ABSTRACT
Macroautophagy/autophagy plays an essential role in hematopoietic stem cell (HSC) differentiation. However, the role of autophagy during monocytic and granulocytic differentiation remains poorly understood. Hence, we first represented global transcriptomic analysis for temporal expression of autophagy genes during monocytic and granulocytic differentiation by combining RNA-Seq data with monocytic and granulocytic induction in CD34+ hematopoietic stem and progenitor cells. According to a self-organizing map (SOM) algorithm, our results show temporal expression patterns of autophagy genes during monocytic and granulocytic differentiation. More importantly, 22 autophagy genes present significantly divergent roles during monocytic and granulocytic differentiation, indicating these autophagy genes could be important factors involved in the differentiation of myeloid progenitors into monocytes and granulocytes.
Introduction
Cell differentiation across myeloid lineage provides an excellent model for understanding the intricate gene regulatory mechanisms that confer identity and function of immune cells. During myelopoiesis, hematopoietic stem and progenitor cells (HSPCs) constantly differentiate into monocytes and granulocytes via a distinct differentiation program that depends on the interplay of a variety of factors, although many factors are involved in the differentiation of granulocyte-monocyte progenitors into granulocytes and monocytes, only SPI1/PU.1 and CEBPE/C/EBPϵ ware confirmed to favor the commitment to one lineage over another.
Autophagy is crucial for maintaining cellular homeostasis and is involved in the regulation of various types of stem cells, in particular HSPCs. Recent studies have indicated that autophagy has a more general role in shaping the identity and function of myeloid cells. Monocytes and granulocytes are the most common myeloid cell populations in the blood. However, there was no systematic transcriptomic analysis on the role of autophagy during monocytic and granulocytic differentiation. Hence, we have systematically examined the temporal expression patterns of autophagy genes during monocytic and granulocytic differentiation by combining RNA-Seq data with myeloid induction in CD34+ HSPCs, providing a critical insight into the mechanistic understanding of the physiological connections between autophagy and monocytic/granulocytic differentiation.
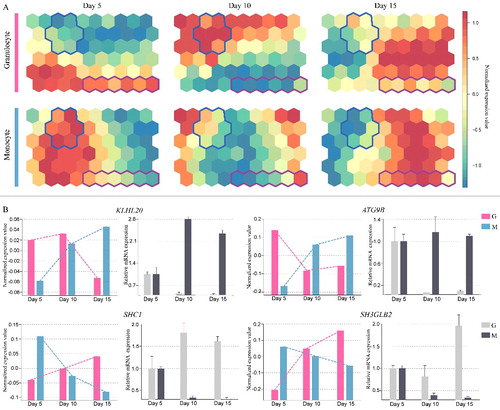
Temporal expression pattern of autophagy genes during monocytic and granulocytic differentiation
According to a global RNA-Seq analysis of RNA isolated from human cold blood CD34+ HSPCs at 5, 10 and 15 d along the monocytic and granulocytic differentiation process, we examined temporal gene expression of 747 human autophagy genes retrieved from Autophagy Database, Autophagy Regulatory Network (ARN) and Human Autophagy Database (HADb). By using a SOM algorithm, our results first showed a global temporal expression of autophagy genes during human monocytic and granulocytic differentiation.
Next, we focused on autophagy genes that presented divergent roles during monocytic and granulocytic differentiation. Interestingly, according to distance correlation statistics, our analysis led to the identification of 22 autophagy genes with significant divergent changes during monocytic and granulocytic differentiation as shown by a blue or pink solid line in SOM components (A). Among them, the increased expression of 13 genes (AP1G1, ARNT, CAMK2B, CAMKK1, CDKL1, EXOC7, GABARAPL2, MAP4K3, MYLK3, PIK3CA, SHC1, SH3GLB2, TNS2) coordinates granulocytic differentiation; however, their levels are decreased in monocytic differentiation. By contrast, the other 9 genes (ACTA2, ATG9B, KLHL20, MAP4K5, NBEA, PIK3R4, SIK1, SLC7A6, TRAPPC2) show gradual elevated expression during monocytic differentiation, but reduced expression in granulocytic differentiation. Furthermore, as shown in B, qRT-PCR analysis was performed to validate the expression level of ATG9B, KLHL20, SHC1 and SH3GLB2 from RNA-Seq results, and experimental results confirm their expression pattern. Namely, these 4 genes exhibit a consistent change in temporal expression in agreement with the RNA-seq data during monocytic and granulocytic differentiation.
Figure 1. Temporal expression pattern of autophagy genes during monocytic and granulocytic differentiation. (A) Temporal expression of autophagy genes represented by a self-organizing map algorithm; divergent expression patterns of autophagy genes are bounded by a blue or pink solid line in SOM components. CD34+ HSPCs were differentiated into either the monocyte or granulocyte lineage, and cells were collected at 5, 10 and 15 d for RNA-Seq analysis. (B) The application of qRT-PCR analysis following RNA-Seq data analysis reveals a divergent expression pattern of ATG9B, KLHL20, SHC1 and SH3GLB2 at 5, 10 and 15 d during CD34+ HSPCs monocytic (M) and granulocytic (G) differentiation.
Conclusions
Autophagy is required for keeping the balance between quiescence, self-renewal and differentiation in HSPCs. However, the underlying mechanism and regulation of autophagy during monocytic and granulocytic differentiation is largely unknown. By using bioinformatics analysis tools, our data revealed that 22 autophagy genes present significantly divergent roles during monocytic and granulocytic differentiation, suggesting these genes could be important regulators involved in the differentiation control of granulocyte-monocyte progenitors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We are grateful to Dr. Daniel J. Klionsky for help in editing the manuscript.
