ABSTRACT
Actin filament, also known as microfilament, is one of two major cytoskeletal elements in plants and plays important roles in various biological processes. Like in animal cells, actin filaments have been thought to participate in autophagy in plants. However, surprisingly, in this study we found that actin filaments are dispensable for the occurrence of autophagy in plants. Disruption of actin filaments by short term treatment with actin polymerization inhibitors, cytochalasin D and latrunculin B, or transient overexpression of Profilin 3 in Nicotiana benthamiana had no effect on basal autophagy as well as the upregulation of nocturnal autophagy and salt stress-induced autophagy. Furthermore, anti-microfilament drug treatment affected neither basal nor salt stress-induced autophagy in Arabidopsis. In addition, prolonged perturbation of actin filaments by silencing Actin7 or 24-h treatment with microfilament-disrupting agents in N. benthamiana caused endoplasmic reticulum (ER) disorganization and subsequent degradation via autophagy involving ATG2, 3, 5, 6 and 7. Our findings reveal that, unlike mammalian cells, actin filaments are unnecessary for bulk autophagy in plants.
Abbreviations: ATG: autophagy-related; CD: cytochalasin D; Cvt pathway: cytoplasm to vacuole targeting pathway; DMSO: dimethyl sulfoxide; ER: endoplasmic reticulum; LatB: latrunculin B; Nb: Nicotiana benthamiana; PAS: phagophore assembly site; PRF3: Profilin 3; RER: rough ER; SER: smooth ER; TEM: transmission electron microscopy; TRV: Tobacco rattle virus; VIGS: virus-induced gene silencing; wpi: weeks post-agroinfiltration
Introduction
Autophagy, in company with the ubiquitin–proteasome system (UPS), constitutes the two major cellular self-clearing pathways, which are responsible for the degradation and recycling of macromolecules in eukaryotes [Citation1]. To date, three types of autophagy have been reported, namely, macroautophagy, microautophagy and chaperon-mediated autophagy [Citation2]. Macroautophagy, here referred as autophagy, describes the clearance process during which cellular components such as proteins, carbohydrates, lipids, nucleic acids and organelles are sequestered in the double-membrane autophagosomes and delivered into vacuole (plant and yeast) or lysosome (animal) for degradation [Citation3]. More than 40 autophagy-related (ATG) genes that participate in autophagy have been identified in yeast, among which 18 ATG genes are demonstrated to be crucial for autophagosome formation [Citation4–Citation6]. Most of the key ATG genes have orthologs in plants [Citation4,Citation5]. Mounting evidence indicates that plant autophagy plays important roles in diverse biological processes such as development, leaf senescence, adaption to biotic and abiotic stresses and quality control of proteins and organelles [Citation7–Citation16].
Actin filaments, also known as microfilaments, are polar cytoskeletal structures composed of two coiled chains of actin monomers. They exhibit highly dynamic behaviors characterized by constant assembly and disassembly, which are essential for multiple cellular events, such as cell locomotion, contractility, cell division and intracellular trafficking [Citation17]. During the last decade, investigations in yeast and animal cells have indicated divergent roles of actin filaments in autophagy. In yeast, strikingly, disruption of actin filaments has no effect on bulk autophagy but does influence the occurrence of selective types of autophagy, including the cytoplasm to vacuole targeting (Cvt) pathway, pexophagy and the specific degradation of endoplasmic reticulum (ER) during starvation [Citation18,Citation19]. The molecular links between Atg11, Atg9 and the actin-related protein-2/3 (Arp2/3) complex, a key nucleator of branched actin filaments, raise a possibility that the actin filament-dependent transfer of Atg9 to the phagophore assembly site (PAS) is required for the precise cargo selection during selective autophagy [Citation20,Citation21]. While the participation of the actin cytoskeleton in yeast autophagy is conditional, its roles in autophagy of mammalian cells are ubiquitous and proven to be involved in various steps of autophagy, including autophagosome biogenesis and its subsequent fusion with lysosomes [Citation22]. In particular, branched actin networks have been recently demonstrated to be crucial for shaping autophagosomal membrane [Citation23]. Moreover, the involvement of the actin cytoskeleton in selective autophagy has also been suggested for removing protein aggregates and bacteria [Citation24,Citation25]. In plants, however, our knowledge of microfilaments involvement in autophagy is extremely limited. Recently, AT2G35110/NAP1, a component of SCAR/WAVE complex required for ARP2/3 mediated actin nucleation, is reported to be positively involved in autophagy in Arabidopsis [Citation26], raising a possibility that actin cytoskeleton might have a role in plant autophagy. However, it is still unknown whether actin filaments do participate in autophagic process in plants.
In this study, we investigated the effect of disruption of actin filaments on plant autophagy by using actin polymerization inhibitors, cytochalasin D and latrunculin B, transient overexpressing Profilin 3 (PRF3) or knocking down the expression of Actin7 in N. benthamiana leaves. Short-term disruption of actin filaments by drug treatment or overexpression of PRF3 affected neither the up-regulation of nocturnal autophagy nor salt stress-induced autophagy process. Prolonged disruption of actin filaments by silencing Actin7 in N. benthamiana as well as continuous treatment with actin polymerization inhibitors led to ER disorganization and its degradation via the autophagic pathway, which depends on at least five core autophagy components including ATG2, ATG3, ATG5, ATG6 and ATG7. Taken together, our data reveal a dispensable role of actin filaments in bulk autophagy in plants.
Results
Short-term disruption of actin filaments by anti-microfilament drugs has no effect on nocturnal autophagy or salt stress-induced autophagy
To evaluate the role of actin filaments in plant autophagy, we first investigated the autophagic response to actin filaments disassembly by using anti-microfilament drugs in N. benthamiana. Two well-studied actin filaments-disrupting agents, cytochalasin D (CD) and latrunculin B (LatB), were used here to perturb the actin cytoskeleton, both of which can cause actin filaments depolymerization through their binding to G- or F-actin [Citation27,Citation28]. To visualize actin filaments in plants, a fluorescent probe was generated by fusing yellow fluorescent protein (YFP) to the N terminus of the actin-binding domain 2 of Arabidopsis Fimbrin (FIM1-fABD2) as described [Citation29]. Transient overexpression of YFP-fABD2 in N. benthamiana leaves for 48 h revealed a fine and dense network of actin filaments (Figure S1A, the upper panel). Exposure of N. benthamiana leaves to either 20 μM CD or 25 μM LatB for 0.5 h by infiltration both resulted in aberrant organization of actin cytoskeleton but differed in the appearance. Decoration of actin filaments by YFP-fABD2 showed that CD treatment resulted in disappearance of nearly all the visible filaments and presence of multiple punctate foci labeled by the filament marker, while LatB treatment mainly made filaments sparse, short and fragmented (Figure S1A and B). Furthermore, the inhibitory effect of 20 μM CD and 25 μM LatB on actin polymerization could last for a long time in drug-infiltrated leaves and that of up to 24-h treatment was tested in this study (Figure S1C). We then examined the effect of CD or LatB treatment on nocturnal autophagy, which has been shown in our previous study to undergo an upregulation before midnight and a downregulation by dawn in plants growing under a 16-h light/8-h dark cycle [Citation14]. Actin polymerization inhibitors were infiltrated into leaves at the end of the day (dark 0 h) to allow 4-h treatment followed by checking the cellular microfilament arrays and autophagic activities by confocal laser scanning microscopy at midnight. 4-h treatment of N. benthamiana cells with CD or LatB resulted in severe disorganization of actin networks as indicated by YFP-fABD2-labeling (Figure S1B). Meanwhile, the autophagic activity indicated by CFP-ATG8f-labeled structures was analyzed to investigate the impact of microfilament disruption on autophagy. As expected, a lot of autophagic structures labeled by CFP-ATG8f appeared in mesophyll cells subjected to darkness for 4 h (Figure 1(a)). However, astonishingly, no significant difference was observed between autophagic activities of control cells and CD or LatB-treated cells in the absence of protease inhibitor E-64d ( and ). Similar result was obtained in mesophyll cells treated with or without microfilaments-disrupting agents even in the presence of E-64d, which blocked the vacuolar degradation of proteins and significantly increased the number of CFP-ATG8f-labeled structures in all the tested cells ( and ), suggesting that autophagic flux occurs normally in the microfilament-disrupted cells. Meanwhile, GFP-ATG8 (green fluorescent protein fused to ATG8) processing assay was performed to monitor autophagy, as indicated by the appearance of free GFP cleaved from GFP-ATG8. In accordance with the confocal observation, GFP-ATG8f was processed in a comparable manner both in control or drug-treated cells during nocturnal autophagy. Furthermore, protein level of GFP-ATG8 was significantly increased in E-64d treated samples, which indicated that the vacuolar processing of GFP-ATG8 could be dramatically blocked by this protease inhibitor ()). These results reveal that autophagic flux is normal in the microfilament-disrupted samples ()). In addition, we also examined the basal autophagic activity at the end of the day in the CD- or LatB-pretreated cells for 4 h and found that this treatment had no effect on the basal level of autophagy (Figure S2A and B).
Figure 1. Treatment of leaves with anti-microfilament drugs for 4 h has no influence on nocturnal autophagy. (a) Representative confocal images of CFP-ATG8f-labeled autophagic structures in leaves subjected to treatment with 20 μM CD or 25 μM LatB for 4 h in the absence (left) or the presence (right) of E-64d. 100 μM E-64d was pre-infiltrated into leaves to allow 12-h inhibition of vacuolar proteases before imaging in the middle of the dark cycle (dark 4 h, short for D4). CFP-ATG8f is in cyan and chloroplasts are in red. Scale bars: 10 μm. (b) Relative autophagic activity in leaves. The autophagic activity in non-drug-treated leaves (control) without E-64d was set to 1.0. More than 200 cells were quantified in each treatment. Values represent means ± SE were from 5 independent experiments. Student’s t test was used to determine the significant difference (***, p < 0.001). (c) GFP-ATG8 processing assay showing autophagy level of each treatment. ProATG8f::GFP-ATG8f was overexpressed for 48 h. GFP-ATG8f and GFP were detected using Anti-GFP antibody. Open and closed arrowheads indicate the GFP and GFP-ATG8f band, respectively. Large subunit of RuBisCO (RBCL) stained by Ponceau indicates equal loading (lower panel). (d) Representative images showing colocalizations of ATG5-GFP-labeled phagophore with RFP-ATG8f-labeled autophagic structures in leaves. Images were taken in the middle of the dark cycle (dark 4 h). ATG5-GFP is in green and RFP-ATG8f is in red. Colocalized structures are indicated by magenta arrowheads. Scale bars: 10 μm. (e) Ratio of ATG5-ATG8-colocalized structures to RFP-ATG8f-labeled autophagic structures. More than 100 cells were quantified in each treatment. Values represent means ± SE from 3 independent experiments. n.s means no significant difference (Student’s t test, p > 0.05).
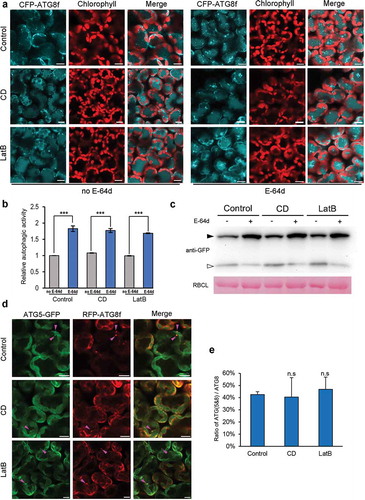
To further assess the engagement of actin filaments in autophagosome formation process, we tested the effect of actin depolymerization on phagophore formation by monitoring the colocalized autophagic structures labeled by red fluorescent protein (RFP)-tagged ATG8f (RFP-ATG8f) and green fluorescent protein (GFP)-tagged ATG5 (ATG5-GFP), which has been shown to unambiguously label phagophores during autophagosome biogenesis [Citation30]. No significant difference was observed in the ratio of ATG5- and ATG8-colocalized structures to RFP-ATG8f-labeled autophagic structures, ATG(5&8)/ATG8, between drug-treated cells and control cells ( and ), suggesting that disruption of actin filaments does not affect phagophore formation during the upregulation of nocturnal autophagy.
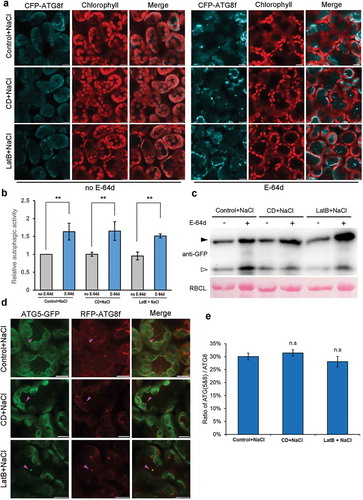
We also investigated the effect of microfilament disruption on autophagy induction by salt stress, which has been reported to induce autophagy rapidly in Arabidopsis [Citation31]. Similarly, 1-h treatment of N. benthamiana leaves with 150 mM NaCl infiltration markedly induced the formation of CFP-ATG8f-labeled autophagic structures (Figure S3). 150 mM NaCl was co-infiltrated into CFP-ATG8f-expressing leaves with anti-microfilaments drugs, 20 μM CD or 25 μM LatB for 1-h treatment to test the effect of microfilament disruption on the occurrence of autophagy. Confocal observation of CFP-ATG8f-labeled structures showed that the salt stress-induced autophagosomes formed normally in CD- or LatB-treated cells and there was no difference in the number of autophagic structures between leaves treated with NaCl and that of NaCl plus anti-microfilaments drugs ( and ). In addition, the presence of E-64d further increased the number of autophagic structures in the above-mentioned leaf samples, suggesting that autophagic flux proceeds normally irrespective of the presence of anti-microfilaments drugs ( and b)). This result was further confirmed by GFP-ATG8 processing assay ()). In addition, we also checked whether the phagophore formation was affected by these actin polymerization inhibitors during salt stress-induced autophagy. The ratio of ATG (5&8)/ATG8 showed no significant difference in leaves treated with NaCl only or that with NaCl plus CD or LatB ( and ), indicating a normal phagophore formation process in actin filaments-disrupted leaves during salt stress-induced autophagy.
Figure 2. Treatment of leaves with anti-microfilament drugs for 1 h has no effect on salt stress-induced autophagy. (a) Representative confocal images of CFP-ATG8f-labeled autophagic structures in salt stressed-leaves subjected to treatment with 20 μM CD or 25 μM LatB for 1 h in the absence (left) or the presence (right) of E-64d. 100 μM E-64d was pre-infiltrated into leaves to allow 12-h inhibition of vacuolar proteases. Autophagy was induced by infiltration of 150 mM NaCl into leaves for 1-h treatment. CFP-ATG8f is in cyan and chloroplasts are in red. Scale bars: 10 μm. (b) Relative autophagic activity in leaves treated with NaCl and NaCl plus anti-microfilaments drugs (with or without E-64d). The autophagic activity in leaves treated solely with 150 mM NaCl (control) without E-64d was set to 1.0. More than 200 cells were quantified in each treatment. Values represent means ± SE were from 3 independent experiments. Student’s t test was used to determine the significant difference (**, 0.01 < p < 0.05). (c) GFP-ATG8 processing assay in each treatment. Open and closed arrowheads indicate the GFP moiety band and GFP-ATG8f band respectively. GFP-ATG8f and GFP were detected using Anti-GFP antibody. RBCL stained by Ponceau indicates equal loading (lower panel). (d) Representative images showing colocalizations of ATG5-GFP-labeled phagophore with RFP-ATG8f-labeled autophagic structures in salt-stressed leaves. ATG5-GFP is in green and RFP-ATG8f is in red. Colocalized structures are indicated by magenta arrowheads. Scale bars: 10 μm. (e) Ratio of ATG5- and ATG8-colocalized structures to RFP-ATG8f-labeled autophagic structures during salt stress-induced autophagy. More than 120 cells were quantified in each treatment. Values represent means ± SE from 3 independent experiments. n.s means no significant difference (Student’s t test, p > 0.05).
Taken together, these results suggest that short-term disruption of actin filaments by application of anti-microfilament drugs has no effect on nocturnal autophagy or salt stress-induced autophagy.
Disorganization of actin filaments by transiently overexpressing Profilin 3 does not affect autophagy
It is reported that overexpression of AT5G56600/AtProfilin 3 (AtPRF3), an essential actin binding protein involved in regulation of actin polymerization, can destabilize actin filaments in hypocotyl cells in Arabidopsis [Citation32]. Here, we tested the effect of actin network disorganization on autophagy by transiently expressing NbPRF3, the homologue of AtPRF3 in N. benthamiana, tagged with MYC (PRF3-MYC) to disturb actin filaments network in N. benthamiana leaves. Agrobacterium-mediated overexpression of PRF3-MYC and its corresponding control construct cLUC-MYC was confirmed by western blot using Anti-MYC antibody (Figure S4A). Confocal microscopic observation using microfilament reporter YFP-fABD2 revealed that actin filaments were largely disorganized and increasing short filamentous segments or actin filament ends in the form of foci appeared when PRF3-MYC was overexpressed in leaves after 48 h post-infiltration, while little perturbation of actin filaments were observed in control leaves where cLUC-MYC was overexpressed (Figure S4B). Furthermore, overexpression of PRF3-MYC did not affect the number of CFP-ATG8f-labeled autophagic structures formed at levels of basal autophagy (Figure S4C and D). Subsequently, we examined the autophagy level and autophagic flux in PRF3-MYC-overexpressing leaves during nocturnal autophagy. Quantification of autophagic structures labeled by CFP-ATG8f showed no difference between autophagy levels occurring in control and PRF3-MYC-overexpressing cells after 4 h of exposure to darkness, irrespective of the presence of E-64d, indicating that nocturnal autophagy is normal in cells with microfilaments-disruption caused by PRF3-MYC expression ( and ). In addition, in GFP-ATG8 processing assay, the amount of free GFP derived from GFP-ATG8 in PRF3-MYC overexpressing cells was similar to that in control cells in the absence of E-64d and full GFP-ATG8 level was further increased by E-64d treatment ()). These results indicate that disorganization of actin filaments by transient overexpression of PRF3 does not affect nocturnal autophagy.
Figure 3. Overexpression of PRF3-MYC does not affect nocturnal or salt stress-induced autophagy. (a) Representative confocal images of CFP-ATG8f-labeled autophagic structures in cLUC-MYC- or PRF3-MYC-expressing leaves with (lower panel) or without (upper panel) E-64d during nocturnal autophagy. CFP-ATG8f was co-expressed with cLUC-MYC (control) or PRF3-MYC in leaves for 2 days before imaging at the time point D4. 100 μM E-64d was infiltrated into leaves for 12 h to block vacuolar degradation. Scale bars: 10 μm. (b) Relative autophagic activity in PRF3-MYC-expressing leaves (with or without E-64d) during nocturnal autophagy upregulation indicated by CFP-ATG8f. The autophagic activity in cLUC-MYC-expressing leaves without E-64d was set to 1.0. (c) GFP-ATG8 processing assay in control and PRF3-MYC-expressing leaves with or without E-64d during nocturnal autophagy. ProATG8f::GFP-ATG8f was coexpressed with cLUC-MYC or PRF3-MYC for 48 h. Open and closed arrowheads indicate the GFP band and GFP-ATG8f band respectively. RBCL stained by Ponceau indicates equal loading (lower panel). (d) Representative images of CFP-ATG8f-labeled autophagic structures in cLUC-MYC- or PRF3-MYC-expressing leaves with (lower panel) or without (upper panel) E-64d during salt stress-induced autophagy. CFP-ATG8f was co-expressed with cLUC-MYC (control) or PRF3-MYC for 2 days before imaging.150 mM NaCl was infiltrated into leaves for 1 h to induce autophagy. 100 μM E-64d was infiltrated into leaves for 12 h. Scale bars: 10 μm. (e) Relative autophagic activity in PRF3-MYC-expressing leaves (with or without E-64d) during salt stress-induced autophagy. Autophagic activity in cLUC-MYC-expressing leaves without E-64d was set to 1.0. More than 200 cells were quantified in each treatment. Values are means ± SE from 3 independent experiments. Different letters indicate significant difference (ANOVA, p < 0.05). (f) GFP-ATG8 processing assay in control and PRF3-MYC-expressing leaves with or without E-64d during salt stress-induced autophagy. Open and closed arrowheads indicate the GFP band and GFP-ATG8f band respectively. RBCL stained by Ponceau indicate equal loading (lower panel).
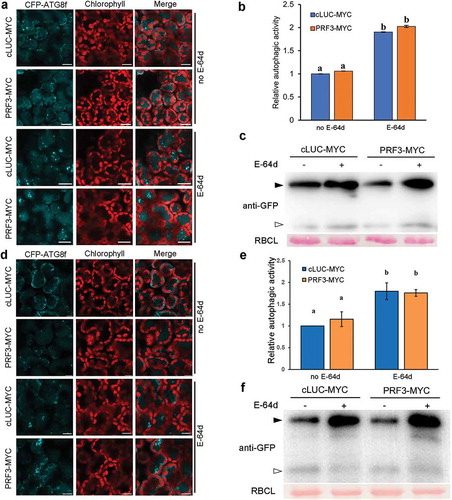
Figure 4. Treatment of ProUBQ10::GFP-ATG8a transgenic Arabidopsis seedlings with anti-microfilament drugs has no effect on salt stress-induced autophagy. (a) Representative confocal images of GFP-ATG8a-labeled autophagic structures in drug-treated root cortex cells. To block autophagic flux, roots were treated with or without 1 μM Con A for 12 h. Roots were treated with 150 mM NaCl plus anti-microfilament drugs for 1 h. Scale bars: 20 μm. (b) Number of autophagic structures per field in each treatment. More than 40 root cortex cells were quantified in each treatment. Values represent means ± SE from 3 independent experiments. Different letters indicate significant difference (ANOVA, p < 0.05). (c) GFP-ATG8 processing assay of seedling in each treatment. Open and closed arrowheads indicate the GFP band and GFP-ATG8a band respectively. Ponceau staining indicates equal loading (lower panel). (d) Quantification of the GFP/GFP-ATG8a ratio shown in (c). Values represent means ± SE from 3 independent experiments. No significant difference was detected among these samples (ANOVA, p > 0.05).
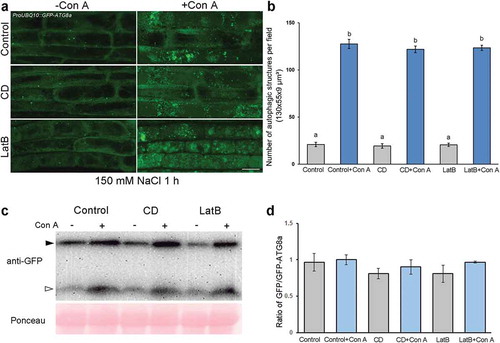
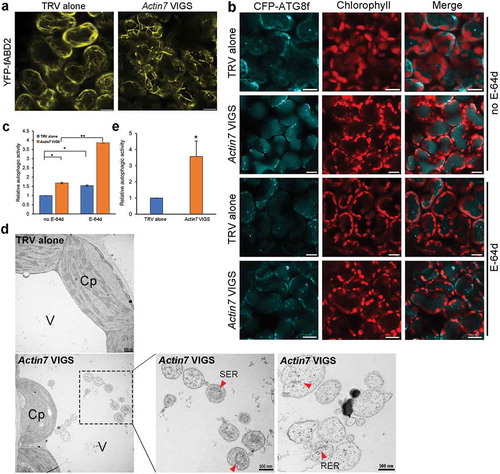
Figure 5. Autophagy is enhanced in Actin7-silenced plants. (a) Silencing of Actin7 (Actin7 VIGS) severely disrupted actin filaments. YFP-fABD2 is in yellow and chloroplasts are in red. Scale bars: 10 μm. (b) Representative confocal images of autophagic structures indicated by CFP-ATG8f in Actin7-silenced plants (Actin7 VIGS) and control plants (TRV alone) with (lower panel) or without (upper panel) E-64d. Confocal images were taken at the end of the day (dark 8 h). Autophagic structures were labeled by CFP-ATG8f. 100 μM E-64d was pre-infiltrated into leaves for 12 h before imaging. Scale bars: 10 μm. (c) Relative autophagic activity in Actin7-silenced plants and control plants. Autophagic activity in TRV alone plants without E-64d treatment was set to 1.0. More than 200 cells were quantified in each treatment. Values are means ± SE from 4 independent experiments. Student’s t test was performed to indicate significant difference (* 0.01 < p < 0.05, ** 0.001 < p < 0.01). (d) Representative TEM images of autophagic bodies in Actin7-silenced plants and control plants. Red arrowheads indicate the autophagic bodies inside the vacuole containing SER or RER. V, vacuole. Cp, chloroplast. SER, smooth ER. RER, rough ER. (e) Relative autophagic activity in Actin7-silenced plants was normalized to that of TRV alone plants. Approximately 30 cells were quantified in each treatment. Student’s t test was performed to indicate significant difference (* 0.01 < p < 0.05).
We also checked the autophagy level and autophagic flux during salt stress in leaves overexpressing PRF3-MYC. In accordance with the observation in nocturnal autophagy process, no significant difference in salt stress-induced autophagic activity was observed in leaves expressing cLUC-MYC and PRF3-MYC, as indicated by CFP-ATG8f ( and ) and GFP-ATG8 processing assay ()). Moreover, normal autophagic flux was observed in salt-stressed leaves overexpressing PRF3-MYC (, and ). These results suggest that overexpression of PRF3 has no effect on salt stress-induced autophagy.
Taken together, these data suggest that disruption of actin filaments by transient overexpression of PRF3 does not affect nocturnal autophagy or salt stress-induced autophagy.
Short-term disruption of actin filaments by anti-microfilament drugs has no effect on autophagy in Arabidopsis
We also tested the effect of microfilament disruption on autophagy in ProUBQ10::GFP-ATG8a transgenic Arabidopsis [Citation33]. Seedlings were subjected to 1 h treatment with 150 mM NaCl for autophagy induction [Citation31] individually or simultaneously with 50 μM CD or 5 μM LatB, which is reported significantly disrupts actin filaments in Arabidopsis [Citation34,Citation35]. Confocal observation of ATG8-labeled autophagic structures and GFP-ATG8 processing assay were then used to monitor the occurrence of autophagy. Application of 50 μM CD or 5 μM LatB individually for 1 h in root cortex cells showed no effect on the number of GFP-ATG8a-labeled autophagic structures (Figure S5A and B), confirming that basal autophagy could occur normally in drug-treated cells. Next, we examined the autophagy level in ProUBQ10::GFP-ATG8a seedlings treated with anti-microfilament drugs after exposure to salt stress. Both comparable numbers of GFP-ATG8a puncta and similar amount of free GFP were observed in seedlings treated with salt solely or simultaneously with CD or LatB. Quantification analysis showed that there was no obvious difference in the ratio of free GFP/GFP-ATG8a between that of drug-treated and wild-type samples. Furthermore, Con A treatment further increased the number of ATG8-labeled autophagic structures and protein levels of full GFP-ATG8 and free GFP in all the tested samples, suggesting that autophagic flux was normal in the absence of Con A (). Taken together, in accordance with that in N. benthamiana, application of anti-microfilament drugs has no effect on either basal or salt stress-induced autophagy.
Prolonged absence of the functional actin cytoskeleton activates autophagy in N. benthamiana
We also tested the role of actin filaments in autophagy by silencing Actin in N. benthamiana using Tobacco rattle virus (TRV)-based virus induced gene silencing (VIGS) [Citation36]. Real-time RT-PCR confirmed that expression of Actin7 was markedly decreased at 2 wk post-agroinfiltration of TRV1 and TRV2-Actin7 (Figure S6A). As indicated by the microfilament reporter YFP-fABD2, severe disorganization of actin filaments occurred in Actin7-silenced leaves as characterized by increasing short segments and thick branches (). We then detected the changes of autophagic activity in Actin7-silenced plants. As expected, a small number of CFP-labeled autophagic structures were observed in the TRV control leaves at the end of night. However, strikingly, a number of CFP-ATG8f-labeled autophagic structures accumulated in Actin7-silenced leaves ( and ). Quantitative analysis showed that the relative autophagic activity in Actin7-silenced plants was about 1.7-fold of that in TRV control in the absence of E-64d. In addition, E-64d treatment significantly increased the number of autophagic structures in control cells as well as Actin7-silenced cells ( and ). These results suggest autophagic flux proceeds normally and autophagy is activated in Actin7-silenced plants, which was further supported by ultrastructural observations with transmission electron microscopy (TEM). In accordance with confocal observations, TEM studies showed that the number of autophagosome and autophagic body was significantly increased in Actin7-silenced plants ( and ).
The above data reveal that disruption of actin filaments by silencing Actin7 in N. benthamiana leads to enhanced basal level of autophagy, which is inconsistent with the effect of short-term interference in actin filaments organization caused by actin filaments-disrupting agents on autophagy. We speculated that the discrepancy might be due to prolonged disruption of actin filaments. To verify this hypothesis, we analyzed the autophagic response in mesophyll cells after the prolonged treatment of 20 μM CD and 25 μM LatB for up to 24 h. YFP-fABD2-labeling observations showed that, compared to the 4-h treatment, this prolonged exposure to anti-microfilament drugs resulted in more severe destruction of actin filaments, evidenced by disappearance of all the filamentous structures and leaving only actin-containing foci (Figure S1 B and C). Then we determined the basal level of autophagy by visualizing CFP-ATG8f-labeled autophagic structures and found that the number of autophagic structures in leaves subjected to 24-h treatment of CD and LatB was conspicuously higher than that in control leaves ( and ). Meanwhile, the number of CFP-ATG8f-labeled structures was further increased by the application of E-64d ( and ), indicating the autophagic flux is normal in leaves treated with CD and LatB for up to 24 h.
Figure 6. 24-h treatment of leaves with 20 μM CD or 25 μM LatB activates autophagy. (a) Representative confocal images of autophagy level in 20 μM CD and 25 μM LatB 24 h-treated leaves with (right) or without (left) E-64d. Images were taken at the end of the day (dark 8 h). Autophagic structures were labeled by CFP-ATG8f. 100 μM E-64d was pre-infiltrated at 12 h ahead of imaging. Scale bars: 10 μm. (b) Relative autophagic activity in drug-treated leaves with or without E-64d. Autophagic activity in control plants without E-64d treatment was set to 1.0. More than 150 cells were quantified in each treatment. Values are means ± SE from 3 independent experiments. Student’s t test was performed to indicate significant difference (* 0.01 < p < 0.05, ** 0.001 < p < 0.01, *** p < 0.001).
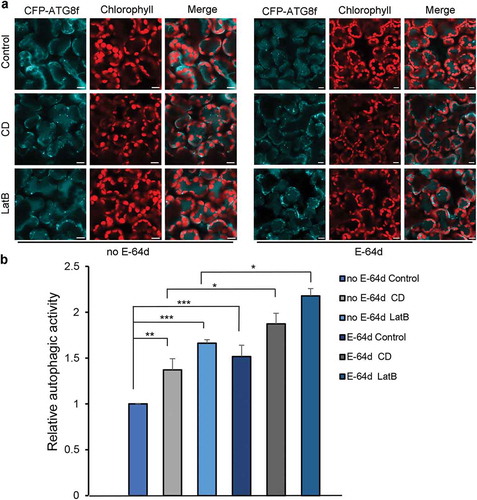
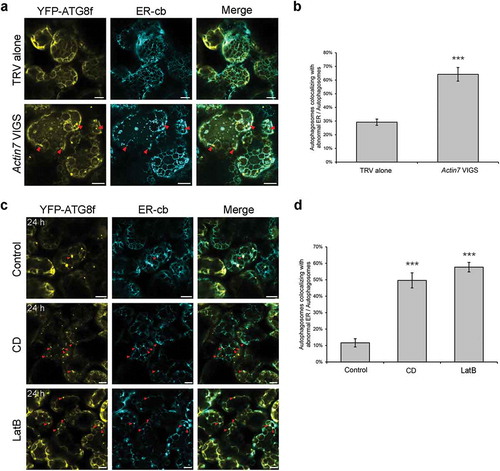
Figure 7. Punctate ER remnants colocalize with autophagic structures in Actin7-silenced plants and 24 h anti-microfilament drug-treated leaves. (a) Representative confocal images showing colocalizations of ER remnants with YFP-ATG8f-labeled autophagic structures (yellow) in Actin7-silenced plants and control plants. ER network was labeled by ER-cb (cyan). Red arrowheads indicate the colocalized structures. Scale bars: 10 μm. (b) Ratio of autophagic structures colocalized with punctate ER remnants to total autophagic structures in Actin7-silenced plants. More than 150 cells were quantified in each treatment. Values are means ± SE from 3 independent experiments. Student’s t test was performed to indicate significant difference (*** p < 0.001). (c) Representative confocal images showing colocalizations of ER remnants with YFP-ATG8f-labeled autophagic structures (yellow) in leaves treated with 20 μM CD or 25 μM LatB for 24 h. Red arrowheads indicate the colocalized structures. Scale bars: 10 μm. (d) Ratio of autophagic structures colocalized with punctate ER remnants to total autophagic structures in drug-treated leaves. More than 100 cells were quantified in each treatment. Values are means ± SE from 3 independent experiments. Student’s t test was performed to indicate significant difference (*** p < 0.001).
Taken together, these results reveal that prolonged absence of the functional actin cytoskeleton in N. benthamiana leads to autophagy activation rather than autophagy deficiency, further supporting that actin filaments are unnecessary for the onset of plant autophagy.
Long-term disruption of actin filaments leads to ER disorganization and ER degradation via autophagy
Ultrastructural analysis of Actin7-silenced plants revealed surprisingly that a number of autophagic bodies containing smooth ER (SER)- and rough ER (RER) fragments-like structures appeared in the central vacuole of Actin7-silenced mesophyll cells but not the nonsilenced control, suggesting possible autophagic degradation of ER structures ()). Moreover, it is reported that cortical ER network overlies the actin filaments in plants and its network formation, remodeling and movement are all largely dependent on actin filaments [Citation37,Citation38]. These findings prompted us to investigate ER network status in actin filaments-disrupted cells and the possible link between ER organization and autophagy induction in N. benthamiana lacking the actin cytoskeleton integrity.
We first checked whether ER morphology is influenced by the integrity of the actin cytoskeleton. A soluble ER marker, named ER-cb, which is composed of a signal peptide of AtWAK2 at the N-terminus, CFP and the ER retention signal HDEL at the C-terminus, was used to label cortical ER network [Citation39,Citation40]. Transient expression of the ER-cb in wild-type N. benthamiana leaves enabled visualization of the intact, polygonal ER network consisting of interconnected tubules and sheet-like cisternae (Figure S7A). However, the cortical ER network was severely disorganized in Actin7-silenced plants, displaying decreased polygons, fragmentation of ER tubules, and increased remnants appearing as segments and punctate structures (Figure S7A). To further confirm the onset of autophagic degradation of ER observed in TEM study ()), confocal microscopy was performed to visualize the subcellular localization of both ER and autophagic structures, indicated by their respective markers, in Actin7-silenced leaves. In the control cells, few autophagic structures and ER fragments appeared and thus rare colocalization between them was observed. In contrast, lots of CFP-ATG8f-labeled autophagic structures formed in Actin7-silenced mesophyll cells and some of them colocalized with the punctate ER remnants (). Quantification analysis showed that, 64% ATG8-labeled autophagic structures overlapped with ER-cb-labeled remnants in Actin7-silenced plants, which is significantly higher than that of control cells (29%) ()). These results reveal that perturbation of the actin cytoskeleton by silencing Actin7 results in ER disorganization and ER degradation via autophagy pathway.
Since disruption of actin filaments by prolonged drug treatment for 24 h also caused autophagy induction, we reasoned that similar autophagic degradation of ER might occur in these cells. Visualization of ER morphology in drug-treated cells showed that 24-h treatment with 20 μM CD and 25 μM LatB led to drastic disorganization of cortical ER network, characterized by disappearance of the most polygonal tubules and increased punctate ER remnants (Figure S7B). Colocalization assay showed a large portion of the autophagic structures contained ER remnants as substrates in drug-treated cells (CD ~50%, LatB ~58%), which was apparently higher than that of control cells (~12%) ( and ), suggesting that autophagic degradation of ER is evident in actin filaments-depolymerized cells undergoing 24-h treatment with CD and LatB. In addition, we also examined the colocalization of autophagic structures with other organelles in Actin7-silenced plants, including mitochondria, Golgi, peroxisomes and chloroplasts, however, rare colocalization was observed (Figure S8), suggesting that the uptake of ER fragments by autophagic structures is probably the specific event.
Taken together, long-term disruption of actin filaments in N. benthamiana leads to disorganization of cortical ER network and subsequent degradation via autophagy.
ATG2, 3, 5, 6 and 7 are required for autophagy induction in Actin7-silenced plants
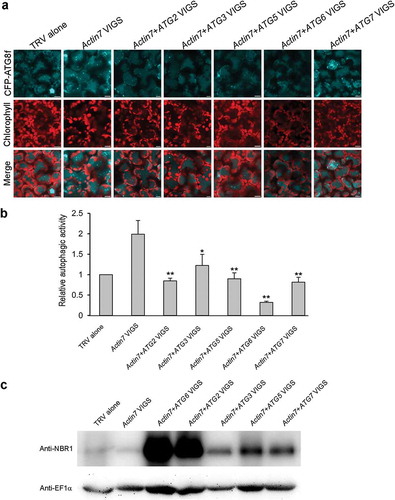
To evaluate the involvement of the core ATG genes in autophagy induction in Actin7-silenced plants, we co-silenced Actin7 with ATG2, ATG3, ATG5, ATG6 or ATG7, respectively, in N. benthamiana (Figure S6A and B) and checked the autophagy onset in these plants by using the autophagy marker CFP-ATG8f. Confocal observations revealed that the number of autophagic structures was significantly decreased in all the Actin7/ATG co-silenced plants, compared to that in Actin7-silenced plants ( and ). Furthermore, we determined autophagy levels by detecting the accumulation of Joka2/NBR1, a functional hybrid homolog of two mammalian selective autophagy cargo receptors, p62 and NBR1, in these co-silenced plants, which has been established to be a selective autophagy substrate and also a suitable autophagy marker for autophagic flux in plants [Citation41,Citation42]. In all the Actin7/ATG co-silenced leaves, the accumulation of Joka2 increased dramatically compared to that in TRV control and Actin7-silenced leaves ()), suggesting that autophagic flux is blocked in plants silenced for all the tested ATG gene in this study. Together, these data indicate that the autophagy activation in Actin7-silenced plants is dependent on ATG2, 3, 5, 6 and 7.
Figure 8. ATG2, 3, 5, 6 and 7 are required for autophagy induction in Actin7-silenced plants. (a) Representative confocal images showing autophagy level in Actin7-silenced and Actin7/ATG-cosilenced plants treated with E-64d. Autophagic structures were labeled by CFP-ATG8f. 100 μM E-64d was pre-infiltrated at 12 h ahead of imaging. CFP-ATG8f is in cyan and chloroplasts are in red. Scale bars: 10 μm. (b) Relative autophagic activity in Actin7-silenced and Actin7/ATG-cosilenced plants treated with E-64d. Autophagic activity in TRV alone plants was set to 1.0. More than 180 cells were quantified in each treatment. Values are means ± SE from 3 independent experiments. Student’s t test was performed to indicate significant difference (* 0.01 < p < 0.05, ** 0.001 < p < 0.01). (c) Joka2 was accumulated in Actin7/ATG-cosilenced plants. Joka2 was detected using Anti-NBR1 antibody. EF1α level was used here to indicate equal loading.
Discussion
Roles of actin filaments in autophagy
Autophagy is a multi-step process mainly involving induction, vesicle nucleation, phagophore expansion, autophagosome closure, autolysosome formation and substrate degradation. In the case of selective autophagy, there is an additional step, cargo recognition, ahead of autophagosome formation [Citation43]. Investigations on the regulation mechanisms of autophagy in the past decade have suggested that cytoskeletal components, such as microtubules and actin filaments, could participate in autophagy [Citation21,Citation22]. In yeast, actin filament is involved in several selective types of autophagy, including Cvt pathway, pexophagy and starvation-induced ER degradation, probably through mediating cargo selection, but is unnecessary for bulk autophagy [Citation18,Citation19]. Mounting evidence reveals that, in animal cells, the participation of the actin cytoskeleton is throughout steps of autophagosome formation and its maturation into autolysosome, irrespective of the selectivity of autophagy [Citation22]. Thus, the roles of actin filaments may vary between different model systems. Recently, Arabidopsis NAP1, a component of SCAR/WAVE complex responsible for actin nucleation in plants, is reported to form puncta colocalizing with ATG8 upon applied pressure, and its corresponding mutant is defective in starvation-induced autophagy, raising a possibility that actin filaments might participate in plant autophagy [Citation26]. However, it is unclear whether the engagement of NAP1 in autophagy is indeed dependent on its actin nucleation activity. In this study, we directly disturbed the actin cytoskeleton through several approaches, including application of anti-microfilament drugs, transient overexpression of PRF3 and silencing Actin7 in N. benthamiana, analyzed both the short-term and long-term effect of disruption of actin filaments on basal autophagy, nocturnal autophagy and salt stress-induced autophagy, and found no apparent inhibitory impact on bulk autophagic flux in plants (, , , and ). Similarly, in Arabidopsis, application of anti-microfilament drugs had no effect on basal autophagy or salt stress-induced autophagy (). Furthermore, prolonged absence of the functional actin cytoskeleton by knocking down expression of Actin7 or subjecting leaves to 24-h treatment with CD and LatB resulted in autophagy activation involving clearance of disorganized ER structures (–), which further excluded the possibility of positive involvement of microfilaments in plant autophagy. The activation of autophagy in cells lacking actin integrity for long periods of time (24 h for drug treatment and at least 2 weeks for gene silencing) could be an indirect or secondary cellular response to depolymerization of actin filaments. In fact, the cortical ER network was severely disorganized (Figure S7), which could be a trigger of autophagy [Citation44,Citation45]. Therefore, to diminish the secondary effects caused by actin filaments depolymerization as much as possible, it is necessary to shorten the treatment time and study autophagic responses to the depolymerization event in a prompt induction system. It is the reason why nocturnal autophagy and salt stress-induced autophagy were chosen as the model systems, which can be efficiently upregulated within 4 h and 1 h respectively. In this study, treatment of mesophyll cells with 20 μM CD and 25 μM LatB was established to have no effect on the basal level of autophagy (Figure S2), nocturnal autophagy () or salt stress-induced autophagy (). In addition, transient overexpression of PRF3 in N. benthamiana disturbed the actin cytoskeleton but also had no effect on basal level of autophagy (Figure S4), the occurrence of nocturnal autophagy and salt stress-induced autophagy (), which mimics the short-term effects of treatment with CD and LatB. We also analyzed the autophagic response to microfilament disruption in Arabidopsis caused by drug treatment in the stable transgenic marker line ProUBQ10::GFP-ATG8a by microscopic observation and biochemical analysis of cleavage of GFP-ATG8. Little effect of aberrant actin cytoskeleton was observed on salt stress-induced autophagy in Arabidopsis. Taken together, our findings reveal that actin filaments are dispensable for bulk autophagy in plants, which are consistent with the observations in yeast in this aspect but distinct from that in animal cells [Citation21,Citation22]. However, we cannot exclude the possibilities that actin filaments might participate in selective autophagy or other unidentified types of autophagy in plants.
In contrast to the dispensable role of actin filaments in bulk autophagy in plants, intact microtubules have been demonstrated to be important for efficient biogenesis of autophagosomes during autophagy induction in our previous work [Citation15]. The association of ATG6 with microtubules may facilitate efficient recruitment of other ATG proteins to assemble scaffolds for autophagosome biogenesis. Our findings reveal different roles of those two major cytoskeleton components, microtubules and actin filaments, in plant autophagy. With the regard to involvement of cytoskeleton in autophagy, both microtubules and microfilaments in mammals play important roles in autophagy throughout steps of autophagosome formation and its maturation into autolysosome, while, in yeast, microtubules seem to be unnecessary for autophagy, and microfilaments participate in selective autophagy rather than bulk autophagy [Citation21,Citation22]. Accordingly, the contributions of cytoskeleton to autophagy vary in different model systems.
In this study, several routine approaches, including GFP-ATG8 processing assay, was applied to monitor autophagic flux. The GFP-ATG8 processing assay was developed based on the fact that ATG8-coated autophagosomes, during autophagy induction, constantly fuse with the vacuole, in which autophagic substrates as well as ATG8 are degraded by proteases while the degradation of GFP is delayed due to its relative stability to vacuole hydrolases [Citation46]. The increased free GFP levels derived from cleavage of GFP-ATG8 in vacuoles could be an indicator for autophagy occurrence [Citation46–Citation48]. An increased ratio of free GFP/GFP-ATG8 during autophagy induction was also recently introduced by two publications to be indicative of autophagic flux in plants [Citation49,Citation50]. Application of protease inhibitors or compounds that modify lysosomal/vacuole pH is usually recommended to be useful for confirming normal autophagic flux [Citation51]. However, it should be noted that whether the cleavage of GFP-ATG8 occurs or its by-product, free GFP, increases closely depend on the degradative capacity of the lysosomal or vacuolar compartments, which will be influenced by varying degrees when different types of inhibitors are applied to different cell lines under different concentrations [Citation52]. Under saturating concentrations for tested cells, the cleavage of GFP-ATG8 will be completely blocked and no free GFP is released, while, under unsaturating concentrations, a proportion of GFP-ATG8 could still be processed which lead to the appearance of free GFP [Citation52]. Concanamycin A, an inhibitor of vacuolar proton pumps, which inhibits vacuolar acidification and indirectly blocks the activity of vacuolar hydrolases, were previously adopted in Arabidopsis to carefully assess autophagic flux in GFP-ATG8 processing assay. Treatment with 1 μM Con A for 16 h in nitrogen-starved autophagic marker lines ProUBQ10::GFP-ATG8a leads to a further increase of band intensity of both GFP-ATG8 and its cleavage product when compared with starvation treatment in the absence of Con A [Citation47]. The same effect of Con A on protein levels of free GFP and GFP-ATG8 was also observed in our experiment for testing the autophagic response to disruption of microfilaments in the ProUBQ10::GFP-ATG8a Arabidopsis seedlings. Quantification analysis showed that there was no obvious difference between the ratio of GFP/GFP-ATG8 in wild-type and that of drug-treated samples (). As with Con A, E-64d, a cysteine protease inhibitor, was also used to monitor changes of either number of autophagic structures or cleavage of GFP-ATG8 for autophagy assessment in N. benthamiana. Treatment with 100 μM E-64d for 8–12 h in N. benthamiana led to a further increase of both numbers of ATG8-labeled structures and protein levels of full GFP-ATG8 but less consistent change in free GFP (, , ). The degradative capacity of vacuoles resulted from inhibition of cysteine protease in different samples may account for this inconsistence. That is, severe blockage of hydrolase activity will lead to a reduced GFP-ATG8 process, leading to the enhanced accumulation of GFP-ATG8, while less complete suppression makes uncertain proportion of GFP-ATG8 process normally to release GFP to the free GFP pools, thus leading to uncertain level of GFP. Strikingly, however, either the protein level of full GFP-ATG8 or the summed amount of full GFP-ATG8 and free GFP was consistently increased in E-64d-treated cells compared to control cells during autophagy induction (, , and f), which can be a good indicator for the normal autophagic degradation within vacuoles.
Disorganized ER in actin filaments-disrupted cells could be a selective substrate of autophagy
The ER network is a dynamic, interconnected structures composed of tubules and sheet-like cisternae [Citation38]. It is reported that both of the network formation and remodeling of ER require the actin cytoskeleton [Citation38,Citation53]. In plants, either drug-induced actin depolymerization or overexpression of a truncated class XI myosin affects ER network remodeling and produces a more persistent network of ER tubules and larger persistent cisternae [Citation38]. Severe ER disorganization was also observed in Actin7-silenced cells or drug-treated cells undergoing a prolonged treatment with CD and LatB for up to 24 h (Figure S7). More interestingly, a number of ER fragment-like structures sequestered in single-membrane-bound autophagic bodies were observed in the central vacuole of Actin7-silenced cells ()), which was further confirmed by the colocalization between ATG8-labeled autophagic structures and ER-cb-labeled punctate ER remnants ( and ). Similarly, the colocalization of punctate ER remnants with autophagic structures was also observed in cells subjected to prolonged drug-treatment ( and ). These results suggest the occurrence of autophagic degradation of ER in cells lacking actin integrity for long periods of time. We also checked the morphology of other organelles, including mitochondria, Golgi, peroxisomes and chloroplasts, in plant cells undergoing 24-h treatment with anti-microfilament drugs, no obvious morphological changes and also rare colocalizations of them with autophagic structures were observed (Figure S8). Ultrastructural analysis of autophagic substrates in Actin7-silenced cells uncovered that few other organelles but ER fragments were engulfed by membrane-bound autophagic structures (data not shown). These findings indicate that ER malformation and degradation was the specific event in cells lacking intact actin cytoskeleton for long periods of time. It is reported that, in Arabidopsis, ER stress caused morphological changes in ER structure and autophagy induction, which could be delivered to the vacuole by autophagosomes for degradation [Citation45]. Similarly, we speculated that the severe ER disorganization caused by prolonged absence of intact actin network might become a trigger for ER autophagy and its subsequent specific removal. Further investigation uncovers that autophagic degradation of ER caused by prolonged disruption of actin filaments requires ATG2, ATG3, ATG5, ATG6 and ATG7. Taken together, we identified a kind of selective autophagy of ER triggered by prolonged absence of actin integrity, during which actin filaments are obviously not necessary elements.
Methods and materials
Plant materials
N. benthamiana plants were grown in growth rooms at 25°C under a 16-h light/8-h dark cycle. Arabidopsis thaliana seeds were surface-sterilized and germinated in 1/2 Murashige-Skoog (MS; Duchefa, M0222.0050), 1% (w:v) sucrose (Sinopharm Chemical Reagent, 10021418) solid medium (pH 5.8). Seedlings were grown at 20 to 22°C under a 16-h light/8-h dark cycle.
Plasmid constructs
pTRV1 was described previously [Citation36]. YFP-fABD2, YFP-ATG8f and RFP-ATG8f were generated by cloning the respective target genes into pCAMBIA1300 vectors with corresponding tags as described previously [Citation54]. The VIGS vector TRV2-Actin7, TRV2-ATG2+Actin7, TRV2-ATG3+Actin7, TRV2-ATG5+Actin7, TRV2-ATG6+Actin7, TRV2-ATG7+Actin7 were generated by cloning the respective cDNA fragments into pTRV2-LIC vector. All vectors were confirmed by DNA sequencing. Primers used for gene cloning are provided in Table S1.
Salt stress-induced autophagy in N. benthamiana
The first and second unfolded leaves of each plant were used. Agrobacterium carrying CFP-ATG8f or ProATG8f::GFP-ATG8f was infiltrated into the whole leaf for 48–60 h expression. E-64d (100 μM; Sigma-Aldrich, E8640) was pre-infiltrated into half leaf to allow 12-h inhibition. To induce the salt stress and disrupt actin filaments, solution (150 mM NaCl (Sinopharm Chemical Reagent, 10019318)+20 μM CD (Abcam, ab143484), 150 mM NaCl+25 μM LatB (Abcam, ab144291), 150 mM NaCl+0.25% DMSO (Amresco, 0231), H2O+20 μM CD, H2O+25 μM LatB or H2O+0.25% DMSO) was infiltrated into the whole leaf, half of which was pre-treated with E-64d previously, for 1 h. Medical injectors were used for both agrobacterium and drug infiltration. Samples for confocal observation or Western blot detection were collected by 7 mm sized hole puncher.
RT-PCR
Real-time PCR was performed to test the silence of ATG genes and Actin7 in the individual VIGS plants. RT-PCR was performed as previously described [Citation55]. Primers used for RT-PCR are provided in Table S1.
Confocal microscopy
Confocal imaging was acquired using an inverted Zeiss LSM 710 three-channel laser scanning microscope. For the observation of N. benthamiana leaves, CFP-ATG8f, YFP-fABD2, ATG5-GFP, YFP-ATG8f, RFP-ATG8f, ER-cb, G-cb, mt-cb and px-cb was introduced into Agrobacterium tumefaciens GV3101. The agrobacterium harboring the corresponding plasmid was infiltrated into N. benthamiana leaves. After about 40–60 h of expression, the samples were imaged by Zeiss LSM 710 microscope. For Arabidopsis, 10-day-old ProUBQ10::GFP-ATG8a seedlings were immersed in 150 mM NaCl in the presence or absence of anti-microfilament drugs for 1 h. The root cortex cells were imaged by Zeiss LSM 710 microscope. For the observation of CFP-ATG8f, CFP was excited at 405 nm and detected at 450–580 nm. For the observation of YFP-fABD2, YFP was excited at 514 nm and excited at 530–570 nm. For the observation of ATG5-GFP or ProUBQ10::GFP-ATG8a, GFP was excited at 488 nm and detected at 494–584 nm. For the observation of RFP-ATG8f, RFP was excited at 594 nm and detected at 599–639 nm.
TEM observation
TEM observation was described previously [Citation14]. Generally, leaf samples infiltrated with 100 μM E-64d (Sigma-Aldrich, E8640) for 8–12 h were sliced into small pieces (1.5 x 1.5 mm2) and infiltrated with paraformaldehyde (Electron Microscopy Sciences, 157–8)-glutaraldehyde (SPI-CHEM, 02607-BA) fixative solution (2 wt.%/2.5 wt.%, pH 7.2) in 0.1 M phosphate buffer for fixation. After staining with uranyl acetate and lead citrate, samples were observed by Hitachi H-7650 electron microscope.
VIGS assay
pTRV1 and pTRV2-related vectors was transformed into GV3101. VIGS assay was performed as described previously [Citation16].
Western blot
Total proteins from leaves were extracted in 2 × loading buffer containing 5% 2-Mercaptoethanol (Sigma-Aldrich, M3148) and used for western blotting. Anti-MYC antibody (Abmart, M20002L) was used at 1:5000 to test the expression of PRF3-MYC and cLUC-MYC. Anti-NBR1 (Agrisera, AS14 2805) antibody was used at 1:3000 to verify the endogenous protein level of Joka2. For GFP-ATG8f processing, construct ProATG8f::GFP-ATG8f was used and expressed in leaves for 48 h before sampling. Anti-GFP (Chromotek, 3h9-100) antibody was used at 1:5000 to verify the protein level of GFP-ATG8 and GFP.
Supplemental Material
Download Zip (20.2 MB)Acknowledgments
We thank Taijoon Chung for providing transgenic ProUBQ10::GFP-ATG8a Arabidopsis seeds.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary material for this article can be accessed here.
Additional information
Funding
References
- Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem. 2017 Jun 20;86:193–224. PubMed PMID: 28460188.
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007 Oct;9(10):1102–1109. PubMed PMID: 17909521.
- Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010 Dec 3;330(6009):1344–1348. PubMed PMID: 21127245; PubMed Central PMCID: PMCPMC3010857.
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001 Mar;2(3):211–216. PubMed PMID: 11265251.
- Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. PubMed PMID: 22242963.
- Marshall RS, Vierstra RD. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 2018 Apr 29;69:173–208. PubMed PMID: 29539270.
- Avin-Wittenberg T, Fernie AR. At long last: evidence for pexophagy in plants. Mol Plant. 2014 Aug;7(8):1257–1260. PubMed PMID: WOS:000340438200001.
- Bassham DC. Plant autophagy-more than a starvation response. Curr Opin Plant Biol. 2007 Dec;10(6):587–593. PubMed PMID: WOS:000251635800008.
- Doelling JH, Walker JM, Friedman EM, et al. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002 Sep 6;277(36):33105–33114. PubMed PMID: WOS:000177859000092.
- Hanaoka H, Noda T, Shirano Y, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002 Jul;129(3):1181–1193. PubMed PMID: 12114572; PubMed Central PMCID: PMCPMC166512.
- Haxim Y, Ismayil A, Jia Q, et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife. 2017 Feb 28;6. PubMed PMID: WOS:000397642200001. DOI:10.7554/eLife.23897.
- Ishida H, Izumi M, Wada S, et al. Roles of autophagy in chloroplast recycling. Biochim Biophys Acta. 2014 Apr;1837(4):512–521. PubMed PMID: 24269172.
- Liu YM, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009 Oct 1;5(7):954–963. PubMed PMID: WOS:000272691100005.
- Wang Y, Yu B, Zhao J, et al. Autophagy contributes to leaf starch degradation. Plant Cell. 2013 Apr;25(4):1383–1399. PubMed PMID: 23564204; PubMed Central PMCID: PMC3663275.
- Wang Y, Zheng X, Yu B, et al. Disruption of microtubules in plants suppresses macroautophagy and triggers starch excess-associated chloroplast autophagy. Autophagy. 2015;11(12):2259–2274. PubMed PMID: 26566764; PubMed Central PMCID: PMCPMC4835195.
- Liu Y, Schiff M, Czymmek K, et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121(4):567–577. PubMed PMID: 15907470
- Winder SJ, Ayscough KR. Actin-binding proteins. J Cell Sci. 2005 Feb 15;118(Pt 4):651–654. PubMed PMID: 15701920.
- Reggiori F, Monastyrska I, Shintani T, et al. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005 Dec;16(12):5843–5856. PubMed PMID: 16221887; PubMed Central PMCID: PMCPMC1289426.
- Hamasaki M, Noda T, Baba M, et al. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005 Jan;6(1):56–65. PubMed PMID: 15569245.
- Monastyrska I, He C, Geng J, et al. Arp2 links autophagic machinery with the actin cytoskeleton. Mol Biol Cell. 2008 May;19(5):1962–1975. PubMed PMID: 18287533; PubMed Central PMCID: PMCPMC2366845.
- Monastyrska I, Rieter E, Klionsky DJ, et al. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc. 2009 Aug;84(3):431–448. PubMed PMID: 19659885; PubMed Central PMCID: PMCPMC2831541.
- Kast DJ, Dominguez R. The cytoskeleton-autophagy connection. Curr Biol. 2017 Apr 24;27(8):R318–R326. PubMed PMID: 28441569; PubMed Central PMCID: PMCPMC5444402.
- Mi N, Chen Y, Wang S, et al. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat Cell Biol. 2015 Sep;17(9):1112–1123. PubMed PMID: 26237647.
- Lee JY, Koga H, Kawaguchi Y, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo J. 2010 Mar 3;29(5):969–980. PubMed PMID: 20075865; PubMed Central PMCID: PMC2837169.
- Mostowy S, Sancho-Shimizu V, Hamon MA, et al. p62 and NDP52 proteins target intracytosolic shigella and listeria to different autophagy pathways. J Biol Chem. 2011 Jul 29;286(30):26987–26995. PubMed PMID: 21646350; PubMed Central PMCID: PMCPMC3143657.
- Wang P, Richardson C, Hawes C, et al. Arabidopsis NAP1 regulates the formation of autophagosomes. Curr Biol. 2016 Aug 8;26(15):2060–2069. PubMed PMID: 27451899.
- Baluska F, Jasik J, Edelmann HG, et al. Latrunculin B-induced plant dwarfism: plant cell elongation is F-actin-dependent. Dev Biol. 2001 Mar 1;231(1):113–124. PubMed PMID: 11180956.
- Palevitz BA. Cytochalasin-induced reorganization of actin in allium root-cells. Cell Motil Cytoskel. 1988;9(4):283–298. PubMed PMID: WOS:A1988N533200001.
- Sheahan MB, Staiger CJ, Rose RJ, et al. A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 2004 Dec;136(4):3968–3978. PubMed PMID: 15557099; PubMed Central PMCID: PMCPMC535829.
- Le Bars R, Marion J, Le Borgne R, et al. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun. 2014 Jun 20;5:4121. PubMed PMID: 24947672.
- Luo LM, Zhang PP, Zhu RH, et al. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front Plant Sci. 2017 Aug 22;8. PubMed PMID: WOS:000408210500001. DOI:10.3389/fpls.2017.01459.
- Fan TT, Zhai HH, Shi WW, et al. Overexpression of profilin 3 affects cell elongation and F-actin organization in Arabidopsis thaliana. Plant Cell Rep. 2013 Jan;32(1):149–160. PubMed PMID: WOS:000313009000013.
- Kim J, Lee H, Lee HN, et al. Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell. 2013 Dec;25(12):4956–4966. PubMed PMID: 24368791; PubMed Central PMCID: PMC3903998.
- Kandasamy MK, Meagher RB. Actin-organelle interaction: association with chloroplast in arabidopsis leaf mesophyll cells. Cell Motil Cytoskeleton. 1999 Oct;44(2):110–118. PubMed PMID: 10506746.
- Grebe M, Xu J, Mobius W, et al. Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol. 2003 Aug 19;13(16):1378–1387. PubMed PMID: 12932321.
- Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002 Sep;31(6):777–786. PubMed PMID: 12220268.
- Boevink P, Oparka K, Santa Cruz S, et al. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998 Aug;15(3):441–447. PubMed PMID: 9750355.
- Sparkes I, Runions J, Hawes C, et al. Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell. 2009 Dec;21(12):3937–3949. PubMed PMID: 20040535; PubMed Central PMCID: PMCPMC2814503.
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007 Sep;51(6):1126–1136. PubMed PMID: 17666025.
- Wang S, Xie K, Xu G, et al. Plant G proteins interact with endoplasmic reticulum luminal protein receptors to regulate endoplasmic reticulum retrieval. J Integr Plant Biol. 2018 Jul;60(7):541–561. PubMed PMID: 29573168.
- Zhou J, Wang J, Cheng Y, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9(1):e1003196. PubMed PMID: 23341779; PubMed Central PMCID: PMC3547818.
- Xu G, Wang S, Han S, et al. Plant bax inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy. 2017 Jul 3;13(7):1161–1175. PubMed PMID: 28537463; PubMed Central PMCID: PMCPMC5529081.
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004 Apr;6(4):463–477. PubMed PMID: 15068787.
- Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006 Dec;26(24):9220–9231. PubMed PMID: 17030611; PubMed Central PMCID: PMCPMC1698520.
- Liu Y, Burgos JS, Deng Y, et al. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell. 2012 Nov;24(11):4635–4651. PubMed PMID: 23175745; PubMed Central PMCID: PMCPMC3531857.
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004 Jul 16;279(29):29889–29894. PubMed PMID: 15138258; PubMed Central PMCID: PMCPMC1712665.
- Shin KD, Lee HN, Chung T. A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells. 2014 May;37(5):399–405. PubMed PMID: 24805779; PubMed Central PMCID: PMCPMC4044311.
- Bassham DC. Methods for analysis of autophagy in plants. Methods. 2015 Mar;75:181–188. PubMed PMID: 25239736.
- Marshall RS, Li FQ, Gemperline DC, et al. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/Ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015 Jun 18;58(6):1053–1066. PubMed PMID: WOS:000360986700017.
- Ustun S, Hafren A, Liu Q, et al. Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell. 2018 Mar;30(3):668–685. PubMed PMID: 29500318; PubMed Central PMCID: PMCPMC5894834.
- Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012 Apr;8(4):445–544. PubMed PMID: 22966490; PubMed Central PMCID: PMC3404883.
- Ni HM, Bockus A, Wozniak AL, et al. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011 Feb;7(2):188–204. PubMed PMID: 21107021; PubMed Central PMCID: PMCPMC3039769.54.
- Ueda H, Yokota E, Kutsuna N, et al. Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc Natl Acad Sci U S A. 2010 Apr 13;107(15):6894–6899. PubMed PMID: 20351265; PubMed Central PMCID: PMCPMC2872430.
- Zhao JP, Liu Q, Zhang HL, et al. The rubisco small subunit is involved in tobamovirus movement and Tm-2(2)-mediated extreme resistance. Plant Physiol. 2013 Jan;161(1):374–383. PubMed PMID: WOS:000312964000030.
- Liu Y, Burch-Smith T, Schiff M, et al. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J Biol Chem. 2004 Jan 16;279(3):2101–2108. PubMed PMID: 14583611.
