ABSTRACT
Mitophagy, the clearance of surplus or damaged mitochondria or mitochondrial parts by autophagy, is important for maintenance of cellular homeostasis. Whereas knowledge on programmed and stress-induced mitophagy is increasing, much less is known about mechanisms of basal mitophagy. Recently, we identified SAMM50 (SAMM50 sorting and assembly machinery component) as a receptor for piecemeal degradation of components of the sorting and assembly machinery (SAM) complex and mitochondrial contact site and cristae organizing system (MICOS) complexes. SAMM50 interacts directly with Atg8-family proteins through a canonical LIR motif and with SQSTM1/p62 to mediate basal piecemeal mitophagy. During a metabolic switch to oxidative phosphorylation (OXPHOS), SAMM50 cooperates with SQSTM1 to mediate efficient piecemeal mitophagy.
Mitochondria play crucial roles in several cellular activities including energy and fat metabolism, signaling, innate immunity, apoptosis, calcium storage and thermogenesis. Mitochondrial quality control is vital to cellular homeostasis in particular and to organismal health in general. As part of this quality control, entire mitochondria, or parts of mitochondria, that become damaged or surplus, are removed and degraded in the lysosome through a selective autophagy processes called mitophagy. This way, dysfunctional mitochondria are effectively removed to prevent apoptosis and disease. Basal mitophagy occurs constitutively to recycle whole (wholesale mitophagy) or parts (piecemeal mitophagy) of mitochondria. Conversely, programmed mitophagy occurs under certain cellular events including cellular development and differentiation. Upon excessive stress and damage, mitophagy is induced to removed damaged mitochondria and protect the cell. Stress- or damaged-induced mitophagy is mainly regulated by the PINK1 (PTEN induced kinase 1)- and the E3-ubiquitin ligase PRKN/Parkin-mediated pathway. Most studies in mitophagy have been focused on understanding how programmed and stress-induced mitophagy is executed but relatively little is known about mechanisms of basal mitophagy.
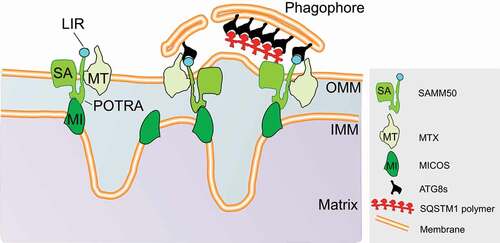
During the course of our studies of the soluble selective autophagy receptor SQSTM1/p62, we immunoprecipitated endogenous SQSTM1 and used mass spectrometry to identify interactors. Several mitochondrial proteins were identified including SAMM50, the pore-forming component of the SAM complex [Citation1]. CRISPR-Cas9-mediated knockdown of SAMM50 in HeLa cells results in reduced levels of a number of mitochondrial proteins and disorganized mitochondrial cristae. The latter finding supports previous reports of the role of SAMM50 in assembly and integration of β-barrel proteins in the mitochondrial outer membrane and maintenance of normal cristae morphology. We also discovered that the N-terminal polypeptide transport associated (POTRA) domain in SAMM50 is dispensible for its role in β-barrel proteins asssembly. From protease-protection and split-fluorescent self-complementation assays, we concluded that the N-terminal segment preceeding the POTRA domain is oriented toward the cytosol. This feature differentiates human SAMM50 from its yeast and bacterial homologs.
Because SAMM50 interacts directly with SQSTM1, we examined whether it has any role in mitophagy. Indeed, using bafilomycin A1 to block lysosomal degradation in wild-type (WT) and SAMM50-depleted cells we discovered that the degradation of a subset of proteins including the SAM complex members MTX1 (metaxin 1) and MTX2, the MICOS complex proteins CHCHD3/MIC19 and IMMT/MIC60, TOMM complex proteins TOMM40 and TOMM20 and the fission and fusion GTPases MFN1, MFN2 and DNM1L/DRP1 are dependent on SAMM50. Using cells stably expressing CHCHD3/MIC19-EGFP and CHCHD3/MIC19-Keima, we could confirm colocalization with, and degradation of, mitochonrial parts by the lysosome and showed that this form of basal mitophagy is executed in a peicemeal fashion. Reconstitution of SAMM50-depleted cells with WT SAMM50 rescues basal mitophagy whereas SAMM50 lacking the N-terminal segment (amino acids 1–40) does not rescue, pointing to the requirement of the SAMM50 N-terminal segment in basal mitophagy.
SQSTM1 is involved in SAMM50-mediated basal mitophagy, but is dispensible. Because several mitophagy receptors depend on human Atg8-family proteins to facilitate mitophagy, we hypothesized that Atg8-family proteins may be required for SAMM50-dependent basal piecemeal mitophagy. We made knockout HeLa cells of all six functional members of the Atg8 family and found that they are indeed necessary for basal mitophagy. By an unbiased peptide array scan we identified a single canonical LC3-interacting region (LIR) motif in the SAMM50 N-terminal region. Structure determination by x-ray crystallography of the SAMM50 LIR bound to GABARAP and to GABARAPL1 demonstrates canonical LIR interactions with the LIR docking sites of these two Atg8-family proteins which bind with highest affinity as determined by biolayer inferometry and GST affinity-isolation assays. Reconstitution studies showed that the LIR motif of SAMM50 is required for basal piecemeal mitophagy.
Normal cells often generate ATP by mitochondrial oxidative phosphorylation (OXPHOS) while cancer cells usually rely on aerobic glycolysis. However, when normal or cancer cells are grown in glucose-free media, they upregulate OXPHOS or undergo a metabolic switch from aerobic glycolysis to OXPHOS, respectively. This leads to an increase in basal mitophagy required to maintain mitochondrial integrity and cellular homeostasis. SQSTM1 plays role in OXPHOS-induced mitophagy and in expression of key OXPHOS enxymes. We hypothesized that SQSTM1 cooperates with SAMM50 during OXPHOS-induced mitophagy. Consistent with this notion, a metaoblic switch leads to increased recruitment of SQSTM1 to mitochondria in a SAMM50-dependent manner and the interaction between SAMM50 and SQSTM1 increases when OXPHOS is induced. Using a tanden-tagged COX8-EGFP-mCHERRY mitophagy reporter, we discovered that knockout of Sqstm1 in mouse embryonic fibroblast (MEF) cells blocks OXPHOS-induced mitophagy. Reconstitution of kncokout cells with WT SQSTM1, but not with a SAMM50-interaction-deficient mutant, rescues basal mitophagy in these MEF cells. A significant reduction in both ATG13 and GABARAP puncta in sqstm1 knockout MEF cells is seen compared to WT. These results show that SQSTM1 may function beyond recruitment of cargos, as a platform for autophagosome assembly on mitochondria during OXPHOS. This hypothesis is supported by the fact that mitochondrial parts to be degraded are first recruited to SQSTM1-positive punta before they are shuttled to the lysosome. The exclusive requirement of SQSTM1 during OXPHOS-induced, but not normal, basal piecemeal mitophagy may arise due to processivity and efficiency of the basal mitophagy process required to maintain mitochondrial quality control during OXPHOS.
In conclusion, our work has revealed that SAMM50 acts as a receptor for basal piecemeal mitophagy of selected mitochondrial components through a LIR-mediated interaction with human Atg8-family proteins. During a metabolic switch from aerobic glycolysis to OXPHOS, SAMM50 depends on SQSTM1 to mediate basal piecemeal mitophagy and recycle key components required for efficient ATP production ().
Figure 1. Role of SAMM50 in piecemeal basal mitophagy of the SAM and MICOS complex components
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Reference
- Abudu YP, Shrestha BK, Zhang W, et al. SAMM50 acts with p62 in piecemeal basal- and OXPHOS-induced mitophagy of SAM and MICOS components. J Cell Biol. 2021;220:e202009092.
