to the necrotrophic pathogens B. cinerea and Alternaria brassicicola [Citation26,Citation27]. The positive role of autophagy in plant resistance to necrotrophic pathogens is associated with the activity of autophagy in promoting plant cell survival and positively modulating jasmonic acid signaling [Citation26], which is required for plant defense response to necrotrophic pathogens. There is also a role of autophagy in virus-induced RNA silencing, either as an antiviral collaborator for targeted degradation of viral RNA silencing suppressors or an accomplice of viral RNA silencing suppressors for targeted degradation of key components of plant cellular RNA silencing machinery [Citation28,Citation29].
The broad roles of autophagy are mediated not simply by nonselective bulk degradation of intracellular contents but by selective clearance of specific cellular structures or proteins targeted by autophagy receptors [Citation30,Citation31]. Autophagy receptors recognize specific autophagy substrates, on the one hand, and interact with the autophagosomal marker protein ATG8, on the other hand, thereby facilitating delivery of captured autophagy cargoes to phagophores (the precursors to autophagosomes) for degradation [Citation30,Citation31]. A substantial number of autophagy receptors from nonplant organisms have been identified that mediate the selective autophagosomal degradation of a variety of cargoes including protein aggregates, signaling complexes, mitochondria and bacterial pathogens [Citation30,Citation31]. In plants, autophagy receptors have also been identified and characterized. Arabidopsis protein AT2G47770/TSPO (outer membrane tryptophan-rich sensory protein) is an ATG8-interacting heme-binding protein that targets the degradation of porphyrins through autophagy [Citation32]. TSPO also interacts with the aquaporin AT4G35100/PIP2;7 and mediates its delivery from the plasma membrane to the vacuole for degradation, thereby reducing the levels of the aquaporin to regulate water permeability under conditions of heat and drought stress [Citation33]. Plant NBR1 is a functional hybrid of the human NBR1 and SQSTM1/p62 selective autophagy receptors that act in aggrephagy [Citation34]. Arabidopsis nbr1 mutants are highly compromised in tolerance to a range of abiotic stresses including heat, drought and salt stresses but are normal in resistance to necrotrophic pathogens and in age- and starvation-induced senescence [Citation19]. The compromised heat tolerance of Arabidopsis atg5, atg7 and nbr1 mutants is associated with increased accumulation of insoluble, highly ubiquitinated protein aggregates [Citation19]. Arabidopsis AT4G24690/NBR1, which contains a ubiquitin-binding domain, is also increasingly accumulated along with insoluble proteins in the autophagy mutants under heat stress [Citation19]. These results indicate that NBR1 is a selective autophagy receptor that targets degradation of ubiquitinated protein aggregates generated under stress conditions. AT2G45980/ATI1 and AT4G00355/ATI2 are 2 closely related, plant-specific ATG8-interacting proteins partially associated with the endoplasmic reticulum (ER) under normal conditions but become mainly associated with a new type of spherical compartments under carbon starvation [Citation35]. ATI1 is also located on bodies associated with plastids under carbon starvation and is involved in autophagy-dependent trafficking of plastid proteins to the vacuole [Citation36]. Selective autophagy also targets degradation of the 26S proteasome when the proteasome is inhibited chemically. This chemically induced proteaphagy is dependent on the proteasome subunit AT4G38630/RPN10 [Citation37]. RPN10 interacts with ATG8 and is increasingly associated with the proteasome when it is ubiquitinated, supporting its role as a selective autophagy receptor mediating the delivery of the proteasome to the vacuole [Citation37].
In the present study, we report identification and characterization of 3 related ATG8-interacting proteins (ATI3A, ATI3B and ATI3C) from Arabidopsis. Close homologs of ATI3 proteins are found only in dicot plants. Disruption of ATI3 does not alter plant growth or development but compromises both heat tolerance and resistance to the necrotrophic fungal pathogen B. cinerea. ATI3 interacts with UBAC2A (UBA domain containing 2A) and UBAC2B, a group of conserved ER proteins implicated in ER-associated degradation (ERAD) in human. Like ati3 mutants, ubac2 mutants are compromised in both heat tolerance and resistance to B. cinerea. Furthermore, increased levels of UBAC2 induced the formation of ATG8- and ATI3-labeled autophagosomes. The ati3 and ubac2 mutants were significantly compromised in sensitivity to the ER stress-inducing agent tunicamycin (TM) but were normal in autophagy-dependent ER degradation under ER stress when using an ER lumenal GFP marker for detection. Based on these findings, we propose that ATI3 and UBAC2 mediate a selective autophagy pathway with an important role in plant stress responses in part through selective autophagy of specific unknown ER components.
Results
Identification of a new family of plant-specific ATG8-interacting proteins
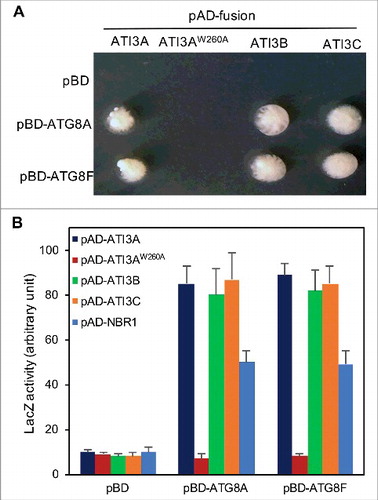
We previously reported the identification of more than 20 positive clones in yeast 2-hybrid screens using Arabidopsis AT4G21980/ATG8A and AT4G16520/ATG8F as baits [Citation19]. The proteins encoded by these positive clones include AT2G44140/ATG4, ATI1, ATI2 and NBR1. These ATG8-interacting proteins have been characterized for their roles in autophagosome formation and selective autophagy. Among the remaining positive clones, 4 match the Arabidopsis loci AT1G17780. Due to its interaction with ATG8 proteins, the corresponding protein product of AT1G17780 was named ATI3A (ATG8-interacting protein 3A). Based on both the prototrophy for His3 and LacZ reporter gene expression (), ATI3A is a strong interactor of ATG8 proteins. In fact, quantitative assays of β-galactosidase activity for LacZ reporter gene expression revealed that the interaction of ATG8A and ATG8F with ATI3A was substantially stronger than their interaction with NBR1 ((B)).
Figure 1. Interaction of Arabidopsis ATI3 and ATG8 proteins in yeast cells. The activation domain (AD) fusions of ATI3 genes were cotransformed with the empty Gal4 DNA binding domain vector (pBD) and fusions of ATG8 genes into yeast cells and grown in the absence of leucine, tryptophan and histidine (A). Proteins were isolated from the yeast cells and assayed for β-galactosidase activity using o-nitrophenyl-β-d-galactopyranose (ONPG) as substrate (B). Data represent means and standard errors (n = 5).
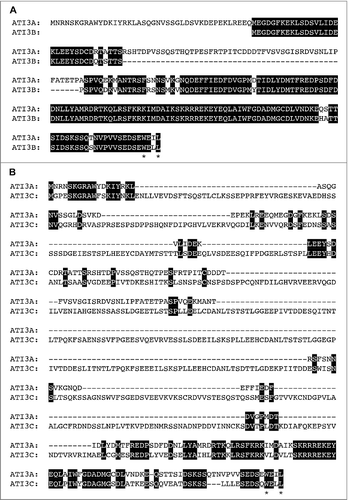
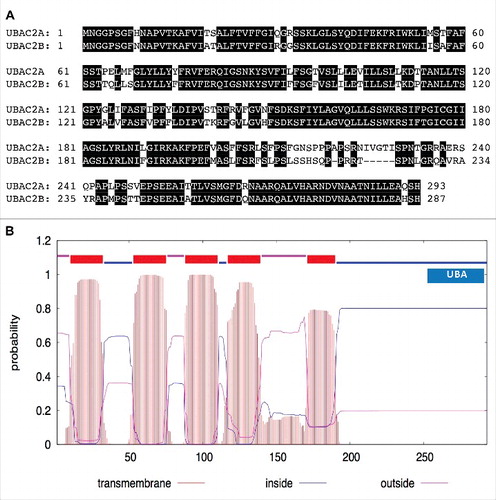
ATI3A is a protein of 263 amino acids (aa) with 2 homologs in Arabidopsis: ATI3B (AT2G16575) and ATI3C (AT1G73130). The protein sequence of ATI3B (170 aa) is highly similar to that of ATI3A but 2 segments of 42 and 51 aa at the N-terminal and middle regions of ATI3A, respectively, are absent in ATI3B (A). ATI3C is a longer protein of 646 aa and its sequence similarity to ATI3A and ATI3B is limited to the C-terminal region whereas its N-terminal region is highly divergent from those of ATI3A and ATI3B (B). The 3 proteins contain no known functional domain but all have a WxxL LIR motif at the C terminus (). Blast search of annotated genomes including those green plants on Phytozome 11 (www.phytozome.jgi.doe.gov) revealed that homologs of Arabidopsis ATI3 proteins are found only in dicots, but not in other organisms including monocots such as rice and maize. Most dicots contain one to 3 ATI3 gene homologs. These ATI3 homologs are divergent in their amino acid sequences at the N-terminal region but share high sequence homology in the C-terminal region of approximately 100 amino acids (Figure S1). At the C-terminal region, one stretch of about 30 amino acids contains a large proportion of charged residues and is highly conserved among the ATI3 homologs from different plants (Figure S1). Most importantly, all these ATI3 homologs from different dicots contain a putative WxxL LIR motif at their C terminus (Figure S1).
Figure 2. Protein sequences and motifs of Arabidopsis ATI3A, ATI3B and ATI3C. Sequence comparisons are between ATI3A and ATI3B (A) and between ATI3A and ATI3C (B). Amino acid residues identical between the 2 compared proteins are in black background and the conserved W and L residues in the putative WxxL LIR motifs at their C termini are indicated with asterisks.
To determine whether other ATI3 proteins are also ATG8-interacting proteins, we tested interaction of Arabidopsis ATI3B and ATI3C with ATG8A and ATG8F in yeast cells (). Coding sequences for Arabidopsis ATI3B and ATI3C were cloned into the pAD-Gal4 prey vector and cotransformed with the pBD-ATG8A and pBD-ATG8F bait vectors into yeast cells. Both the prototrophy for His and LacZ reporter gene expression confirmed that ATI3B and ATI3C also interacted strongly with ATG8A and ATG8F (). To determine whether the WxxL LIR motif at the C terminus of ATI3 proteins is required for interaction with ATG8, we changed the tryptophan residue at amino acid position 260 of ATI3A in the canonical WxxL LIR motif into an alanine residue (ATI3AW260L). The gene encoding the ATI3AW260L mutant was cloned into the prey vector and cotransformed into yeast cells with the ATG8 bait vectors. Unlike wild-type ATI3A, the ATI3AW260A mutant protein failed to interact with ATG8A or ATG8F in yeast cells based on both the prototrophy for His and the LacZ reporter gene expression (). Thus, the WxxL LIR motif at the C terminus of the ATI3 proteins is essential for interaction with ATG8.
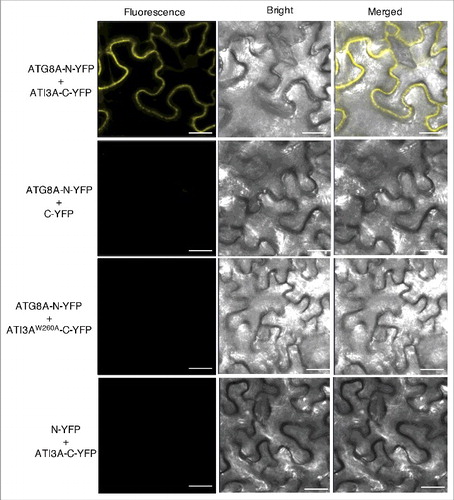
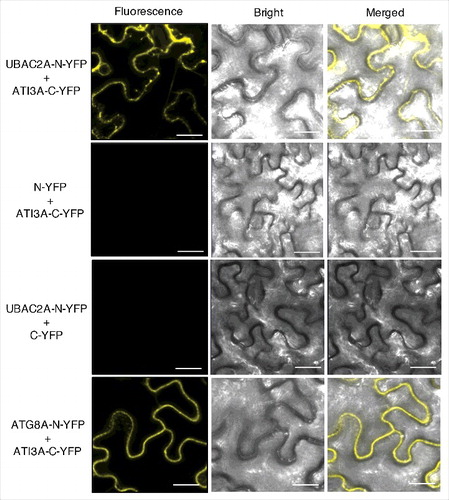
To determine whether ATG8 and ATI3 interact in plant cells, we performed bimolecular fluorescence complementation (BiFC) in Agrobacterium tumefaciens-infiltrated tobacco (N. benthamiana). We fused Arabidopsis ATG8A to the N-terminal yellow fluorescent protein (YFP) fragment (ATG8A-N-YFP) and ATI3A to the C-terminal YFP fragment (ATI3A-C-YFP). When fused ATG8A-N-YFP was co-expressed with ATI3A-C-YFP in tobacco leaves, BiFC signals were detected in transformed cells (). Control experiments in which ATG8A-N-YFP was co-expressed with unfused C-YFP or unfused N-YFP was co-expressed with ATI3A-C-YFP did not show fluorescence (). Furthermore, when we fused the ATI3AW260A mutant protein to the C-terminal YFP fragment (ATI3AW260A-C-YFP) and co-expressed with ATG8A-N-YFP, we observed no fluorescence (). These results indicated that ATI3A interacts with ATG8A in plant cells and this interaction is dependent on the WxxL LIR motif of ATI3A.
Figure 3. BiFC analysis of ATI3A interaction with ATG8A in planta. Fluorescence was observed in the transformed N. benthamiana leaf epidermal cells from complementation of the N-terminal half of YFP fused with ATG8A (ATG8A-N-YFP) by the C-terminal half of YFP fused with ATI3A (ATI3A-C-YFP). No fluorescence was observed when ATG8A-N-YFP was co-expressed with unfused C-YFP or with ATI3AW260A-C-YFP, or when unfused N-YFP was co-expressed with ATI3A-C-YFP. YFP epifluorescence, bright-field and emerged images of the same cells are shown. Bar: 20 μm.
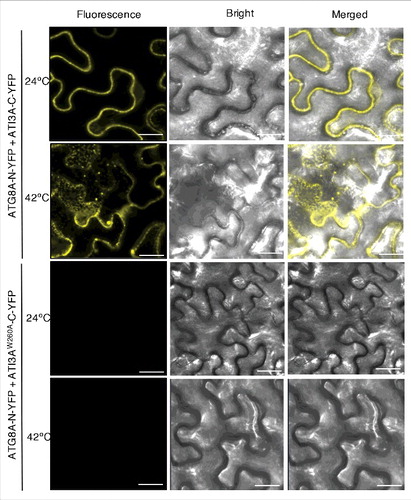
A number of studies using GFP-ATG8 fusion proteins have shown that stress conditions induce formation of ATG8-labeled punctate fluorescent signals in plant cells likely representing phagophores or autophagosomes [Citation19,Citation26,Citation38,Citation39]. We have previously shown that when fused ATG8A-N-YFP is co-expressed with NBR1-C-YFP in tobacco leaves, BiFC signals are detected in transformed cells, including punctate phagophores or autophagosome structures [Citation19]. Interestingly, the detected BiFC signals from the interaction of ATG8A-N-YFP and ATI3A-C-YFP were largely dispersed with few punctate fluorescent structures (). To determine whether formation of puncta labeled by the ATG8A-ATI3A complexes is responsive to stress, we treated infiltrated tobacco plants at 42ºC for 3 h and then placed them at room temperature for 0.5 h. The heat treatment increased the number of ATG8- and ATI3-labeled punctate structures in tobacco cells by almost 10-fold (). As expected, when ATI3AW260A-C-YFP was co-expressed with ATG8A-N-YFP, no BIFC signals or punctate structures were observed after heat treatment in the infiltrated tobacco leaves (). Thus, interaction of ATG8 and ATI3 proteins occurred under normal conditions but heat stress induced formation of ATG8- and ATI3-labeled punctate structures, likely reflecting increased formation of ATG8-labeled phagophores or autophagosomes under heat stress in plant cells.
Figure 4. Heat-induced punctate structures of ATI3-ATG8 complexes. Agrobacterium cells containing the complementing ATG8A-N-YFP and ATI3A-C-YFP BiFC constructs were infiltrated into N. benthamiana leaves for co-expression. One d after the infiltration, tobacco plants were treated at 42°C for 3 h and complemented BiFC signals were observed by confocal fluorescence microscopy. Increased punctate structures of complemented ATG8A-N-YFP-ATI3A-C-YFP BIFC signals were observed after treatment at 42°C. No BiFC signals were observed from co-expression of ATG8A-N-YFP and ATI3AW260A-C-YFP at either 24°C or 42°C. Bar: 20 μm.
Functional analysis of ATI3 genes
To determine the biological functions of the ATI3 proteins, we first isolated T-DNA insertion mutants for ATI3A. The ati3a-1 mutant (Salk_202281) contains a T-DNA insertion in the first exon of the gene (Figure S2A). Quantitative real-time PCR (qRT-PCR) analysis detected little transcript for ATI3A in the mutant, indicating that it is a knockout mutant (Figure S2B). The mutant is normal in growth and development and displayed no detectable changes in morphological phenotypes. We subsequently analyzed the phenotypes of the ati3a-1 mutant in the biological processes in which autophagy is involved. We first compared wild-type (WT) and ati3a mutants for differences in age- and dark-induced senescence and salt tolerance but found no significant alteration in the ati3a mutant when compared with WT plants. Thus, ATI3A does not appear to play a critical role in some of the important processes affected by autophagy.
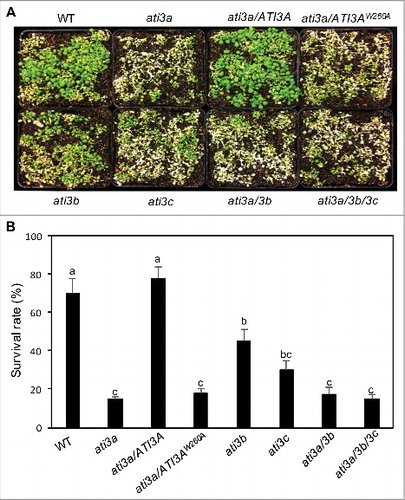
Additional analysis, however, revealed that the heat tolerance of the ati3a-1 mutant was substantially reduced. For testing heat tolerance, we placed 2-wk-old seedlings of WT and ati3a-1 mutant seedlings in a 45°C growth chamber for 10 h and scored for survival rates after recovery for 5 d at room temperature. Approximately 70% of WT seedlings but less than 20% of the ati3a-1 mutant seedlings survived after the heat stress (). To determine whether reduced heat tolerance of the ati3a-1 mutant is caused by disruption of ATI3A, we transformed the wild-type ATI3A coding sequence under the CaMV 35S promoter into the mutant. The ati3a-1 mutant expressing the ATI3A gene was restored in heat tolerance to the WT level (). By contrast, transformation of the ATI3AW260A mutant gene into the ati3a-1 mutant failed to restore the heat tolerance (). These results indicated that ATI3A plays a critical role in plant heat tolerance and this role is dependent on its interaction with ATG8.
Figure 5. Functional analysis of ATI3 genes in heat tolerance. Two-wk-old seedlings of Col-0 WT, ati3 single, double and triple mutants and ati3a mutant complemented with WT ATI3A and the mutant gene encoding ATI3AW260A were placed in a 45°C growth chamber for 10 h. The heat-treated plants were then moved to a 22°C growth chamber for recovery. The picture was taken after 3 d of recovery at 22°C (A). The survival rates were determined after 5 d of recovery at 22°C following heat treatment at 45°C for 10 h (B). Data represent means and standard errors calculated from 5 replicates (each with approximately 100 seedlings for each genotype). According to Duncan's multiple range test (P = 0.05), means of survival rates do not differ significantly if they are indicated with the same letter.
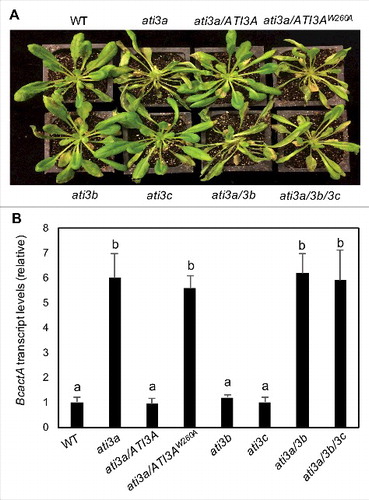
A number of plant ATG8-interacting proteins have been previously identified and mutants for some of the putative selective autophagy receptors have altered phenotypes in abiotic stress tolerance [Citation19,Citation21,Citation36]. However, there have been no reported studies demonstrating critical roles of selective autophagy receptors in plant disease resistance. To assess the involvement of ATI3A in plant disease resistance, we compared the ati3a-1 with WT for resistance to the necrotrophic fungal pathogen B. cinerea. In WT plants, the majority of leaves remained green with only a limited amount of necrosis and chlorosis at 4 d post inoculation (dpi). In the ati3a mutant plants, the necrotic spots and chlorosis were more extensive with a substantial portion being macerated by 4 dpi (A). qRT-PCR analysis indicated that the B. cinerea actA gene transcript levels in ati3a-1 were about 6-fold higher than those in WT at 5 dpi (B). Thus, both disease symptoms and fungal growth indicated that the ati3a mutant was compromised in resistance to the necrotrophic pathogen. Transformation of the WT ATI3A, ATI3A-GFP or ATI3A-C-YFP gene, but not the mutant gene encoding ATI3AW260A, into the ati3a-1 mutant restored the disease resistance to the wild-type levels (; Figure S3(A)). These results supported the idea that ATI3A plays a critical role in plant resistance to the necrotrophic fungal pathogen and this role is also dependent on its interaction with ATG8.
Figure 6. Functional analysis of ATI3 genes in plant resistance to B. cinerea. Six-wk-old Col-0 WT, ati3 single, double and triple mutants and ati3a mutant complemented with WT ATI3A and the mutant gene encoding ATI3AW260A were inoculated by spraying spore suspension at a density of 2.5 × 105 spores/mL and kept at high humidity. Photographs of representative plants were taken 4 d after inoculation (A). Total RNA was isolated from the plants 4 d after inoculation and the transcript levels of the B. cinerea actA gene were determined using qRT-PCR with the Arabidopsis AT3G18780/ACT2 gene as an internal control (B). Data represent means and standard errors (n = 5). According to Duncan's multiple range test (P = 0.05), means of B. cinerea actA gene transcript levels do not differ significantly if they are indicated with the same letter.
We have also isolated T-DNA insertion mutants for the 2 Arabidopsis ATI3A homologs. The ati3b-1 mutant (Salk_056098) contains a T-DNA insertion in the gene promoter whereas the ati3c-1 mutant (Sail_754_D05) contains a T-DNA insertion in the third exon of the gene (Figure S2(A)). qRT-PCR analysis indicated that the transcript levels of ATI3B were reduced to less than 10% of WT levels in ati3b-1 and the transcript levels for ATI3C were reduced to less than 5% of WT levels in ati3c-1 (Figure S2(B)). Like ati3a-1, the T-DNA insertion mutants for ATI3B and ATI3C were normal in growth and development. They were also indistinguishable from WT in dark- and age-induced senescence and in salt tolerance. On the other hand, the survival rates of the ati3b-1 and ati3c-1 mutant seedlings were substantially lower than those of WT seedlings but significantly higher than the ati3a-1 mutant seedlings after heat stress at 45ºC (). Reduced heat tolerance of the ati3b-1 and ati3c-1 mutants could be restored by transformation of the mutants with the WT ATI3B and ATI3C genes, respectively (Figure S3B). Thus, ATI3B and ATI3C also play a significant role in plant heat tolerance. Unlike ati3a, the ati3b and ati3c mutants were as resistant as WT plants to B. cinerea (). To determine possible functional redundancy among the 3 ATI3 genes, we have generated ati3a ati3b double and ati3a ati3b ati3c triple mutants. These double and triple mutants were again unchanged in growth and development under normal conditions but displayed the phenotypes of compromised heat tolerance and disease resistance largely similar to those of the ati3a-1 single mutant ( and ).
Identification of UBAC2 as ATI3A-interacting proteins
Based on their interaction with ATG8 proteins and the critical roles in plant heat tolerance and disease resistance, plant ATI3 proteins probably act as selective autophagy receptors that target specific cellular components during the plant stress response. However, apart from the WxxL LIR motif required for interaction with ATG8, ATI3 proteins contain no known functional domains and, therefore, the potential cellular components targeted by ATI3-mediated autophagy are unclear. To gain insights into the action of ATI3, we tried to identify ATI3-interacting proteins using yeast 2-hybrid screens. We initially tried the Gal4 system but found strong transcription-activating activity of ATI3A in yeast cells. Therefore, we used the split-ubiquitin yeast 2-hybrid system using ATI3AW260A as bait to identify proteins other than ATG8 that interact with ATI3A. After screening 7 × 106 independent transformants of an Arabidopsis cDNA prey library, we isolated 15 clones but subsequent tests using the empty bait vector confirmed that 14 of them were false positive. The single positive clone was sequenced and found to match the sequence of the loci AT3G56740. Blast search revealed that the ATI3A-interacting protein is closely related to UBAC2 (Ubiquitin-associated [UBA] protein 2) found in other eukaryotic organisms (Figure S4) and therefore is named Arabidopsis UBAC2A because it has a close homolog (AT2G41160/UBAC2B) with ∼80% identical residues (A). Arabidopsis UBAC2 proteins contain an N-terminal transmembrane domain and a C-terminal domain of ∼200 residues with a ubiquitin-association (UBA) domain at the C terminus (B). This domain structure is also conserved in UBAC2 proteins from other organisms (Figure S4).
Figure 7. Protein sequences and structures of UBAC2. The amino acid sequence of UBAC2A was compared with that of UBAC2B (A). Amino acid residues identical between the 2 compared proteins are in black background. The transmembrane helices and orientation in the UBAC2A protein were predicted using the TMHMM method based on a hidden Markov model (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (B). A ubiquitin-association domain (UBA) at the C-terminal region of UBAC2A is also indicated. The size of the protein is indicated by the number of amino acid residues shown at the bottom.
To determine whether ATI3A and UBAC2 interact in plant cells, we performed BiFC in Agrobacterium-infiltrated tobacco. We fused Arabidopsis UBAC2A to the N-terminal yellow fluorescent protein (YFP) fragment (UBAC2A-N-YFP) and co-expressed it with ATI3A-C-YFP in tobacco leaves. BiFC signals were detected in transformed cells (). Control experiments in which UBAC2A-N-YFP was co-expressed with unfused C-YFP, or unfused N-YFP was co-expressed with ATI3A-C-YFP did not show fluorescence (). Similar results with UBAC2B were also obtained for interaction with ATI3A in plant cells (data not shown). Interestingly, BiFC showed that while in vivo interaction between ATI3A and ATG8A produced largely dispersed signals under normal growth conditions, in vivo interaction between ATI3A and UBAC2 generated a substantial number of punctate fluorescent signals ().
Figure 8. BiFC analysis of ATI3-UBAC2 interactions. BiFC fluorescence was observed in the transformed N. benthamiana leaf epidermal cells from complementation of the N-terminal half of the YFP fused with UBAC2A (UBAC2A-N-YFP) by the C-terminal half of the YFP fused with ATI3A (ATI3A-C-YFP). No fluorescence was observed when UBAC2A-N-YFP was co-expressed with unfused C-YFP or when unfused N-YFP was co-expressed with ATI3A-C-YFP. YFP epifluorescence, bright-field and emerged images of the same cells are shown. BiFC fluorescence from complemented ATG8A-N-YFP and ATI3A-C-YFP is also shown for comparison of punctate fluorescent structures. Bar: 20 μm.
UBAC2 proteins are targeted to the ER
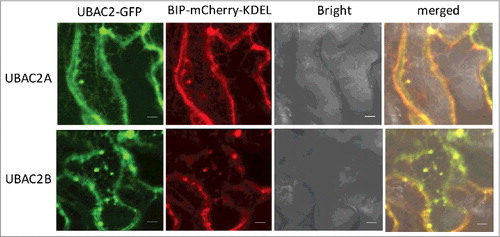
UBAC2 has been implicated in ERAD [Citation40], a cellular process that targets misfolded ER proteins for ubiquitination, retrotranslocation into the cytosol and degradation by the proteasome. As a component of ERAD, UBAC2 is localized to the ER membrane in mammalian cells and may be involved in recognition of ubiquitinated nonnative ER proteins through its cytosolic UBA domain at the C terminus [Citation40]. Like UBAC2 from other organisms, both Arabidopsis UBAC2A and UBAC2B contain a large N-terminal rhomboid-like transmembrane domain and a UBA domain at their C terminus (). To determine the subcellular localization of Arabidopsis UBAC2, we generated a UBAC2-GFP fusion construct and co-expressed it with the AT5G42020/BIP-mCherry-KDEL ER marker in N. benthamiana. As expected, the red fluorescent signals of BIP-mCherry-KDEL were present as numerous networks with blobs throughout the cells (). The green fluorescent signals of UBAC2-GFP were also present as typical ER networks with extensive overlapping with those of BIP-mCherry-KDEL (). This result indicates that Arabidopsis UBAC2 proteins are localized in the ER.
Figure 9. Subcellular localization of UBAC2 proteins. The UBAC2–GFP fusion genes were co-expressed with the BIP-mCherry-KDEL ER marker in N. benthamiana. The GFP, mCherry, bright field, and merged images are shown. Co-expressed BIP-mCherry-KDEL ER marker and UBAC2-GFP generated networks of fluorescent signals with blobs typical of ER structures throughout the cells and the BIP-mCherry-KDEL signals extensively overlapped those of UBAC2-GFP signals. Bar: 10 µm.
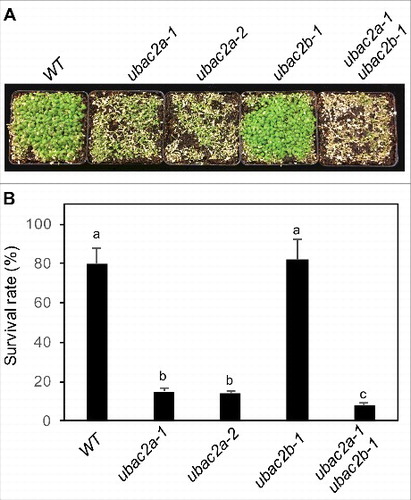
Functional analysis of UBAC2 in plant heat tolerance and disease resistance
To determine the biological functions of UBAC2 proteins, we isolated 2 T-DNA insertion mutants for UBAC2A and 1 T-DNA insertion mutant for UBAC2B. The ubac2a-1 (WiscDsLoxHs094_04C) and ubac2a-2 (WiscDsLox495E11) mutants contained a T-DNA insertion in the third exon and the second intron of UBAC2A, respectively (Figure S5A). The ubac2b-1 mutant (Salk_021788C) contained a T-DNA insertion in the fourth intron of UBAC2B (Figure S5A). qRT-PCR analysis revealed that these mutants contained less than 5% of the transcripts for the disrupted gene when compared to those in WT and, therefore, are likely to be knockout mutants (Figure S5B). To overcome possible functional redundancy, we also generated ubac2a ubac2b double mutants. The ubac2 single and double mutants for the 2 genes were unchanged in growth and development under normal conditions. Because of the critical role of ERAD and autophagy in plant heat tolerance, we first examined the responses of the ubac2 mutant seedlings to heat stress. After treatment at 45°C for 10 h and recovery at room temperature for 5 d, almost 80% of WT seedlings survived but less than 20% of the 2 ubac2a single mutants survived (). However, the ubac2b-1 mutant was normal in survival rate after heat stress but the ubac2a-1 ubac2b-1 double mutant had an even lower survival rate (∼10%) than those of the ubac2a single mutants (). These results indicate that UBAC2A and UBAC2B are partially redundant with UBC2A playing a relatively more important role in plant heat tolerance.
Figure 10. Functional analysis of UBAC2 in heat tolerance. Two-wk-old seedlings of Col-0 WT, ubac2a and ubac2b single and double mutants were placed in a 45°C growth chamber for 10 h. The heat-treated plants were then moved to a 22°C growth chamber for recovery. The picture was taken after 3 d of recovery at 22°C (A). The survival rates were determined after 5 d of recovery at 22°C following heat treatment at 45°C for 10 h (B). Data represent means and standard errors calculated from 5 replicates (each with approximately 100 seedlings for each genotype). According to Duncan's multiple range test (P = 0.05), means of survival rates do not differ significantly if they are indicated with the same letter.
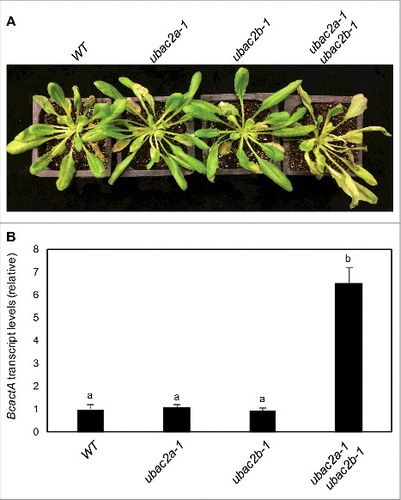
To assess the involvement of UBAC2 in plant disease resistance, we compared the ubac2 mutants with WT plants for resistance to B. cinerea. We observed no significant alteration of the ubac2a and ubac2b single mutants in disease symptom development when compared to that of WT (A). qRT-PCR also indicated levels of fungal growth in the 2 ubac2 single mutants to be similar to those in the WT plants (B). Conversely, the ubac2a ubac2b double mutant developed increased disease symptoms, which were correlated with increased fungal growth when compared to those in WT plants (). Reduced resistance of the ubac2a ubac2b double mutants to B. cinerea could be restored by transformation of the mutant with the UBAC2A-GFP and UBAC2B-GFP genes (Figure S6). Thus, like ATI3, UBAC2 proteins have an important role not only in plant heat tolerance but also in plant resistance to the necrotrophic fungal pathogen.
Figure 11. Functional analysis of UBAC2 genes in plant resistance to B. cinerea. Six-wk-old Col-0 wild-type (WT), ubac2a and ubac2b single and double mutants were inoculated by spraying spore suspension at a density of 2.5 × 105 spores/mL and kept at high humidity. Photographs of representative plants were taken 4 d after inoculation (A). Total RNA was isolated from the plants 4 d after inoculation and the transcript levels of the B. cinerea actA gene were determined using qRT-PCR with the Arabidopsis ACT2 gene as an internal control (B). Data represent means and standard errors (n = 5). According to Duncan's multiple range test (P = 0.05), means of the B. cinerea actA gene transcript levels do not differ significantly if they are indicated with the same letter.
Since UBAC2 has been implicated in ERAD, we compared the phenotypes of the ubac2a ubac2b double mutants with a T-DNA knockout mutant for Arabidopsis AT1G18260/HRD3A, an ERAD-associated E3 ubiquitin ligase component. First, we compared the ubac2a ubac2b and hrd3a mutants for heat tolerance. As shown in Figure S7A and S7B, the hrd3a mutant was more heat sensitive than the ubac2a ubac2b double mutant based on the lower seedling survival rates after treatment for 10 h at 45°C. However, unlike the ubac2a ubac2b mutant with compromised resistance to B. cinerea, the hrd3a mutant responded normally to the necrotrophic fungal pathogen (Figure S7C and S7D). Thus, the phenotypes of the ubac2a ubac2b and hrd3a mutants did not completely overlap.
Effect of heat and UBAC2 on ATI3- and ATG8-labeled punctate structures
Under stress conditions, misfolded proteins in the ER increase, causing ER stress. To overcome the problem, the unfolded protein response is activated for increased production of specific ER-resident proteins including the BIP (binding protein) family of molecular chaperones, which assist in protein folding and refolding [Citation41,Citation42]. As part of the unfolded protein response, ERAD is activated to remove misfolded proteins by delivery to the cytoplasm for proteasomal degradation [Citation40]. Previous studies have shown that ER stress induces autophagy and portions of the ER are also delivered to the vacuole by autophagy during ER stress in plant cells as evident from the localization of a soluble ER marker in autophgosomes, and accumulation in the vacuole upon inhibition of vacuolar proteases due to neutralization of acidic pH in the vacuole by concanamycin A (concA), an inhibitor of the vacuolar-type H+-translocating ATPase [Citation39,Citation43-45]. Given their physical interaction and the similar mutant phenotypes in compromised heat tolerance and resistance to B. cinerea, UBAC2 proteins could potentially function as receptors that recognize ubiquitinated ER proteins through its C-terminal UBA domain and target their degradation by ATI3- and ATG8-mediated selective autophagy during plant stress responses. To examine these possibilities, we analyzed the colocalization of ATI3A and UBAC2A with ATG8A before and after heat treatment. Using Agrobacterium infiltration, we co-expressed ATI3A-mCherry with GFP-ATG8A fusion genes in tobacco leaves. At 24ºC, we observed a low number of cytoplasmic punctate structures from co-expressed tobacco leaf cells (Figure S8A). After treatment at 42ºC for 3 h, the number of punctate fluorescent signals of ATI3A-mCherry increased substantially and a majority of these puncta were also labeled by GFP-ATG8A (Figure S8A). Similarly, we co-expressed ATG8-mCherry with UBAC2A-GFP (Figure S8(B)) or UBAC2B-GFP (Figure S8B) and found that the numbers of punctate fluorescent signals of UBAC2A- and UBAC2B-GFP also increased after heat treatment. Again, a majority of heat-induced punctate fluorescent signals of UBAC2A- and UBAC2B-GFP were labeled by ATG8A-mCherry (Figure S8B and S8C). Thus, heat stress induced the formation of ATG8-labeled ATI3 and UBAC2 punctate structures, which are likely to be phagophores or autophagosomes that contain ATI3 and UBAC2.
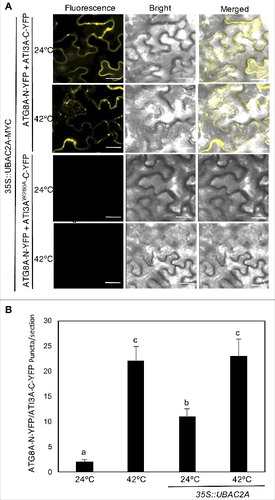
From the BiFC experiments for testing protein interactions in plant cells, we found that while the ATI3A-ATG8 BIFC signals in transformed tobacco leaf cells were largely dispersed (), the UBAC2-ATI3A BiFC signals were enriched in punctate structures even under normal growth conditions (). The increased punctate structures of the UBAC2-ATI3A BiFC signals could represent increased formation of ATI3A-labeled autophagosomes because of elevated levels of UBAC2 in the transformed tobacco cells. To test this possibility, we analyzed the effect of UBAC2A overexpression on the patterns of the ATI3A-ATG8A BiFC signals. Without UBAC2A overexpression, co-expression of ATG8A-N-YFP and ATI3A-C-YFP constructs in tobacco plants resulted in largely dispersed BiFC signals with few puncta under normal growth conditions (24°C) (, and ). Conversely, co-overexpression of UBAC2A with the ATG8 and ATI3A BiFC constructs increased the number of punctate ATG8-ATI3A BIFC signals by more than 5 fold at 24°C (). Heat treatment at 42°C could further increase the number of punctate ATG8-ATI3 BIFC signals by another 2 fold (). No dispersed or punctate BiFC signals were observed from the co-expression of ATG8A-N-YFP and ATI3AW260A-C-YFP with or without UBAC2A overexpression (). Thus, delivery of the ATG8-ATI3 complexes to phagophores, as indicated from the punctate BiFC signals, was promoted by UBAC2 overexpression. These observations suggest UBAC2 proteins may act as both cargo receptors and inducers of an ATI3-mediated selective autophagy pathway.
Figure 12. Overexpression of UBAC2 promoted formation of ATI3- and ATG8-labeled punctate structures. BiFC fluorescence was observed in transformed N. benthamiana leaf epidermal cells from complementation of the N-terminal half of the YFP fused with UBAC2A (UBAC2A-N-YFP) by the C-terminal half of the YFP fused with ATI3A (ATI3A-C-YFP) (A). No fluorescence was observed when UBAC2A-N-YFP was co-expressed with unfused C-YFP or when unfused N-YFP was co-expressed with ATI3A-C-YFP (A). YFP epifluorescence images, bright-field images and merged images of the same cells are shown. BiFC fluorescence from complemented ATG8A-N-YFP and ATI3A-C-YFP is also shown for comparison of punctate fluorescent structures. The numbers of formed ATG8A- and ATI3A-labeled puncta per microscopy section were determined in response to heat treatment or overexpression of UBAC2A (B). Data represent means and standard errors calculated from 10 microscopy sections. According to Duncan's multiple range test (P = 0.05), means do not differ significantly if they are indicated with the same letter. Bar: 20 μm.
Significant increase in sensitivity of ati3 and ubac2 to ER stress
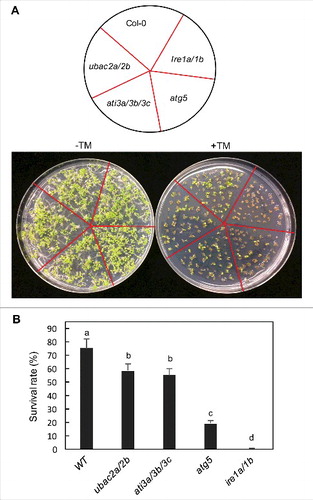
Extensive evidence has indicated that misfolded ER proteins are targeted for degradation not only by the proteasome but also by autophagy [Citation39,Citation43-45]. Therefore, it is possible that ATI3 and UBAC2 mediate a selective autophagy pathway in plants that targets ER degradation under ER stress. To test this possibility, we compared ati3a ati3b ati3c triple and ubac2a ubac2b double mutants with WT for tolerance to ER stress by testing the seedling growth and survival in the presence of the ER stress agent TM, an inhibitor of N-linked glycosylation that disturbs the protein-folding machinery and causes accumulation of unfolded proteins in the ER [Citation43,Citation45]. As a positive control, we included the autophagy deficient atg5-1 mutant [Citation26] and the ire1a-2 irelb-1 double mutant in the assays [Citation46]. In the absence of TM, WT and all these mutants grew similarly well (). In the presence of 0.1 mg/L TM, about 75% of WT seedlings survived but only about 20% of atg5-1 mutant seedlings survived after 2 wk (). Conversely, none of the ire1a-2 irelb-1 double-mutant seedlings survived in the presence of 0.1 mg/L TM after 2 wk (). Thus, the atg5 and ire1a ire1b mutants displayed increased sensitivity to the ER stress agent as previously reported [Citation45,Citation46]. The survival percentages of the ati3a ati3b ati3c triple and ubac2a ubac2b double-mutant seedlings were 55 and 58%, respectively, after 2 wk in the presence of TM (). These survival percentages were substantially higher than those of the atg5 and ire1 mutants but were significantly lower than that of WT. Thus, disruption of ATI3 and UBAC2 genes led to significantly increased sensitivity to ER stress agent TM.
Figure 13. Response of ati3 and ubac2 mutants to ER stress agents. Arabidopsis seeds of WT and mutant seeds were surface sterilized and sown on ½ strength MS media supplemented without (-TM) or with 0.1 mg/L TM (+TM). Pictures were taken 2 wk post germination (A). Seedling surviving rates were determined 2 wk post germination (B). Means and standard errors were calculated from 5 replicates (each with approximately 80 seedlings for each genotype). According to Duncan's multiple range test (P = 0.05), means do not differ significantly if they are indicated with the same letter.
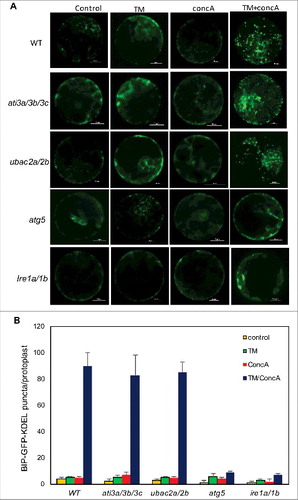
Competence of ati3 and ubac2 in autophagy-dependent ER degradation
To further analyze whether ATI3 proteins act as selective autophagy receptors that target ER through UBAC2 receptors for degradation in the vacuole during ER stress, we examined the delivery and accumulation of punctate autophagosome-like structures labeled by a fluorescent lumenal ER marker in the vacuole of leaf protoplasts. In control WT leaf protoplasts transfected with a BIP-GFP-KDEL lumenal ER marker-encoding gene, we observed a typical cytoplasmic ER pattern (). A similar cytoplasmic ER pattern was observed in WT leaf protoplasts after treatment with the vacuolar-type H+-translocating ATPase inhibitor concA, which blocks acid-dependent vacuolar protein turnover (). To induce ER stress, we treated the leaf protoplasts with TM and again observed largely cytoplasmic ER patterns of the fluorescent ER marker with relatively few puncta (). However, in the presence of both TM and concA, we observed that the number of the ER GFP punctate structures greatly increased in WT leaf protoplasts (). In the leaf protoplasts of the ati3a ati3b ati3c triple mutant and the ubac2a ubac2b double mutant, we observed the typical cytoplasmic ER pattern of the GFP-KDEL ER marker with or without incubation with TM or concA (). In the presence of both TM and concA, we observed a great increase in the ER GFP punctate structures in the leaf protoplasts of the mutants, similar to those in WT protoplasts (). By contrast, incubation of both TM and concA did not trigger a significant increase in the ER GFP punctate structures in the leaf protoplasts of mutants for ATG5 and IRE1 proteins required for ER stress-induced autophagy (). Thus, ati3 and ubac2 mutants were fully competent in autophagy- and IRE1-dependent ER degradation under ER stress when the BIP-GFP-KDEL lumenal protein was used as an ER marker for detection.
Figure 14. Role of ATI3 and UBAC2 in ER stress-induced autophagy-dependent ER degradation. Leaf protoplasts obtained from 4-wk-old plants were transformed with a BIP-GFP-KDEL construct. The final concentrations of TM and concA used in the experiments were 5 μg/mL and 1 μM, respectively. DMSO was used as a solvent control. Protoplasts were incubated at room temperature in darkness for 12 h, with 40 rpm orbital shaking. Confocal microscopy was used to visualize the GFP fluorescence (A). Bar: 10 μm. The numbers of BIP-GFP-KDEL puncta per leaf protoplast in response to TM-induced ER stress were determined (B). Data represent means and standard errors calculated from 3 replicates (each with about 10 protoplasts for each genotype).
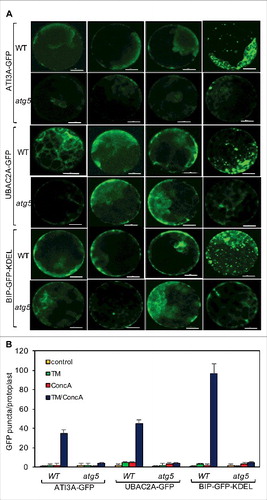
Autophagy-dependent delivery of ATI3 and UBAC2 to the vacuole
If ATI3 and UBAC2 proteins mediate a selective autophagy pathway that targets specific cargo for degradation, the ATI3 and UBAC2 proteins would likely be delivered as part of autophagosomes to the vacuole for degradation. To test this, we examined the delivery and accumulation of punctate autophagosome-like structures labeled by the ATI3A-GFP or UBAC2A-GFP maker in the vacuole of leaf protoplasts under ER stress. In untreated protoplasts or protoplasts treated with TM or concA alone, we observed largely cytoplasmic ER patterns of the ATI3A-GFP or UBAC2A-GFP signals (). In the presence of both TM and concA, we observed substantial increases in the ATI3- and UBAC2A-GFP punctate structures in the leaf protoplasts (). The average numbers of the ATI3- and UBAC2A-GFP punctate structures per leaf protoplast were approximately 40–50% of those of the punctate structures from the BIP-GFP-KDEL marker in the presence of both TM and concA (B). In the autophagy-deficient atg5 leaf protoplasts, we observed largely cytoplasmic patterns with few punctate structures for all 3 GFP makers even in the presence of both TM and concA (). Thus, ATI3 and UBAC2 proteins are delivered to the vacuole under ER stress in an autophagy-dependent manner.
Figure 15. Delivery of ATI3A and UBAC2A into the vacuole under ER stress. Leaf protoplasts obtained from 4-wk-old Col-0 and atg5 plants were transformed with a ATI3A-GFP or UBAC2A-GFP construct. The final concentrations of TM (5 μg/mL) and concA used in the experiments were 5 μg/mL and 1 μM, respectively. DMSO was used as a solvent control for all treatments. Protoplasts were incubated at room temperature in darkness for 12 h, with 40 rpm orbital shaking. Confocal microscopy was used to visualize the GFP fluorescence (A). Bar: 10 μm. The numbers of ATI3A-GFP and UBAC2A-GFP puncta per leaf protoplast in response to TM-induced ER stress were determined (B). Data represent means and standard errors calculated from 3 replicates (each with about 10 protoplasts for each genotype).
Discussion
Selective autophagy has been increasingly recognized as a critical process with a role in intracellular homeostasis by degrading specific cargo materials such as aggregated proteins, damaged or excess organelles and invading pathogens. In plants, several ATG8-interacting proteins including AT2G47770/TSPO, AT4G24690/NBR1, AT2G45980/ATI1, AT4G00355/ATI2 and AT4G38630/RPN10 have been identified and found to mediate selective autophagy of specific cellular components during plant responses to different environmental stresses. In this study, we identified a new ATG8-interacting protein, ATI3A, from Arabidopsis from yeast 2-hybrid screens and confirmed by BiFC the interaction of ATI3A with ATG8 through its C-terminal WxxL LIR motif ( and ). ATI3A has 2 homologs (ATI3B and ATI3C) that also contain a C-terminal WxxL LIR motif and interact with ATG8 ( and ). Further characterization of the ATG8-interacting ATI3 proteins revealed novel characteristics that are not found in previously identified ATG8-interacting proteins from plants. First, an extensive database search revealed that homologs of ATI3 with the conserved C-terminal WxxL LIR motif are present only in dicot plants but not in monocots or other organisms. The absence of the ATI3 homologs in monocots is intriguing and could be because these genes either have been lost or never developed in monocots. If lost in monocots, ATI3 homologs might still be present in ancestral species including green alga, mosses, ferns, gymnosperms or basal angiosperms. However, database searches of a range of sequenced ancestral species including Chlamydomonas reinhardtii (green algae), Physcomitrella patens (moss), Selaginella moellendorffii (fern), as well as species of gymnosperms and basal angiosperms failed to identify close ATI3 homologs with the conserved C-terminal WxxL LIR motif. Thus, it appears that ATG8-interacting ATI3 proteins may have been developed only in dicots, although further analysis with additional plant species is necessary to conclusively establish their dicot-specific nature.
Second, Arabidopsis ATI3 genes have novel biological functions not observed with other reported ATG8-interacting proteins from plants. Previously characterized plant selective autophagy receptors including TSPO, NBR1, ATI1, ATI2 and RPN10 play a critical role in plant tolerance to abiotic stresses including heat, drought and salinity. Autophagy also plays a critical role in plant immune responses to microbial pathogens based on the compromised disease resistance of mutants for the genes required for the core processes of autophagy [Citation26,Citation27]. However, no such compromised disease resistance has been reported from mutants of previously reported plant genes for the selective autophagy receptors. Arabidopsis ATI3 proteins interact strongly with ATG8 and increasingly form punctate structures with ATG8 at high temperature (; Figure S8), most likely representing stress-induced autophagosomes. Importantly, ati3a was compromised not only in heat tolerance but also in resistance to the necrotrophic fungal pathogen B. cinerea ( and ). The critical role of ATI3 proteins in plant heat tolerance and disease resistance is dependent on interaction with ATG8 ( and ). Therefore, ATI3 proteins could potentially mediate a novel selective autophagy pathway important for plant responses to both biotic and abiotic stresses. The role of the ATG8-interacting ATI3A in plant resistance to B. cinerea makes it particularly valuable for the analysis of the role and action of autophagy in plant immune responses.
Apart from the WxxL LIR motif at the C terminus required for interaction with ATG8, ATI3 proteins have no other known functional domain. Both Y2H and BiFC assays confirmed that ATI3 interacted with ER-localized UBAC2 (), which belong to a conserved family of proteins implicated in ERAD in human [Citation40]. Like ati3 mutants, ubac2 mutants had compromised phenotypes in both heat tolerance and in disease resistance (, , and ). Conversely, even though the ERAD-defective hrd3a mutant was also severely compromised in heat tolerance as expected, it was largely normal in response to B. cinerea (Figure S7). Thus, the phenotypes of the ubac2 mutants were more similar to those of the ati3 mutants than to those of hrd3a mutant plants. These findings raised the possibility that ATI3 proteins mediate a selective autophagy that targets ER proteins through UBAC2 acting as a receptor for recognition of ubiquitinated ER cargo and subsequent degradation in the vacuole. Consistent with this possibility, both UBAC2A-GFP and ATI3A-GFP were delivered to the vacuole in punctate forms under ER stress in an autophagy-dependent manner (). This is also consistent with the finding that the sensitivity of both ati3 and ubac2 mutants to the ER stress agent TM was significantly increased when compared with that of WT (). Furthermore, overexpression of UBAC2 in plant cells induced formation of ATG8- and ATI3-labeled punctate structures likely representing increased formation of phagophores or autophagosomes (). Thus, UBAC2 may function both as receptors for recognition of ER cargo but also as inducers of the ATI3-mediated selective autophagy pathway. Intriguingly, the sensitivity of the ati3 and ubac2 mutants to the ER stress agent TM, although significantly higher than that of WT, were substantially lower than that of the autophagy-deficient atg5-1 mutant (). In addition, ati3 and ubac2 mutants were largely competent in autophagy-mediated ER degradation under TM-induced ER stress when using a lumenal ER marker for detection (). These seemingly inconsistent or even conflicting results suggest a complex relationship between the role of ATI3 and UBAC2 in plant stress responses and their involvement in ERAD and autophagy-dependent ER degradation, which merits further consideration.
The significant increase in the sensitivity of both ati3 and ubac2 mutants to TM and the association of ATI3 proteins with ATG8 through the C-terminal WxxL LIR motif support the proposed role of the interacting ATI3 and UBAC2 proteins in mitigating ER stress through a selective autophagy pathway. The smaller increase in the sensitivity to ER stress in the ati3 and ubac2 mutants than in the autophagy-deficient atg5 mutant could be explained by the existence of additional autophagy pathways that target ER components and protect cells against ER stress in plants. Proteins destined for the plasma membrane, the extracellular space and other secretory compartments all undergo folding and maturation within the ER. Studies from different organisms including plants have shown that there are multiple ER protein quality control and ERAD pathways for different ER proteins. Studies in both yeast and mammalian cells have identified multiple ubiquitin E3 ligases that mediate distinct ERAD pathways through targeting different types of ER proteins [Citation47,Citation48]. In yeast, 2 ubiquitin E3 ligases (Hrd1 and Ssm4/Doa10) play functional and organizational roles in degradation of ERAD substrates. In mammalian cells, 3 ubiquitin E3 ligases (AMFR/gp78, HRD1 and MARCH6/TEB4 share similar domain and topological organization with yeast Hrd1 (ortholog of AMFR and HRD1) and Ssm4 (ortholog of MARCH6). Yeast proteins with misfolded ER-lumenal domains are degraded by the ERAD-L pathway mediated by the Hrd1-Hrd3 E3 ligase, which forms a membrane core complex with Der1 through the linker protein Usa1 [Citation49,Citation50]. Degradation of proteins with misfolded intramembrane domains is dependent on an ERAD-M pathway that is independent of Usa1 and Der1 [Citation49,Citation50]. Membrane proteins with misfolded cytosolic domains require the ERAD-C pathway mediated by the Ssm4 ubiquitin ligase [Citation49,Citation50]. In Arabidopsis, mutants for specific ER protein quality control components are defective in the biogenesis of some but not all pattern recognition receptors [Citation51–54]. If ATI3- and UBAC2-mediated autophagy targets a specific group or type of ER proteins for degradation by autophagy, its role in mitigating ER stress would be more limited than the entire autophagy process, and consequently the phenotype of the ati3 and ubac2 mutants in sensitivity to a general ER stress inducer would not be as strong as an autophagy-deficient mutant such as atg5-1. In this case, detection of ATI3- and UBAC2-depednent ER degradation would also rely on the use of correct ER markers that are targeted by the ATI3-UBAC2-mediated autophagy pathway. Therefore, the failure to detect a phenotype in TM-induced ER degradation in the ati3 and ubac2 mutants (, Figure S6), despite their increased sensitivity to the ER stress agent, could be due to the use of a BIP-GFP-KDEL lumenal ER marker, which may not necessarily be a target of the ATI3- and UBAC2-mediated autophagy pathway.
It has been previously shown that accumulation of unfolded ER proteins is a trigger for induction of autophagy in an AT5G24360/IRE1B-dependent manner under ER stress [Citation45]. The chemical chaperones sodium 4-phenylbutyrate and tauroursodeoxycholic acid reduce tunicamycin- or dithiothreitol-induced autophagy. 4-Phenylbutyrate also inhibits heat-induced autophagy [Citation45]. Similarly, overexpression of an ER heat shock protein, AT3G12580/HSP70, reduces autophagy [Citation45]. Conversely, expression in Arabidopsis of the misfolded protein mimics zeolin or a mutated form of AT3G10410/carboxypeptidase Y induces autophagy [Citation45]. How unfolded ER proteins trigger autophagy in plant cells is unclear. Interestingly, we found that overexpression of UBAC2A increased punctate structures of ATG8-ATI3A BiFC fluorescent signals, likely reflecting increased formation of autophagosomes (; Figure S2). The UBAC2 proteins are ER membrane proteins but contain an extended cytosolic C-terminal region with a conserved UBA domain at the C terminus (). The cytosolic UBA domain of UBAC2 could function in the recognition of ubiquitinated unfolded ER proteins that are retrotranslocated from the ER lumen or from membrane proteins with misfolded cytosolic domains. In addition, we observed that UBAC2A-GFP was delivered to the vacuole under conditions of ER stress in an autophagy-dependent manner, suggesting that UBAC2 proteins might themselves be potential cargo proteins of selective autophagy (). Therefore, increased formation of ATG8-labeled autophagosome-like structures in UBAC2A-overexpressing cells could be due to increased levels of misfolded UBAC2 proteins or UBAC2 proteins complexed with other ubiquitinated ER proteins that act as autophagy inducers.
In summary, we have identified a new group of ATG8-interacting proteins that are unique to dicots. We have further discovered that ATI3 interacts with UBAC2, an evolutionarily conserved protein implicated in ERAD. Genetic analysis revealed that both the ATI3 and UBAC2 genes play important roles in plant heat tolerance and resistance to Botrytis. ATI3 and UBAC2 proteins could mediate a new selective autophagy pathway with an important role in plant stress responses in part through selective autophagy of specific ER components. Significantly increased sensitivity of the ati3 and ubac2 mutant to the ER stress agent TM is consistent with the proposed role of ATI3 and UBAC2 proteins in plant stress responses and stress-induced autophagy. However, future experiments are needed to identify the specific targets of the ATI3- and UBAC2-mediated autophagy pathway in the ER compartment, and to understand the regulation of the autophagy pathway during plant stress responses.
Materials and methods
Arabidopsis genotypes and growth conditions
The Arabidopsis mutants and WT plants are all in the Co-0 background. The atg5-1 single and ire1a-2 irelb-1 double mutants have been previously described [Citation26,Citation46]. Homozygous ati3a-1 (Salk_202281C), ati3b-1 (Salk_074382), ati3c-1 (Salk_056098C), ubac2a-1 (WiscDsLoxHs094_04C), ubac2a-2 (WiscDsLox495E11), ubac2b-1 (Salk_021788C) and hrd3a (Salk_109430) mutants were identified by PCR using gene-specific primers flanking the T-DNA/transposon insertions (Table S1). The transcripts of Arabidopsis ATI3 and UBAC2 genes in the corresponding mutants were determined by qRT-PCR using gene-specific primers (Table S1). The Arabidopsis ACT2 gene was used as internal control as previously described [Citation55]. Arabidopsis plants were grown in growth chambers at 22°C, 120 μE m−2 light on a photoperiod of 12-h light and 12-h dark.
Yeast 2-hybrid screen and assays
Arabidopsis ATI3A was identified as an ATG8-interacting protein from the Gal4-based yeast 2-hybrid screens as previously described [Citation19]. For assays of ATI3B- and ATI3C-ATG8 interaction in yeast cells, full-length ATI3B and ATI3C coding sequences were PCR-amplified using gene-specific primers (ATI3B: 5’-agcgaattcatggagggggatggttttaa-3’/5’-gctctcgagttacaaaagctcccattcaga-3’; ATI3C: 5’-agcgaattcatggagggggatggttttaa-3’/5’-gctctcgagttacaaaagctcccattcagaatcttctg-3’) and cloned into a pAD-Gal4 vector (Agilent Technologies, 978000). The mutant gene encoding ATI3AW260A was generated by overlapping PCR and cloned into pAD-Gal4. Various combinations of bait and prey constructs were cotransformed into yeast cells and interactions were analyzed by plating onto selection medium lacking Trp, Leu and His or by β-galactosidase activity assays using o-nitrophenyl-β-D-galactopyranose (Sigma-Aldrich, N1127) as substrate.
ATI3A-interacting proteins were identified using the split-ubiquitin complementation screens from a normalized Arabidopsis cDNA library purchased from Dualsystems Biotech. Full-length ATI3A coding sequence was amplified by PCR using gene-specific primers (5’-cgcggccattacggccatgaatagaaattccaaaggaaga-3’/ 5’-catggccgaggcggccaacgcaatctccgcctcagaatcttctgatacaa-3’) and cloned into the bait vector. The bait plasmid and the cDNA library were used to transform yeast cells. Yeast transformants were plated onto selection medium and confirmed by β-galactosidase activity assays.
BiFC assay
The BiFC vectors pFGC-N-YFP and pFGC-C-YFP have been previously described [Citation56]. The full-length ATI3A and UBAC2A sequences were PCR-amplified using gene-specific primers (ATI3A: 5’-agcgagctcatgaatagaaattccaaaggaagag-3’ and 5’- agctctagacaaaatctcccactcagaatctt-3’; UBAC2A: 5’-agcgagctcatgaacggcggtccctcc-3’ and 5’-agctctagagtgggactgtgcttcgaga-3’) and cloned into pFGC-C-YFP or pFGC-N-YFP, as appropriate. The plasmids were introduced into A. tumefaciens (strain GV3101) and infiltration into N. benthamiana was performed as described previously [Citation57]. BiFC fluorescent signals in infected tissues were analyzed at ∼24 h after infiltration by a Zeiss LSM710 confocal microscope with appropriate filter sets (excitation 514 nm, emission 525–555 nm). The images were superimposed with Zeiss LSM710 software.
Analysis of tolerance to heat, salt and the ER stress agent TM
Testing of heat and salt tolerance of Arabidopsis seedlings was performed as previously described [Citation19,Citation58]. For assays of sensitivity to TM, WT and mutant seeds were sterilized and germinated on solid half-strength MS medium supplemented with or without 0.1 mg/L TM (Cayman Chemical, 11445). Survival rates were determined after 2 wk.
Botrytis infection
Culture and inoculation of B. cinerea were performed as previously described [Citation26,Citation59]. Biomass of the fungal pathogen was quantified by qRT-PCR of total RNA isolated from inoculated plants for the B. cinerea actA gene transcript levels as described previously [Citation59].
Subcellular localization
Full-length UBAC2 and ATI3 genes were fused to the GFP and mCherry genes, respectively, behind the CaMV 35S promoter in a pFGC5941-based plant transformation vector [Citation60]. The UBAC2-GFP fusion gene was co-expressed with a BIP-mCherry-KDEL ER marker gene [Citation60]. or ATG8A-mCherry autophagosome marker [Citation19,Citation21]. in N. benthamiana. The ATI3A-mCherry fusion gene was co-expressed with an ATG8A-GFP marker gene in N. benthamiana. Imaging of co-expressed GFP and mCherry signals was performed with standard confocal laser microscopy with appropriate filter sets: GFP (excitation 488 nm, emission 500–550 nm) and mCherry (excitation 561 nm, emission 570–620 nm).
Preparation and transfection of leaf protoplasts
Arabidopsis leaf protoplasts were prepared using the Tape-Arabidopsis-Sandwich procedure [Citation61]. Approximately 5 × 104 protoplasts were used in transfection using 20 μg BIP-GFP-KDEL, ATI3A-GFP or UBAC2A-GFP plasmid DNA as previously described [Citation62]. TM (5 μg/mL) and concA (Cayman Chemical, 11050) were added to transformed protoplasts at the final concentrations of 5 μg/mL and 1 μM, respectively. DMSO was used as a solvent control for all treatments. Protoplasts were incubated at room temperature in darkness for 12 h, with 40 rpm orbital shaking. Confocal microscopy was performed with a Leica confocal microscope using a × 63 Leica oil immersion objective.
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
KAUP_A_1422856_suppl.zip
Download Zip (12.9 MB)Acknowledgements
We would like to thank the Arabidopsis Resource Center at the Ohio State University for providing the Arabidopsis mutants. We also thank Dr. Lan-Ying Lee (Purdue University, USA) for advices in Arabidopsis leaf protoplast isolation, Dr. Inhwan Hwang (Pohang University of Science and Technology, Korea) for the florescent ER marker constructs and Dr. Nozomu Koizumi (Nara Institute of Science and Technology, Japan) for the ire1a-2/irelb-1 mutant.
Additional information
Funding
References
- Mizushima N. Autophagy. FEBS Lett. 2010;584:1279.
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi:10.1101/gad.1599207. PMID:18006683
- Klionsky DJ. Autophagy. Curr Biol. 2005;15:R282–R283.
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi:10.1038/nature09782. PMID:21248839
- Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009;149:220–234. doi:10.1104/pp.108.126714. PMID:18790996
- Kwon SI, Park OK. Autophagy in plants. J Plant Biol. 2008;51:313–320. doi:10.1007/BF03036132.
- Liu Y, Schiff M, Czymmek K, et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi:10.1016/j.cell.2005.03.007. PMID:15907470
- Shin JH, Yoshimoto K, Ohsumi Y, et al. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol Cells. 2009;27:67–74. doi:10.1007/s10059-009-0006-2. PMID:19214435
- Su W, Ma H, Liu C, et al. Identification and characterization of two rice autophagy associated genes, OsAtg8 and OsAtg4. Mol Biol Rep. 2006;33:273–278. doi:10.1007/s11033-006-9011-0. PMID:17082902
- Zhuang X, Chung KP, Cui Y, et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci USA. 2017;114:E426–E435. doi:10.1073/pnas.1616299114. PMID:28053229
- Thompson AR, Doelling JH, Suttangkakul A, et al. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi:10.1104/pp.105.060673. PMID:16040659
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42:535–546. doi:10.1111/j.1365-313X.2005.02397.x. PMID:15860012
- Doelling JH, Walker JM, Friedman EM, et al. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277:33105–33114. doi:10.1074/jbc.M204630200. PMID:12070171
- Hanaoka H, Noda T, Shirano Y, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi:10.1104/pp.011024. PMID:12114572
- Ishida H, Yoshimoto K, Izumi M, et al. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 2008;148:142–155. doi:10.1104/pp.108.122770. PMID:18614709
- Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. doi:10.4161/auto.5.7.9290. PMID:19587533
- Xiong Y, Contento AL, Bassham DC. Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy. 2007;3:257–258. doi:10.4161/auto.3847. PMID:17312382
- Xiong Y, Contento AL, Nguyen PQ, et al. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–299. doi:10.1104/pp.106.092106. PMID:17098847
- Zhou J, Wang J, Cheng Y, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9:e1003196. doi:10.1371/journal.pgen.1003196. PMID:23341779
- Zhou J, Wang J, Yu JQ, et al. Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci. 2014;5:174. doi:10.3389/fpls.2014.00174. PMID:24817875
- Zhou J, Zhang Y, Qi J, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in Plant stress responses. PLoS Genet. 2014;10:e1004116. doi:10.1371/journal.pgen.1004116. PMID:24497840
- Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–27. doi:10.4161/auto.5056. PMID:17932459
- Hofius D, Schultz-Larsen T, Joensen J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi:10.1016/j.cell.2009.02.036. PMID:19450522
- Yoshimoto K, Jikumaru Y, Kamiya Y, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi:10.1105/tpc.109.068635. PMID:19773385
- Zhou J, Yu JQ, Chen Z. The perplexing role of autophagy in plant innate immune responses. Mol Plant Pathol. 2014. doi:10.1111/mpp.12118.
- Lai Z, Wang F, Zheng Z, et al. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. The Plant J Cell Mol Biol. 2011;66:953–968. doi:10.1111/j.1365-313X.2011.04553.x. PMID:21395886
- Lenz HD, Haller E, Melzer E, et al. Autophagy controls plant basal immunity in a pathogenic lifestyle-dependent manner. Autophagy. 2011;7:773–774. doi:10.4161/auto.7.7.15535. PMID:21460628
- Derrien B, Baumberger N, Schepetilnikov M, et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Nat Acad Sci USA. 2012;109:15942–15946. doi:10.1073/pnas.1209487109. PMID:23019378
- Nakahara KS, Masuta C, Yamada S, et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Nat Acad Sci USA. 2012;109:10113–10118. doi:10.1073/pnas.1201628109. PMID:22665793
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi:10.4161/auto.7.3.14487. PMID:21189453
- Shaid S, Brandts CH, Serve H, et al. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi:10.1038/cdd.2012.72. PMID:22722335
- Vanhee C, Zapotoczny G, Masquelier D, et al. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23:785–805. doi:10.1105/tpc.110.081570. PMID:21317376
- Hachez C, Veljanovski V, Reinhardt H, et al. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell. 2014;26:4974–4990. doi:10.1105/tpc.114.134080. PMID:25538184
- Svenning S, Lamark T, Krause K, et al. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi:10.4161/auto.7.9.16389. PMID:21606687
- Honig A, Avin-Wittenberg T, Ufaz S, et al. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of arabidopsis plants to carbon starvation. Plant Cell. 2012;24:288–303. doi:10.1105/tpc.111.093112. PMID:22253227
- Michaeli S, Honig A, Levanony H, et al. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014;26:4084–4101. doi:10.1105/tpc.114.129999. PMID:25281689
- Marshall RS, Li F, Gemperline DC, et al. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/Ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015;58:1053–1066. doi:10.1016/j.molcel.2015.04.023. PMID:26004230
- Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42:598–608. doi:10.1111/j.1365-313X.2005.02396.x. PMID:15860017
- Liu Y, Bassham DC. Degradation of the endoplasmic reticulum by autophagy in plants. Autophagy. 2013;9:622–623. doi:10.4161/auto.23559. PMID:23360963
- Christianson JC, Olzmann JA, Shaler TA, et al. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi:10.1038/ncb2383.
- Fu XL, Gao DS. Endoplasmic reticulum proteins quality control and the unfolded protein response: the regulative mechanism of organisms against stress injuries. Biofactors. 2014;40:569–585. doi:10.1002/biof.1194. PMID:25530003
- Pluquet O, Pourtier A, Abbadie C. The unfolded protein response and cellular senescence. A review in the theme: cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. Am J Physiol Cell Physiol. 2015;308:C415–C425.
- Liu Y, Burgos JS, Deng Y, et al. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012;24:4635–4651. doi:10.1105/tpc.112.101535. PMID:23175745
- Pu Y, Bassham DC. Links between ER stress and autophagy in plants. Plant Signal Behav. 2013;8:e24297. doi:10.4161/psb.24297. PMID:23603973
- Yang X, Srivastava R, Howell SH, et al. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 2016;85:83–95. doi:10.1111/tpj.13091. PMID:26616142
- Nagashima Y, Mishiba K, et al. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep. 2011;1:29. doi:10.1038/srep00029. PMID:22355548
- Lemus L, Goder V. Regulation of Endoplasmic Reticulum-Associated Protein Degradation (ERAD) by Ubiquitin. Cells. 2014;3:824–847. doi:10.3390/cells3030824. PMID:25100021
- Thibault G, Ng DT. The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb Perspect Biol. 2012;4:a013193. doi:10.1101/cshperspect.a013193. PMID:23209158
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi:10.1016/j.cell.2006.05.043. PMID:16873066
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi:10.1038/nrm2546. PMID:19002207
- Li J, Zhao-Hui C, Batoux M, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA. 2009;106:15973–15978. doi:10.1073/pnas.0905532106. PMID:19717464
- Liu Y, Li J. Endoplasmic reticulum-mediated protein quality control in Arabidopsis. Front Plant Sci. 2014;5:162. doi:10.3389/fpls.2014.00162. PMID:24817869
- Nekrasov V, Li J, Batoux M, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi:10.1038/emboj.2009.262. PMID:19763086
- Saijo Y, Tintor N, Lu X, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi:10.1038/emboj.2009.263. PMID:19763087
- Huang J, Gu M, Lai Z, et al. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi:10.1104/pp.110.157370. PMID:20566705
- Kim KC, Lai Z, Fan B, et al. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi:10.1105/tpc.107.055566. PMID:18776063
- Cui X, Fan B, Scholz J, et al. Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell. 2007;19:1388–1402. doi:10.1105/tpc.107.051375. PMID:17468259
- Wang F, Yang Y, Wang Z, et al. A critical role of lyst-interacting Protein5, a positive regulator of multivesicular body biogenesis, in plant responses to heat and salt stresses. Plant Physiol. 2015;169:497–511. doi:10.1104/pp.15.00518. PMID:26229051
- Lai Z, Li Y, Wang F, et al. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell. 2011;23:3824–3841. doi:10.1105/tpc.111.090571. PMID:21990940
- Chi Y, Yang Y, Li G, et al. Identification and characterization of a novel group of legume-specific, Golgi apparatus-localized WRKY and Exo70 proteins from soybean. J Exp Bot. 2015;66:3055–3070. doi:10.1093/jxb/erv104. PMID:25805717
- Wu FH, Shen SC, Lee LY, et al. Tape-Arabidopsis Sandwich – a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009;5:16. doi:10.1186/1746-4811-5-16. PMID:19930690
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi:10.1038/nprot.2007.199. PMID:17585298
