ABSTRACT
Membrane protein recycling is a fundamental process from yeast to humans. The lysosome (or vacuole in yeast) receives membrane proteins from the secretory, endocytic, and macroautophagy/autophagy pathways. Although some of these membrane proteins appear to be recycled, the molecular mechanisms underlying this retrograde trafficking are poorly understood. Our recent study revealed that the transmembrane autophagy protein Atg27 is recycled from the vacuole membrane using a 2-step recycling process. First, the Snx4 complex recycles Atg27 from the vacuole to the endosome. Then, the retromer complex mediates endosome-to-Golgi retrograde transport. Thus, 2 distinct protein complexes facilitate the sequential retrograde trafficking for Atg27. As far as we know, Atg27 is the first physiological substrate for the vacuole-to-endosome retrograde trafficking pathway.
Summary
Membrane protein recycling is a fundamental process from yeast to humans. The lysosome (or vacuole in yeast) receives membrane proteins from the secretory, endocytic, and macroautophagy/autophagy pathways. Although some of these membrane proteins appear to be recycled, the molecular mechanisms underlying this retrograde trafficking are poorly understood. Our recent study revealed that the transmembrane autophagy protein Atg27 is recycled from the vacuole membrane using a 2-step recycling process. First, the Snx4 complex recycles Atg27 from the vacuole to the endosome. Then, the retromer complex mediates endosome-to-Golgi retrograde transport. Thus, 2 distinct protein complexes facilitate the sequential retrograde trafficking for Atg27. As far as we know, Atg27 is the first physiological substrate for the vacuole-to-endosome retrograde trafficking pathway.
Eukaryotic cells have a clever system to recycle membrane proteins. The best known example is recycling of the transmembrane protein receptor for the precursor form of Prc1/carboxypeptidase Y/CPY, Pep1/Vps10, studied in budding yeast. Pep1/Vps10 sorts the p2 precursor form of Prc1 (p2-Prc1) into vesicles at the Golgi. After p2-Prc1-containing vesicles are transported to the endosome, p2-Prc1 is released from the receptor.
The endosome matures and then fuses with the vacuole, delivering soluble p2-Prc1 to the vacuole lumen. Unlike p2-Prc1, Pep1/Vps10 is not delivered to the vacuole. Pep1/Vps10 is recycled from the endosome back to the Golgi by the retromer complex, before the endosome fuses with the vacuole. The retromer is a highly conserved protein complex responsible for membrane protein retrieval. The retromer is composed of the Vps5, Vps17, Pep8/Vps26, Vps29, and Vps35 proteins in yeast. This complex deforms the endosomal membrane to form cargo-containing recycling tubules/vesicles. These tubules/vesicles give rise to recycling vesicles that fuse with the Golgi, delivering Pep1/Vps10 to the Golgi where it can be reused for another round of p2-Prc1 sorting. In mammalian cells, the retromer mediates retrograde traffic of M6PR (mannose-6-phosphate receptor, cation dependent), which is responsible for the Golgi-to-endosome anterograde transport of lysosomal enzymes. In retromer mutants, M6PR fails to recycle, causing the mislocalization of lysosomal enzymes. A mutation in VPS35 is associated with an autosomal dominant form of familial Parkinson disease.
The retromer complex mediates retrograde trafficking from the endosome to the Golgi. However, how membrane proteins are recycled from the lysosome membrane is not well understood. The lysosome (or vacuole in yeast and plants) is responsible for degradation of cellular proteins delivered by autophagy or the endocytic pathway, and also serves as a reservoir for nutrient storage (i.e,. amino acids, ions, sugars, and fatty acids). Because lysosomes fuse with AP-3 vesicles, autophagosomes, and endosomes, they constantly receive new membrane proteins. To maintain lysosome identity and function, this exogenous membrane and membrane proteins have to be degraded or recycled.
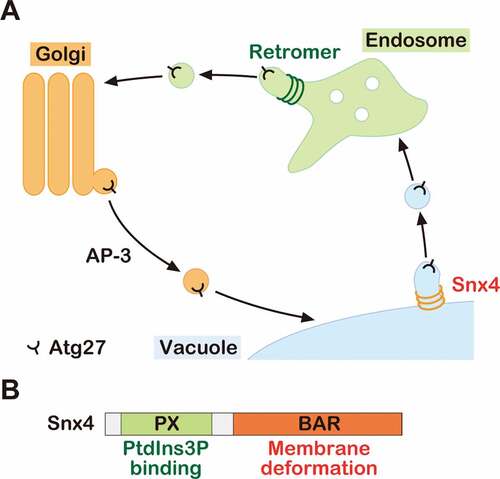
We recently revealed that the transmembrane protein Atg27 is delivered to the vacuole membrane via the AP-3 pathway and then recycled back to the Golgi by a 2-step recycling process (). First, Atg27 is delivered from the vacuole membrane to the endosome via the Snx4 complex, and then from the endosome to the Golgi by the retromer complex. Snx4 is an evolutionarily conserved protein from yeast to humans (also called SNX4 in humans), which comprises an N-terminal PX domain and a C-terminal BAR domain, required for PtdIns3P binding and membrane deformation, respectively (). During vacuole-to-endosome retrograde trafficking, Snx4 complexes assembly on the vacuole membrane in a PtdIns3P-dependent manner and recognize specific residues in the cytoplasmic tail of Atg27. The Snx4 BAR domains deforms the vacuole membrane to form recycling vesicles/tubules containing Atg27. We proposed that the Snx4 complex plays a critical role in membrane protein retrieval from the vacuole/lysosome membrane.
Figure 1. The model of Atg27 trafficking. (A) Schematic model of Atg27 trafficking. Atg27 is delivered to the vacuole membrane via the AP-3 pathway. After delivery, it is recycled from the vacuole membrane to the endosome by the Snx4 complex. Then, it is retrieved from the endosome by the retromer complex. (B) Schematic of Snx4 domains
Although the vacuole fuses with other membrane compartments (i.e., autophagosomes and endosomes, etc.), its size remains fairly constant. This fact suggests that there is a mechanism to maintain vacuole size. Exogenous membrane might be trafficked in a retrograde manner from the vacuole to the other organelles. Unfortunately, snx4Δ cells do not exhibit abnormal vacuole morphology, implying that the Snx4 complex is not essential for this process. An undefined molecular complex might facilitate this membrane recycling. Future studies are required to define these machineries. The other possibility is that excess membrane is degraded. Recently, we revealed that the ESCRTs directly invaginate the membrane not only at the endosome but also at the vacuole. This raises the possibility that the ESCRTs could play a role in degrading the excess membrane.
Recycling of membrane and key proteins (SNAREs, receptors, etc.) allows cells to maintain organelle identity. Several other sorting nexins might also function in this vacuole-to-endosome retrograde transport. It will be important to uncover the role for each sorting nexin to understand how cells maintain the composition and function of each organelle.
Acknowledgments
This work was supported by a Cornell University Research Grant to S.D.E. Sho Suzuki is supported by JSPS research fellowship (SPD) and JSPS Postdoctoral Fellowships for Research Abroad.
Disclosure statement
No potential conflict of interest was reported by the authors.
