ABSTRACT
Aged and damaged mitochondria can be selectively degraded by specific autophagic elimination, termed mitophagy. Defects in mitophagy have been increasingly linked to several diseases including neurodegenerative diseases, metabolic diseases and other aging-related diseases. However, the molecular mechanisms of mitophagy are not fully understood. Here, we identify PRPF8 (pre-mRNA processing factor 8), a core component of the spliceosome, as an essential mediator in hypoxia-induced mitophagy from an RNAi screen based on a fluorescent mitophagy reporter, mt-Keima. Knockdown of PRPF8 significantly impairs mitophagosome formation and subsequent mitochondrial clearance through the aberrant mRNA splicing of ULK1, which mediates macroautophagy/autophagy initiation. Importantly, autosomal dominant retinitis pigmentosa (adRP)-associated PRPF8 mutant R2310K is defective in regulating mitophagy. Moreover, knockdown of other adRP-associated splicing factors, including PRPF6, PRPF31 and SNRNP200, also lead to ULK1 mRNA mis-splicing and mitophagy defects. Thus, these findings demonstrate that PRPF8 is essential for mitophagy and suggest that dysregulation of spliceosome-mediated mitophagy may contribute to pathogenesis of retinitis pigmentosa.
Introduction
Selective autophagy mediates cellular quality control by delivering intracellular material to lysosomes for degradation. Damaged or unwanted mitochondria can be selectively removed by autophagy, termed mitophagy. The most studied mitophagy pathway involves PINK1 (PTEN induced putative kinase 1) and PRKN/PARK2 (parkin RBR E3 ubiquitin protein ligase). PINK1 specifically accumulates on damaged mitochondria, recruiting and phosphorylating PRKN, to activate the mitophagic progress. Mutations in PINK1 and PRKN are involved in neurodegenerative diseases such as Parkinson disease [Citation1,Citation2]. Given the critical roles of the PINK1-PRKN pathway in regulating mitophagy, several genome-wide RNAi screenings based on the cytosol-to-mitochondria translocation of PRKN have been performed to identify new regulators of mitophagy [Citation3–Citation8]. In addition to the PINK1-PRKN pathway, PRKN-independent mitophagy has been reported to be involved in various pathophysiological processes [Citation9–Citation12]. For example, hypoxia induces mitophagy in a PRKN-independent manner in multiple pathophysiological processes such as angiogenesis [Citation13], tumors [Citation14], neurodegenerative diseases [Citation15,Citation16] and metabolic disorders [Citation17]. However, the molecular mechanisms of hypoxia-induced mitophagy and its role in the pathophysiology of diseases are not well understood.
mt-Keima is a mitochondrial-targeted form of Keima [Citation18], which is a coral-derived, lysosomal degradation resistant and pH-sensitive fluorescent protein. We chose to depict the mt-Keima fluorescence signal from 561-nm laser excitation (in lysosomes) in red and the signal from 458-nm laser excitation (in neutral cytoplasm) in green. This property of mt-Keima allows rapid and sensitive determination as to whether the mitochondrion is in the cytoplasm or in the lysosome, and the ratio of Keima-derived fluorescence (561 nm:458 nm) can indicate mitophagic flux and qualitatively assess the mitophagy level [Citation18]. Thus, mt-Keima has been increasingly utilized to measure mitophagy in vitro and in vivo [Citation19–Citation23]. In this study, we performed an RNAi screening based on the mt-Keima reporter for novel regulators of hypoxia-induced mitophagy. We identified PRPF8 (pre-mRNA processing factor 8) as a novel regulator of mitophagy.
The spliceosome is a large multi-component RNA-protein machinery that catalyzes precursor messenger RNA (pre-mRNA) to form a functional mRNA in the nucleus via removing the non-coding introns and ligating the exons [Citation24]. Splicing of pre-mRNA is a central step in the post-transcriptional regulation of gene expression [Citation25], and misregulation of splicing is a common feature of many human diseases, such as retinitis pigmentosa, amyotrophic lateral sclerosis and spinal muscular atrophy [Citation26]. Accumulating evidence suggests that splicing events affect many cellular processes [Citation27]. It has been reported that several autophagy-related genes, such as BECN1/Beclin1 and GABARAPL2 (GABA type A receptor associated protein like 2), have alternative splicing variants, suggesting that the spliceosome could modulate autophagy [Citation28,Citation29]. However, mechanistic details underlying deregulated splicing in mitophagy, especially hypoxia-induced mitophagy are still limited. In this study, we demonstrate that the autosomal dominant retinitis pigmentosa-associated gene PRPF8, a core component of the spliceosome, is essential for hypoxic mitophagy through ensuring proper mRNA splicing of ULK1 (unc-51 like autophagy activating kinase 1), which mediates mitophagy initiation.
Results
RNAi screen identifies PRPF8 as a regulator of mitophagy
To identify novel regulators of mitophagy, we established a high-content screening system of mitophagic activity based on the mitochondrial localized fluorescent protein mt-Keima. We depicted the mt-Keima fluorescence signal from 458-nm laser excitation in green and the signal from 561-nm laser excitation in red. We first detected and confirmed the rapid, sensitive and pH-dependent fluorescent properties of mt-Keima by changing medium with different pH (Figure S1A). Then, we accessed mitophagy induced by hypoxia in HeLa cells stably expressing mt-Keima by confocal microscopy. We noted that hypoxia induced a marked rise in the red-to-green fluorescence ratio, indicating mitophagy increased cumulatively over time () and Figure S1B).
Figure 1. RNAi screen identifies PRPF8 as a regulator of mitophagy. (a) Representative confocal images of HeLa cells stably expressing mt-Keima subjected to hypoxia (1% O2) for the indicated times. (b) Graphical representation of the siRNA screen output. The ‘Z-score’ conveys the distribution of mitophagic activity per well across the entire library (see details in Methods). Data ordered from most negative to positive. Dashed lines are screen-specific cutoffs for active siRNA reagents. Blue dots represent positive control siRNAs. (c) mt-Keima imaging in HeLa cells transfected with the indicated siRNAs in a normoxic (21% O2) environment or after 24 h of hypoxia (1% O2). (d) Quantified mitophagy index in (c). Mitophagy index in hypoxia-treated control cells was normalized to ‘1ʹ. n = 3 independent experiments. (e) Western blot analysis of HeLa cells transfected with siRNAs after hypoxia treatment (1% O2, 24 h) with the indicated antibodies. (f) mt-Keima imaging in HeLa cells transfected with PRPF8 siRNA and GFP-PRPF8 siRNA-resistant plasmid after hypoxia treatment (1% O2, 24 h). GFP fluorescence is depicted in gray. Arrows indicate GFP-PRPF8-transfected cells. (g) Quantification of mitophagy index of GFP-positive cells in (f). Mitophagy index in hypoxia-treated control cells was normalized to ‘1ʹ. n = 3 independent experiments. Data are shown as the mean ± SD (One-way ANOVA, *p < 0.05, ***p < 0.001). Scale bars: 10 μm.
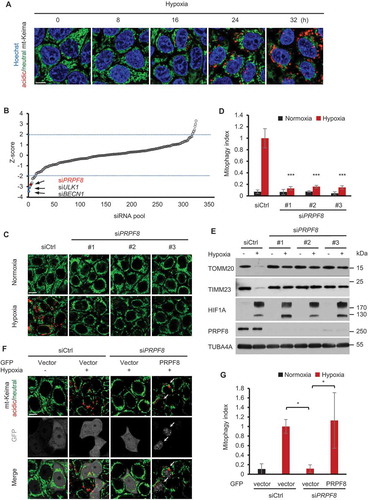
Based on this system, we performed a small interfering RNA (siRNA) screen using a library [Citation30] containing 320 siRNA pools to identify novel regulators of mitophagy under hypoxia conditions. To do so, HeLa cells stably expressing mt-Keima were transfected with siRNA duplexes in 96-well plates. After the siRNA transfection, the cells were subsequently treated with hypoxia for 24 h to induce mitophagy. Mitophagy activities were assessed with green and red mt-Keima signals (see details in Methods) by automated image analysis (Figure S1C). We set initial thresholds for putative hits at −2.0 or 2.0 Z-scores to identify positive or negative regulators of hypoxia-induced mitophagy, respectively. We observed a significant inhibitory effect on mitophagy following the transfection with positive-control siRNAs against ULK1 or BECN1 compared with non-targeting control siRNA (siCtrl) ()). In the first round of screening, 11 genes were hit below the thresholds at −2 Z-score, suggesting that they are potential positive regulators of mitophagy. We therefore knocked down these genes by siRNAs and assessed their roles in mitophagy. However, among these genes, only PRPF8 was further validated to be required for mitophagy.
PRPF8 (pre-mRNA processing factor 8) is a core component of the spliceosome and is involved in messenger RNA processing. To further evaluate the role of PRPF8 in mitophagy, we first transfected mt-Keima-expressing HeLa cells with 3 siRNAs that target different regions of the PRPF8 mRNA. When the cells were cultured under hypoxia for 24 h, we noted that hypoxia induced a marked rise in mitophagic activity indicated as ‘mitophagy index’ (see details in Methods). In contrast, the induction of mitophagy by hypoxia was significantly inhibited in these PRPF8-knockdown cells (). Consistent with this result, western blotting analysis showed that knockdown of PRPF8 dramatically inhibited mitochondrial protein (TOMM20 and TIMM23) degradation in response to hypoxia ()). In order to rule out the off-target effects of the siRNAs, we transfected plasmid expressing siRNA-resistant GFP-tagged PRPF8 in PRPF8 knockdown cells, and found that the reconstituted GFP-PRPF8 was able to restore the mitophagic activity in these cells (), and Figuer S2). Taken together, these results indicate that PRPF8 is indispensable for hypoxia-induced mitophagy.
PRPF8 is required for hypoxia-induced mitophagosome formation
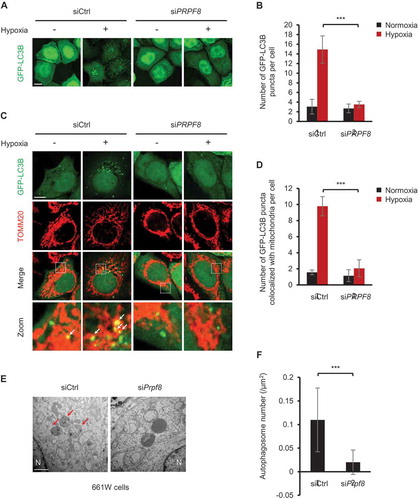
We next sought to explore the mechanism by which PRPF8 regulates mitophagy. MAP1LC3B/LC3B (microtubule associated protein 1 light chain 3 beta) puncta are widely used as autophagosome markers [Citation31]. We found that HeLa cells with PRPF8 knockdown showed less autophagosome formation compared with the control cells after hypoxia treatment as demonstrated by a decrease in the number of GFP-LC3B puncta per cell ()). We next investigated whether PRPF8 is required for mitophagosome formation. As expected, the colocalization of GFP-LC3B puncta and mitochondria was remarkably reduced in PRPF8-knockdown cells under hypoxia by confocal microscopy analysis (). We also used electron microscopy to examine the autophagosomes in murine 661W photoreceptor-derived cells at 36 h after hypoxia. The typical double-membrane autophagosomes (as indicated by arrows) were seen in control cells. Engulfment of entire organelles (possibly mitochondria) was occasionally observed. However, there were few autophagosomes and autolysosomes observed in cells with PRPF8 knockdown (). Collectively, these results suggested that PRPF8 regulates hypoxic mitophagy activity through inhibiting mitophagosome formation.
Figure 2. PRPF8 is required for hypoxia-induced mitophagosome formation. (a) Representative images of HeLa cells expressing GFP-LC3B transfected with the indicated siRNAs following normoxia or hypoxia (1% O2, 12 h) treatment. Scale bar: 10 μm. (b) Quantification of the number of GFP-LC3B puncta per cell in (A). n = 3 independent experiments. Data are shown as the mean ± SD. (c) Representative confocal images of colocalization of GFP-LC3B puncta and mitochondria (red, anti-TOMM20) in HeLa cells transfected with the indicated siRNAs during growth in normoxia or hypoxia (1% O2) for 32 h. Scale bar: 10 μm. White arrows indicate GFP-LC3B puncta colocalized with mitochondria. (d) Quantification of colocalization of GFP-LC3B puncta and mitochondria in (c). n = 3 independent experiments. Data are shown as the mean ± SD. (e) Electron microscopy images of murine 661W photoreceptor-derived cells transfected with the indicated siRNAs after hypoxia (1% O2, 36 h) treatment. N, nucleus. Scale bar: 1 μm. Arrows indicate autophagosomes. (f) Quantification of the number of autophagosomes per μm2 in (E). n = 13 siCtrl cells and 16 siPrpf8 cells. Data are shown as the mean ± SD in (B) and (D) (One-way ANOVA, ***p < 0.001). Data are shown as the mean ± SD in (F) (Mann-Whitney test, ***p < 0.001).
PRPF8 regulates ULK1 gene expression during mitophagy
We next sought to investigate the molecular mechanism of PRPF8 in regulating mitophagosome formation. Although PRPF8 predominantly localizes in the nucleus [Citation24], it was also reported that PRPF8 localizes in other cellular compartments, such as the inner segments and apical connecting cilium of the photoreceptors [Citation32]. We therefore sought to investigate the subcellular localization of PRPF8 during mitophagy in HeLa cells. Subcellular fractionation assays showed that PRPF8 localized in the nucleus in both normoxic and hypoxic conditions ()). To further confirm this observation, we performed immunofluorescence staining with antibodies against PRPF8 and the mitochondria protein HSPD1/HSP60 (heat shock protein family D [Hsp60] member 1). Confocal microscopy analysis showed that PRPF8 predominantly localized in the nuclear speckles and it did not colocalize with HSPD1/HSP60. Again, hypoxia did not alter PRPF8 subcellular localization (). These data indicated that PRPF8 remains localized in nucleus during the mitophagic process, suggesting that its function in the nucleus as a splicing factor might be important for mitophagy.
Figure 3. PRPF8 regulates ULK1 gene expression during mitophagy. (a) Western blot analysis of subcellular fractions of HeLa cells in normoxia or hypoxia (1% O2) for 24 h with the indicated antibodies. WCL, whole-cell lysate. (b) Representative immunofluorescence images of HeLa cells in normoxia or hypoxia (1% O2) for 24 h with the indicated antibodies. Mitochondria are labelled with HSPD1/HSP60 and nuclei are labelled with Hoechst. Scale bar: 5 μm. (c) Quantitative real-time PCR analysis of mRNA levels for the indicated genes in HeLa cells. Data are shown as the mean ± SD from a representative of 3 independent experiments (One-way ANOVA, **p < 0.01). (d) Western blot analysis of ULK1 and other genes involved in mitophagosome formation in HeLa cells transfected with the indicated siRNAs during normoxia or hypoxia (1% O2, 24 h). ‘※’indicates the ATG12–ATG5 conjugate.
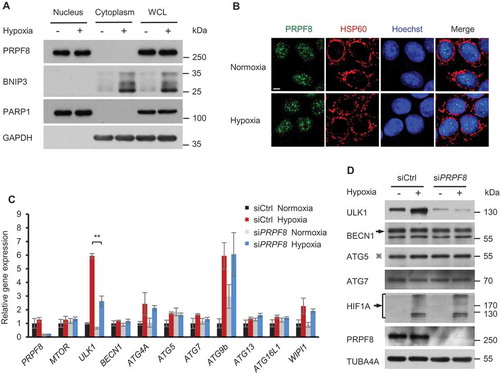
PRPF8 is the ‘master regulator’ of the spliceosome, the molecular machine executing pre-mRNA splicing, which is critical for gene expression [Citation33]. We therefore investigated whether PRPF8 regulates the expression of core autophagy genes that affect the formation of mitophagosomes. Among the genes, we found that the mRNA expression of ULK1, which plays a crucial role in the initiation of mitophagosome formation [Citation34], was significantly increased after hypoxia treatment ()). Interestingly, this increase was inhibited when PRPF8 was knocked down. Other genes, however, remained unchanged in PRPF8 knockdown cells ()). This observation was further confirmed by western blot analysis where PRPF8 knockdown inhibited the elevation of ULK1 protein levels after hypoxia treatment, whereas other proteins that are essential for mitophagosome formation, such as BECN1, ATG5 (autophagy related 5) and ATG7 (autophagy related 7) [Citation35], remained unchanged ()). Together, these data suggested that PRPF8 regulates ULK1 gene expression during mitophagy.
PRPF8 is required for mitophagy through regulating ULK1 mRNA splicing
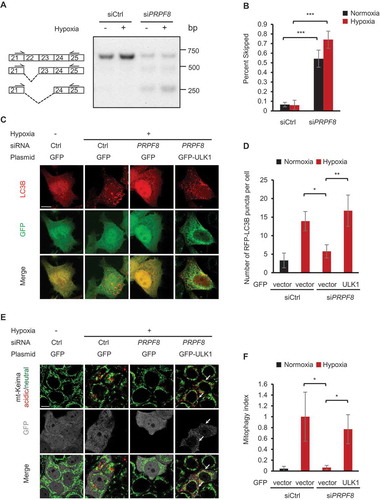
Human ULK1 has 28 exons. To further investigate how ULK1 expression is regulated by PRPF8, we assessed the pre-mRNA splicing of the ULK1 gene in control and PRPF8 knockdown cells using the reverse transcription-polymerase chain reaction (RT-PCR). We found that knockdown of PRPF8 in HeLa cells led to enhanced ULK1 exon 22 and exon 22–23 skipping. We also noted that the level of ULK1 normal spliced transcript was decreased (), and Figure S3), which is consistent with the observation that ULK1 gene expression was reduced in PRPF8 knockdown cells (). Next, to evaluate whether PRPF8 controls mitochondrial clearance under hypoxia depending on its role in regulating ULK1 mRNA splicing, we transfected a GFP-ULK1 plasmid into PRPF8 knockdown cells. We found that the reconstituted GFP-ULK1 was able to restore the number of LC3B puncta under hypoxia (). Similarly, the overexpression of GFP-ULK1 in PRPF8 knockdown cells also rescued the mitophagic activity (). Thus, these results collectively suggest that PRPF8 is required for mitophagy by regulating ULK1 mRNA splicing and expression.
Figure 4. PRPF8 is required for mitophagy through regulating ULK1 mRNA splicing. (a) A representative gel picture shows bands corresponding to normal and aberrant skipping spliced products of ULK1 mRNA in HeLa cells transfected with the indicated siRNAs during normoxia or hypoxia (1% O2, 24 h) in RT-PCR assays. (b) The percentage of the PCR product representing exon 22 and exon 22–23 skipped transcripts in the total transcripts is calculated by band intensity quantification in (a) and is shown as ‘percent skipped’ on the y axis. n = 3 independent experiments. (c) Representative confocal images of RFP-LC3B signals in HeLa cells transfected with the siRNAs and plasmids as shown under normoxia or hypoxia (1% O2) for 12 h. (d) Quantification of the number of RFP-LC3B puncta per cell in (C). n = 3 independent experiments. (e) mt-Keima imaging in HeLa cells transfected with PRPF8 siRNA and GFP-tagged ULK1 plasmid after hypoxia treatment (1% O2, 24 h). GFP fluorescence is depicted in gray. Arrows indicate GFP-ULK1-transfected cells. (f) Quantification of mitophagy index of GFP-positive cells in (E). Mitophagy index in hypoxia-treated control cells was normalized to ‘1ʹ. n = 3 independent experiments. Data are shown as the mean ± SD (One-way ANOVA, **p < 0.01, ***p < 0.001). Scale bars: 10 μm.
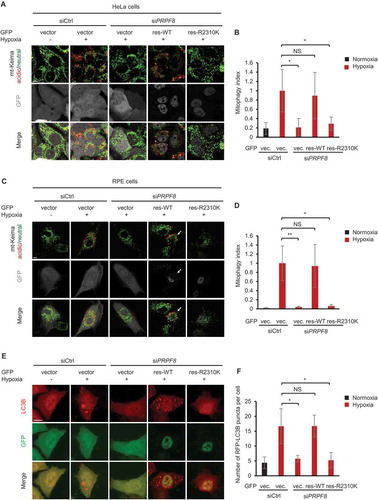
PRPF8 retinitis pigmentosa disease-associated mutant has defects in mitophagy
Mutations in PRPF8 cause progressive degeneration of the retina and finally autosomal dominant retinitis pigmentosa (adRP) [Citation36]. It is known that there is a low oxygen level in the outer retina due to the high oxygen consumption in the inner segments of the photoreceptors [Citation37,Citation38]. In addition, retinal hypoxia induces autophagy [Citation39,Citation40]. Hence, we asked whether PRPF8 retinitis pigmentosa disease-associated mutants have any defects in regulating hypoxic mitophagy. To test this possibility, we compared the effects of siRNA-resistant wild-type (WT) and disease-associated mutation of PRPF8 (R2310K) on mitophagy in HeLa cells as well as retinal pigment epithelium (RPE) cells. Whereas siRNA-resistant WT PRPF8 rescued the mitophagy defect in PRPF8 knockdown cells, the disease-associated mutant PRPF8R2310K was not able to do so in both cell lines ()). Similarly, unlike the WT PRPF8, the PRPF8R2310K was incapable of rescuing the decreased number of LC3 puncta in PRPF8-depleted HeLa and RPE cells () and Figure S4). These data indicate that the adRP-associated PRPF8R2310K mutation is defective in regulating hypoxic mitophagy in RPE and HeLa cells.
Figure 5. PRPF8 retinitis pigmentosa disease-associated mutant has defects in mitophagy. (a) mt-Keima imaging in HeLa cells transfected with PRPF8 siRNA and GFP-tagged PRPF8 siRNA-resistant plasmid after hypoxia treatment (1% O2, 24 h). GFP fluorescence is depicted in gray. (b) Quantification of mitophagy index of GFP-positive cells in (A). Mitophagy index in hypoxia-treated control cells was normalized to ‘1ʹ. n = 4 independent experiments. (c) mt-Keima imaging in RPE cells transfected with PRPF8 siRNA and GFP-tagged PRPF8 siRNA-resistant plasmid after hypoxia treatment (1% O2, 24 h). GFP fluorescence is depicted in gray. Arrows indicate GFP-PRPF8-transfected cells. (d) Quantification of mitophagy index of GFP-positive cells in (c). Mitophagy index in hypoxia-treated control cells was normalized to ‘1ʹ. n = 3 independent experiments. (e) Representative confocal images of RFP-LC3B signals in HeLa cells transfected with the siRNAs and plasmids as shown under normoxia or hypoxia (1% O2) for 12 h. (f) Quantification of the number of RFP-LC3B puncta per cell in (E). n = 3 independent experiments. Data are shown as the mean ± SD (One-way ANOVA, *p < 0.05, **p < 0.01, NS, nonsignificant). Scale bars: 10 μm.
Retinitis pigmentosa-associated spliceosomal proteins regulate mitophagy under hypoxia
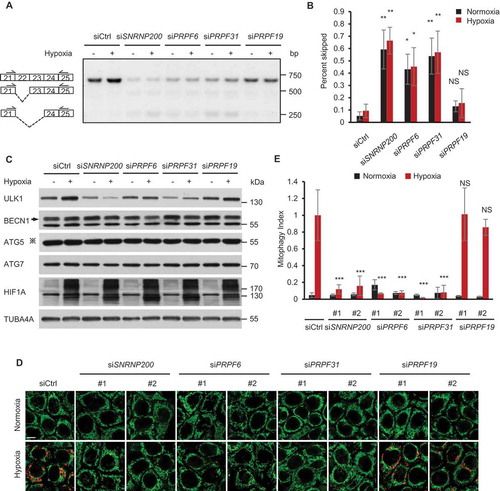
The mRNA splicing process is performed by the spliceosome, which consists of several small nuclear RNAs and splicing factors. We therefore asked whether other splicing factors, such as SNRNP200/BRR2 (small nuclear ribonucleoprotein U5 subunit 200), PRPF6 (pre-mRNA processing factor 6), PRPF31 (pre-mRNA processing factor 31) and PRPF19 (pre-mRNA processing factor 19) are also required for proper ULK1 mRNA splicing under hypoxia. We knocked down these genes by siRNA transfection, and the knockdown efficiency of these genes were tested by a quantitative-PCR assay (Figure S5A). Similar to PRPF8, the knockdown of SNRNP200, PRPF6 and PRPF31 resulted in increased ULK1 exon 22 and exon 22–23 skipping, whereas PRPF19 knockdown did not affect ULK1 mRNA splicing ()). Interestingly, the mutations of SNRNP200, PRPF6 and PRPF31, but not PRPF19, cause adRP [Citation41–Citation44]. Moreover, we tested the protein level of ULK1 following the knockdown of these spliceosomal proteins by western blotting. We found that knockdown of SNRNP200, PRPF6 or PRPF31, but not PRPF19, remarkably reduced the hypoxia-induced ULK1 expression ()). Furthermore, the knockdown of SNRNP200, PRPF6 or PRPF31 but not PRPF19 inhibited hypoxia-induced mitophagy (). This effect was rescued by co-transfection of siRNA-resistant plasmids expressing SNRNP200, PRPF6 or PRPF31 in RPE cells, indicating that the corresponding gene knockdowns were not exerting a nonspecific effect (Figure S5B-D). Thus, these results suggested that retinitis pigmentosa-associated spliceosomal proteins regulate ULK1 splicing and mitophagy under hypoxia.
Figure 6. Retinitis pigmentosa-associated spliceosomal proteins regulate mitophagy under hypoxia. (a) A representative gel picture shows bands corresponding to normal and aberrant skipping spliced products of ULK1 mRNA in HeLa cells transfected with the indicated siRNAs during normoxia or hypoxia (1% O2, 24 h) in RT-PCR assays. (b) The percentage of the PCR product representing exon 22 and exon 22–23 skipped transcripts in the total transcripts is calculated by band intensity quantification in (a) and is shown as ‘percent skipped’ on the y axis. n = 3 independent experiments. (c) Western blot analysis of HeLa cells transfected with siRNAs after hypoxia treatment (1% O2, 24 h) with the indicated antibodies. ‘※’indicates the ATG12–ATG5 conjugate. (d) mt-Keima imaging in HeLa cells transfected as indicated in a normoxic (21% O2) environment or after 24 h of hypoxia (1% O2). Scale bar: 10 μm. (e) Quantified mitophagy index in (d). Mitophagy index in hypoxia-treated control cells was normalized to ‘1ʹ. n = 3 independent experiments. Data are shown as the mean ± SD (One-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001, NS, nonsignificant).
Discussion
Previous studies on the screening of mitophagy were mainly based on PRKN translocation to mitochondria [Citation4–Citation6], however, these strategies are limited to identifying either regulators of PRKN or upstream molecules in mitophagy and cannot be used to examine the entirety of the mitophagic process. In this study, we established a new mitophagy screening system based on the mt-Keima fluorescent reporter to identify novel regulators of mitophagy under hypoxia. Based on this system, we identified the spliceosome component PRPF8 as an indispensable positive regulator of mitophagy. We show that the proper mRNA splicing of ULK1 controlled by PRPF8 and other spliceosome components is a previously unknown key nuclear event for the mitophagic process. Although the components of mitophagy in the cytoplasm have been well studied, the molecular mechanism for the transcriptional and post-transcriptional regulation of mitophagy in the nucleus is poorly understood [Citation45,Citation46]. Our findings therefore contribute to a better understanding of the mitophagic process with a ‘whole-cell view’.
ULK1 is a serine/threonine protein kinase and plays important roles in the initiation of autophagosome formation during autophagy [Citation47,Citation48]. Our study indicates that ULK1 can be regulated by PRPF8 at the post-transcriptional level ()). This post-transcriptional regulation of ULK1 is indispensable particularly in the hypoxia-induced mitophagic process. Given that ULK1 is transcriptionally upregulated, and the ULK1 protein translocates to mitochondria to induce mitophagy initiation under hypoxic conditions [Citation49,Citation50] (), it is therefore possible that PRPF8 is essential to ensure proper splicing of the newly transcribed ULK1 mRNA in order to sustain sufficient mitophagy activity for cells to survive the hypoxic stress. It is worth noting that proper ULK1 mRNA splicing under resting conditions also required PRPF8 (). However, because the basal autophagy level is low, it seems that the aberrant splicing of ULK1 mRNA caused by PRPF8 deletion has less effect on the autophagic process compared to that on hypoxic mitophagy (). Therefore, our findings suggest that the PRPF8-mediated post-transcriptional regulation of ULK1 is critical particularly for hypoxic mitophagy.
Autosomal dominant retinitis pigmentosa is a hereditary degenerative eye disease due to progressive cell death in rod-type and cone-type photoreceptor cells [Citation51]. Mutations in several core spliceosomal components such as PRPF8, SNRNP200, PRPF6 and PRPF31 have been reported to be closely related to adRP [Citation26,Citation41–Citation44]. However, the precise mechanism by which these gene mutations lead to adRP remains unknown. In this study, we find that a series of spliceosome components which are mutated in adRP patients are essential for hypoxia-induced mitophagy. In particular, we demonstrate that PRPF8 adRP disease-associated mutant R2310K has defects in mitophagy. These findings thus for the first time suggest that adRP might be due in part to the impaired mitophagic activity caused by the mutations of spliceosomes and aberrant mRNA splicing in the neurons of retina. In support of this hypothesis, several studies have shown the correlation between the dysregulation of autophagy and RP in vivo [Citation52–Citation55].
The mammalian retina is one of the most metabolically active tissues, consuming oxygen more rapidly than any other tissues [Citation56–Citation60]. The high oxygen consumption rate is not only accompanied with increased oxidative stress and damaged mitochondria, but also leads to local physiological hypoxic conditions in the normal retina [Citation37,Citation38,Citation53,Citation60–Citation63]. In addition, hypoxia could also be caused by pathologies such as central retinal artery occlusion and ischemic central retinal vein thrombosis [Citation64–Citation68]. Whereas it has been shown that hypoxia in the retina is sufficient to induce photoreceptor cell death that results in retinal degeneration diseases [Citation56,Citation69–Citation72], several lines of evidence have also suggested that some hypoxia-induced cellular responses could be protective. For instance, in the hypoxic retina, HIF1A/HIF1α (hypoxia inducible factor 1 subunit alpha) induces some proteins, such as EPO (erythropoietin) [Citation62] and HDAC4 (histone deacetylase 4) [Citation73], which protect against retinal degeneration. Similarly, our observation that the wild-type, but not adRP-associated mutated, PRPF8 induces hypoxic mitophagy () supports a model that the local hypoxia condition would induce PRPF8-mediated mitophagy, which is helpful for retinal functions by eliminating damaged mitochondria to protect against retinitis pigmentosa [Citation74]. Further studies are needed to investigate the roles of dysfunction of the spliceosome in hypoxic mitophagy and the pathogenesis of retinitis pigmentosa in mice or RP patients. Mitophagy could be targeted for prevention and control of adRP and may extend to the pathogenesis of other spliceosome-associated diseases.
Materials and methods
Cell culture and transfection
HeLa and RPE (retinal pigment epithelium) cell lines were obtained from American Type Culture Collection (ATCC, CCL-2; CRL-2302). The 661W photoreceptor cell line was purchased from Oulu biotechnology (Shanghai, XB-3247) and originated from Muayyad Al-Ubaidi (University of Oklahoma Health Science Center). HeLa and 661W cells were maintained in DMEM (Gibco, 41965–039) containing 10% fetal bovine serum (Gibco, 10270) and RPE cells were cultured in DMEM/F12 (Gibco, 10565018) medium containing 10% fetal bovine serum under standard conditions (37°C, 5% CO2). HeLa and RPE cells stably expressing mt-Keima were generated using lentiviral vectors of pLVX-Puro-mtKeima. For transfection, plasmids or siRNAs were transfected into cells with TurboFect (Thermo Fisher Scientific, R0531) or Lipofectamine RNAiMax (Invitrogen, 13778150) following the manufacturer’s instruction.
siRNAs and plasmids
PRPF8, SNRNP200, PRPF6 and PRPF31 siRNA were purchased from Sigma-Aldrich, and the sequences targeting those siRNA duplexes are provided in Table S1. PRPF19 siRNA was described previously [Citation75]. The PRPF8 expression construct was generated by PCR amplification from cDNA derived from HeLa cells. SNRNP200, PRPF6, and PRPF31 expression constructs were generated by PCR amplification from cDNA derived from RPE cells. pLVX-Puro-mtKeima plasmid was kindly provided by Dr. Toren Finkel (Aging Institute of UPMC and the University of Pittsburgh). The LC3B plasmid was a gift from Dr. Li Yu (Tsinghua University) [Citation76] and was subcloned into pEGFP-C1 vector (Clontech, PT3028-5) to generate GFP-LC3B. The ULK1 plasmid was a gift from Dr. Quan Chen (Chinese Academy of Sciences) [Citation49]. Point mutation of PRPF8 was introduced by site-directed mutagenesis (TransGen Biotech, FM111).
High-content image screen
An siRNA library containing 320 siRNA SMART pools was purchased from Dharmacon. Pooled siRNAs at a final concentration of 50 nM was used for transfection in HeLa cells stably expressing mt-Keima in 96-well optical plates. 48 h after the siRNA transfection, cells were treated with 1% O2 for 24 h, and then high-content imaging was acquired using a × 20 objective of an Operreta automated microscope (Perkin Elmer, Waltham, MA, USA). mt-Keima localized within the mitochondria was excited by 410- to 430-nm light and mt-Keima delivered within lysosomes by mitophagy was excited by 560- to 580-nm light and both of them were detected by a 590- to 640-nm filter. Images were quantitatively analyzed using Harmony software 3.5. We calculate the ratio of (560–580 nm):(410–430 nm) fluorescence of each single cell. Ratio values ranged from 0 to 0.32 in cells under normoxia. We defined the cell with a higher ratio as a ‘mitophagic-positive cell’. The percentage of mitophagic-positive cell in each well represents its mitophagic activity. The screen data were normalized using the Z-score method, incorporating the median absolute deviation [Citation77]. The hit cutoff was defined as −2.0 or 2.0.
Fluorescence microscopy
Fluorescence of mt-Keima was imaged in 2 channels via 2 sequential excitations (458 nm, green; 561 nm, red) and using a 609- to 735-nm emission range. We identified the ratio of 561 nm:458 nm fluorescence as the ‘mitophagy index’. For immunofluorescence analysis, the cells were grown on coverslips and were fixed in 4% paraformaldehyde (for TOMM20) or in −20°C methanol (for HSPD1/HSP60 and PRPF8) for 15 min and then permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, X100). Newborn goat serum (3%; Thermo Fisher Scientific, 16210072) in PBST (Thermo Fisher Scientific, 28352) was used for blocking. Fixed cells were incubated overnight at 4°C with the corresponding primary antibodies: rabbit anti-TOMM20 (Santa Cruz Biotechnology, sc-11415; 1:400), mouse anti-HSPD1/HSP60 (Santa Cruz Biotechnology, sc-59567; 1:200) and rabbit anti-PRPF8 (Santa Cruz Biotechnology, sc-30207; 1:200). Coverslips were then washed in phosphate-buffered saline (PBS; Gibco, 10010023) and stained for 45 min with Alexa Fluor 488- or Alexa Fluor 546-conjugated secondary antibodies (Invitrogen, A-11010, A11003 and A27034; 1:400). Coverslips were washed again with PBS and mounted with VectaShield containing Hoechst (Invitrogen, H3570). Confocal images were obtained using a 60× oil lens objective on an inverted fluorescence microscope (Nikon Eclipse Ti-E, Melville, NY USA) or ZEISS LSM 880 (Zeiss, Germany). Images were viewed and analyzed using ZEISS ZEN Imaging Software or Volocity 6.1.1 software.
Western blotting
Cells were collected and resuspended in RIPA lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40 [Santa Cruz Biotechnology, CAS 9016–45-9], 0.5% Na-deoxycholate [Sigma-Aldrich, D6750]) containing certain protease inhibitors (0.1 mM phenylmethanesulfonyl fluoride [Target Molecule Corporation, T0789], 1× EDTA-free Protease Inhibitor Cocktail [ROCHE, 11873580001]) on ice. Antibodies were used at the following concentrations: rabbit anti-PRPF8 (Santa Cruz Biotechnology, sc-030207; 1:1000), mouse anti-TOMM20 (Santa Cruz Biotechnology, sc-17764; 1:1000), mouse anti-TIMM23 (Santa Cruz Biotechnology, sc-514463; 1:1000), mouse anti-BNIP3 (Santa Cruz Biotechnology, sc-56167; 1:1000), rabbit anti-PARP1 (Cell Signaling Technology, 9532s; 1:2000), mouse anti-BECN1 (Santa Cruz Biotechnology, sc-48341; 1:1000), rabbit anti-ULK1 (Cell Signaling Technology, 8054s; 1:1000), rabbit anti-ATG5 (Cell Signaling Technology, 12994s; 1:1000), rabbit anti-ATG7 (Cell Signaling Technology, 8558s; 1:1000), rabbit anti-HIF1A (GeneTex, GTX127309; 1:3000), rabbit anti-PINK1 (Novus Biologicals, D-2; 1:2000), goat anti-SNRNP200/BRR2 (Santa Cruz Biotechnology, sc-68563; 1:1000), rabbit anti-PRPF6 (Santa Cruz Biotechnology, sc-48786; 1:1000), and mouse anti-PRPF31 (Santa Cruz Biotechnology, sc-166792; 1:1000) incubated overnight at 4°C; mouse anti-GAPDH (Proteintech, 60004–1-Ig; 1:5000) and mouse anti-TUBA4A/tubulin (Sigma-Aldrich, T5168; 1:5000) incubated for 1 h at room temperature (RT). Membranes were incubated with HRP-conjugated secondary antibodies (Dako Cytomation, P0447 and P0448) for 1 h at room temperature and immunopositive bands visualized with Supersignal West Femto chemiluminescent substrate (Pierce, 34095).
Reverse transcription-PCR and quantitative real-time PCR
RNA was prepared using TRIzol Reagent (Invitrogen, 15596018). Diluted RNA was reverse-transcribed using random priming and M-MLV Reverse Transcriptase according to the manufacturer’s instructions (Promega Corporation, M1701). For PCR analysis, primers are listed in Table S2. Quantification of all gene expression was carried out by quantitative real-time PCR using the SYBR Premix Ex Taq kit (TaKaRa, RR820A). ACTB was used as internal control. The primers were provided in the Table S3.
Transmission electron microscopy
661W cells were transfected with siCtrl or siPrpf8 (mouse) siRNAs and subjected to hypoxia (1% O2) for 36 h. The cells were washed with PBS and collected by centrifugation at 600 g for 10 min. The cell mass was fixed in 3% glutaraldehyde (Sigma-Aldrich, G5882). Ultrathin sections were examined with the H-7650 transmission electron microscope (Hitachi-Science & Technology, Tokyo, Japan).
Statistical analysis
All data presented as histograms refer to a mean value ± SD of the total number of independent experiments. Statistical comparisons between only 2 groups were carried out using a Mann-Whitney test. One-way ANOVA was used for multiple-group comparisons. Statistical calculations were carried out using GraphPad Prism 6.0. We tested data for normality and variance and considered a P-value of less than 0.05 significant. Quantification of RT-PCR was done by using ImageJ software.
Abbreviations
| adRP | = | autosomal dominant retinitis pigmentosa |
| ATG5 | = | autophagy related 5 |
| ATG7 | = | autophagy related 7 |
| BECN1 | = | beclin 1 |
| GFP | = | green fluorescent protein |
| HSPD1/HSP60 | = | heat shock protein family D (Hsp60) member 1 |
| MAP1LC3/LC3 | = | microtubule associated protein 1 light chain 3 |
| PINK1 | = | PTEN induced putative kinase 1 |
| PRKN/PARK2 | = | parkin RBR E3 ubiquitin protein ligase |
| PRPF6 | = | pre-mRNA processing factor 6 |
| PRPF8 | = | pre-mRNA processing factor 8 |
| PRPF19 | = | pre-mRNA processing factor 19 |
| PRPF31 | = | pre-mRNA processing factor 31 |
| SNRNP200/BRR2 | = | small nuclear ribonucleoprotein U5 subunit 200 |
| ULK1 | = | unc-51 like autophagy activating kinase 1. |
Supplemental Material
Download MS Word (1.4 MB)Acknowledgments
We thank Dr. Toren Finkel, Dr. Quan Chen and Dr. Li Yu for kindly providing plasmids, Dr. Nuo Sun and Dr. Zibing Jin for helpful discussion and Sa Zhang for assistance with electron microscopy.
Disclosure statement
No potential conflict of interest was reported by the authors.
SUPPLEMENTAL DATA
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998 Apr 9;392(6676):605–608. PubMed PMID: 9560156.
- Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004 May 21;304(5674):1158–1160. PubMed PMID: 15087508.
- Orvedahl A, Sumpter R Jr., Xiao G, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011 Dec 1;480(7375):113–117. PubMed PMID: 22020285; PubMed Central PMCID: PMC3229641.
- Hasson SA, Kane LA, Yamano K, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013 Dec 12;504(7479):291–295. PubMed PMID: 24270810.
- Ivatt RM, Sanchez-Martinez A, Godena VK, et al. Genome-wide RNAi screen identifies the Parkinson disease GWAS risk locus SREBF1 as a regulator of mitophagy. Proc Natl Acad Sci U S A. 2014 Jun 10;111(23):8494–8499. PubMed PMID: 24912190; PubMed Central PMCID: PMC4060696.
- Lefebvre V, Du Q, Baird S, et al. Genome-wide RNAi screen identifies ATPase inhibitory factor 1 (ATPIF1) as essential for PARK2 recruitment and mitophagy. Autophagy. 2013 Nov 1;9(11):1770–1779. PubMed PMID: 24005319.
- Sumpter R Jr., Sirasanagandla S, Fernandez AF, et al. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016 May 5;165(4):867–881. PubMed PMID: 27133164; PubMed Central PMCID: PMC4881391.
- Lee MY, Sumpter R Jr., Zou Z, et al. Peroxisomal protein PEX13 functions in selective autophagy. EMBO Rep. 2017 Jan;18(1):48–60. PubMed PMID: 27827795; PubMed Central PMCID: PMC5210156.
- Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008 Jul 10;454(7201):232–235. PubMed PMID: 18454133; PubMed Central PMCID: PMC2570948.
- Sato M, K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011 Nov 25;334(6059):1141–1144. PubMed PMID: 21998252.
- O’Sullivan TE, Johnson LR, Kang HH, et al. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015 Aug 18;43(2):331–342. PubMed PMID: 26253785.
- Altshuler-Keylin S, Shinoda K, Hasegawa Y, et al. Beige adipocyte maintenance is regulated by autophagy-induced mitochondrial clearance. Cell Metab. 2016 Sep 13;24(3):402–419. PubMed PMID: 27568548; PubMed Central PMCID: PMC5023491.
- Kieran MW, Folkman J, Heymach J. Angiogenesis inhibitors and hypoxia. Nat Med. 2003 Sep;9(9):1104; author reply 1104–5. PubMed PMID: 12949515.
- Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006 May 25;441(7092):437–443. PubMed PMID: 16724055.
- Peers C, Dallas ML, Boycott HE, et al. Hypoxia and neurodegeneration. Ann N Y Acad Sci. 2009 Oct;1177:169–177. PubMed PMID: 19845619.
- Correia SC, Moreira PI. Hypoxia-inducible factor 1: a new hope to counteract neurodegeneration? J Neurochem. 2010 Jan;112(1):1–12. PubMed PMID: 19845827.
- Formenti F, Constantin-Teodosiu D, Emmanuel Y, et al. Regulation of human metabolism by hypoxia-inducible factor. Proc Natl Acad Sci U S A. 2010 Jul 13;107(28):12722–12727. PubMed PMID: 20616028; PubMed Central PMCID: PMC2906567.
- Katayama H, Kogure T, Mizushima N, et al. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011 Aug 26;18(8):1042–1052. PubMed PMID: 21867919.
- Murakawa T, Yamaguchi O, Hashimoto A, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015 Jul 6;6:7527. PubMed PMID: 26146385; PubMed Central PMCID: PMC4501433.
- Bingol B, Tea JS, Phu L, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014 Jun 19;510(7505):370–375. PubMed PMID: 24896179.
- Mizumura K, Cloonan SM, Nakahira K, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest. 2014 Sep;124(9):3987–4003. PubMed PMID: 25083992; PubMed Central PMCID: PMC4151233.
- Kageyama Y, Hoshijima M, Seo K, et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014 Dec 1;33(23):2798–2813. PubMed PMID: 25349190; PubMed Central PMCID: PMC4282557.
- Sun N, Yun J, Liu J, et al. Measuring in vivo mitophagy. Mol Cell. 2015 Nov 19;60(4):685–696. PubMed PMID: 26549682; PubMed Central PMCID: PMC4656081.
- Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005 May;11(5):533–557. PubMed PMID: 15840809; PubMed Central PMCID: PMC1370742.
- Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. PubMed PMID: 25784052; PubMed Central PMCID: PMC4526142.
- Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016 Jan;17(1):19–32. PubMed PMID: 26593421.
- Sveen A, Kilpinen S, Ruusulehto A, et al. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016 May 12;35(19):2413–2427. PubMed PMID: 26300000.
- Cheng B, Xu A, Qiao M, et al. BECN1s, a short splice variant of BECN1, functions in mitophagy. Autophagy. 2015 Nov 2;11(11):2048–2056. PubMed PMID: 26649941; PubMed Central PMCID: PMC4824595.
- Liu C, Ma H, Wu J, et al. Arginine68 is an essential residue for the C-terminal cleavage of human Atg8 family proteins. BMC Cell Biol. 2013 May 30;14:27. PubMed PMID: 23721406; PubMed Central PMCID: PMC3686597.
- Wang YB, Tan B, Mu R, et al. Ubiquitin-associated domain-containing ubiquitin regulatory X (UBX) protein UBXN1 is a negative regulator of nuclear factor kappaB (NF-kappaB) signaling. J Biol Chem. 2015 Apr 17;290(16):10395–10405. PubMed PMID: 25681446; PubMed Central PMCID: PMC4400349.
- Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000 Nov 1;19(21):5720–5728. PubMed PMID: 11060023; PubMed Central PMCID: PMC305793.
- Wheway G, Schmidts M, Mans DA, et al. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol. 2015 Aug;17(8):1074–1087. PubMed PMID: 26167768; PubMed Central PMCID: PMC4536769.
- Mayerle M, Raghavan M, Ledoux S, et al. Structural toggle in the RNaseH domain of Prp8 helps balance splicing fidelity and catalytic efficiency. Proc Natl Acad Sci U S A. 2017 May 2;114(18):4739–4744. PubMed PMID: 28416677; PubMed Central PMCID: PMC5422793.
- Itakura E, C, Koyama-Honda I, et al. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci. 2012 Mar 15;125(Pt 6):1488–1499. PubMed PMID: 22275429.
- Antonioli M, Di Rienzo M, Piacentini M, et al. Emerging mechanisms in initiating and terminating autophagy. Trends Biochem Sci. 2017 Jan;42(1):28–41. PubMed PMID: 27765496.
- Daiger SP, Bowne SJ, Sullivan LS. Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007 Feb;125(2):151–158. PubMed PMID: 17296890; PubMed Central PMCID: PMC2580741.
- Yu DY, Cringle SJ. Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Prog Retin Eye Res. 2001 Mar;20(2):175–208. PubMed PMID: 11173251.
- Yu DY, Cringle SJ. Retinal degeneration and local oxygen metabolism. Exp Eye Res. 2005 Jun;80(6):745–751. PubMed PMID: 15939030.
- Kim SH, Munemasa Y, Kwong JM, et al. Activation of autophagy in retinal ganglion cells. J Neurosci Res. 2008 Oct;86(13):2943–2951. PubMed PMID: 18521932.
- Wu BX, Darden AG, Laser M, et al. The rat Apg3p/Aut1p homolog is upregulated by ischemic preconditioning in the retina. Mol Vis. 2006 Oct;26(12):1292–1302. PubMed PMID: 17110912.
- McKie AB, McHale JC, Keen TJ, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet. 2001 Jul 15;10(15):1555–1562. PubMed PMID: 11468273.
- Vithana EN, Abu-Safieh L, Allen MJ, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell. 2001 Aug;8(2):375–381. PubMed PMID: 11545739.
- Zhao C, Bellur DL, Lu S, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009 Nov;85(5):617–627. PubMed PMID: 19878916; PubMed Central PMCID: PMC2775825.
- Tanackovic G, Ransijn A, Ayuso C, et al. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2011 May 13;88(5):643–649. PubMed PMID: 21549338; PubMed Central PMCID: PMC3146730.
- Shin HJ, Kim H, Oh S, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016 Jun 23;534(7608):553–557. PubMed PMID: 27309807; PubMed Central PMCID: PMC5568428.
- Baek SH, Kim KI. Epigenetic control of autophagy: nuclear events gain more attention. Mol Cell. 2017 Mar 2;65(5):781–785. PubMed PMID: 28257699.
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010 Apr;22(2):132–139. PubMed PMID: 20056399.
- Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol. 2016 Apr;39:61–68. PubMed PMID: 26921696; PubMed Central PMCID: PMC4828305.
- Wu W, Tian W, Hu Z, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014 May;15(5):566–575. PubMed PMID: 24671035; PubMed Central PMCID: PMC4210082.
- Li J, Qi W, Chen G, et al. Mitochondrial outer-membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy. Autophagy. 2015;11(8):1216–1229. PubMed PMID: 26018823; PubMed Central PMCID: PMC4590677.
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006 Nov 18;368(9549):1795–1809. PubMed PMID: 17113430.
- Miao G, Zhao YG, Zhao H, et al. Mice deficient in the Vici syndrome gene Epg5 exhibit features of retinitis pigmentosa. Autophagy. 2016 Dec;12(12):2263–2270. PubMed PMID: 27715390; PubMed Central PMCID: PMC5173265.
- Boya P, Esteban-Martinez L, Serrano-Puebla A, et al. Autophagy in the eye: development, degeneration, and aging. Prog Retin Eye Res. 2016 Nov;55:206–245. PubMed PMID: 27566190.
- Rodriguez-Muela N, Hernandez-Pinto AM, Serrano-Puebla A, et al. Lysosomal membrane permeabilization and autophagy blockade contribute to photoreceptor cell death in a mouse model of retinitis pigmentosa. Cell Death Differ. 2015 Mar;22(3):476–487. PubMed PMID: 25501597; PubMed Central PMCID: PMC4326579.
- Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009 Jan;12(1):44–52. PubMed PMID: 19060896; PubMed Central PMCID: PMC3339764.
- Schmidt M, Giessl A, Laufs T, et al. How does the eye breathe? Evidence for neuroglobin-mediated oxygen supply in the mammalian retina. J Biol Chem. 2003 Jan 17;278(3):1932–1935. PubMed PMID: 12409290.
- Boon KL, Grainger RJ, Ehsani P, et al. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol. 2007 Nov;14(11):1077–1083. PubMed PMID: 17934474; PubMed Central PMCID: PMC2584834.
- Cao H, Wu J, Lam S, et al. Temporal and tissue specific regulation of RP-associated splicing factor genes PRPF3, PRPF31 and PRPC8–implications in the pathogenesis of RP. PloS One. 2011 Jan 19;6(1):e15860. PubMed PMID: 21283520; PubMed Central PMCID: PMC3023711.
- Liu MM, Zack DJ. Alternative splicing and retinal degeneration. Clin Genet. 2013 Aug;84(2):142–149. PubMed PMID: 23647439; PubMed Central PMCID: PMC4147722.
- Steinberg RH. Monitoring communications between photoreceptors and pigment epithelial cells: effects of “mild” systemic hypoxia. Friedenwald lecture. Invest Ophthalmol Vis Sci. 1987 Dec;28(12):1888–1904. PubMed PMID: 3316105.
- Anderson B Jr., Saltzman HA. Retinal oxygen utilization measured by hyperbaric blackout. Arch Ophthalmol. 1964 Dec;72:792–795. PubMed PMID: 14205438.
- Grimm C, Wenzel A, Groszer M, et al. HIF-1-induced erythropoietin in the hypoxic retina protects against light-induced retinal degeneration. Nat Med. 2002 Jul;8(7):718–724. PubMed PMID: 12068288.
- Cringle SJ, Yu DY. A multi-layer model of retinal oxygen supply and consumption helps explain the muted rise in inner retinal PO(2) during systemic hyperoxia. Comp Biochem Physiol A Mol Integr Physiol. 2002 May;132(1):61–66. PubMed PMID: 12062192.
- Jang H, Ahn HR, Jo H, et al. Chlorogenic acid and coffee prevent hypoxia-induced retinal degeneration. J Agric Food Chem. 2014 Jan 8;62(1):182–191. PubMed PMID: 24295042.
- Kaur C, Foulds WS, Ling EA. Hypoxia-ischemia and retinal ganglion cell damage. Clin Ophthalmol. 2008 Dec;2(4):879–889. PubMed PMID: 19668442; PubMed Central PMCID: PMC2699791.
- Hayreh SS, Podhajsky PA, Zimmerman MB. Retinal artery occlusion: associated systemic and ophthalmic abnormalities. Ophthalmology. 2009 Oct;116(10):1928–1936. PubMed PMID: 19577305; PubMed Central PMCID: PMC2757505.
- Nickells RW. The cell and molecular biology of glaucoma: mechanisms of retinal ganglion cell death. Invest Ophthalmol Vis Sci. 2012 May 4;53(5):2476–2481. PubMed PMID: 22562845; PubMed Central PMCID: PMC3990459.
- Eliasdottir TS, Bragason D, Hardarson SH, et al. Venous oxygen saturation is reduced and variable in central retinal vein occlusion. Graefe’s archive for clinical and experimental ophthalmology = Albrecht von Graefes Archiv fur klinische und experimentelle. Ophthalmologie. 2015 Oct;253(10):1653–1661. PubMed PMID: 25404523.
- Kurihara T, Westenskow PD, Gantner ML, et al. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. eLife. 2016 Mar 15;5: e14319. PubMed PMID: 26978795; PubMed Central PMCID: PMCPMC4848091.
- Barben M, Schori C, Samardzija M, et al. Targeting Hif1a rescues cone degeneration and prevents subretinal neovascularization in a model of chronic hypoxia. Mol Neurodegener. 2018 Mar 7;13(1):12. PubMed PMID: 29514656; PubMed Central PMCID: PMCPMC5842520.
- Wiedman M, Tabin GC. High-altitude retinopathy and altitude illness. Ophthalmology. 1999 Oct;106(10):1924–1926; discussion 1927. PubMed PMID: 10519586.
- Karakucuk S, Mirza GE. Ophthalmological effects of high altitude. Ophthalmic Res. 2000 Jan-Feb;32(1):30–40. PubMed PMID: 10657753.
- Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009 Jan 9;323(5911):256–259. PubMed PMID: 19131628; PubMed Central PMCID: PMC3339762.
- Wright AF, Chakarova CF, Abd El-Aziz MM, et al. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010 Apr;11(4):273–284. PubMed PMID: 20212494.
- Mu R, Wang YB, Wu M, et al. Depletion of pre-mRNA splicing factor Cdc5L inhibits mitotic progression and triggers mitotic catastrophe. Cell Death Dis. 2014 Mar 27;5:e1151. PubMed PMID: 24675469; PubMed Central PMCID: PMC3973201.
- Rong Y, Liu M, Ma L, et al. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol. 2012 Sep;14(9):924–934. PubMed PMID: 22885770.
- Fisher KH, Wright VM, Taylor A, et al. Advances in genome-wide RNAi cellular screens: a case study using the Drosophila JAK/STAT pathway. BMC Genomics. 2012 Sep 24;13:506. PubMed PMID: 23006893; PubMed Central PMCID: PMC3526451.
