ABSTRACT
Selective disposal of a wide range of cellular entities by macroautophagy/autophagy is achieved through a special class of proteins called autophagy receptors, which link corresponding cargo to the membrane-bound autophagosomal protein Atg8/LC3. In pursuit of novel autophagy receptors and their cargo, we uncovered a previously undescribed autophagy pathway for removal of aberrant clathrin-mediated endocytosis (CME) protein condensates in S. cerevisiae. Of these CME proteins, Ede1 functions as an autophagy receptor, harboring distinct Atg8-binding domains and driving phase separation into condensates. The aberrant CME condensates at the plasma membrane (PM) exhibit a drop-like structure surrounded by a fenestrated ER, which are engulfed in pieces in an Ede1-dependent manner by autophagy. Thus, our work suggests that aberrant CME is a target for autophagic degradation, with the scaffold protein Ede1 serving as a built-in autophagy receptor that monitors the assembly status of the CME machinery.
The ability to degrade cytosolic components ranging from single proteins to large complexes and organelles is essential for cellular homeostasis. Macroautophagy, hereafter referred to as autophagy, is an evolutionarily conserved pathway that can remove a diverse set of cytoplasmic entities by engulfing cargo into a transient double-membrane organelle called a phagophore; this structure subsequently matures into an autophagosome, which later fuses with a degradative compartment. Key to substrate selectivity is a class of specialized proteins, called autophagy receptors, which link the sequestered cargoes to the growing phagophores by binding to the ubiquitin-like protein Atg8. Knowledge of autophagy receptors is crucial to understand the organized degradation of autophagic cargo and allows the dissection of the individual autophagic pathways relevant in health and disease.
In our recent study [Citation1], we analyzed the Atg8 interactome in budding yeast upon autophagy induction using quantitative mass spectrometry, which yielded a list of 225 significantly enriched proteins. The list comprises nearly a complete set of proteins involved in the early steps of CME. Through the action of these proteins clathrin-coated vesicles form at the PM, allowing entry of extracellular molecules into the cell as well as remodeling of the PM protein content. In vitro binding assays with recombinantly purified proteins revealed that among the tested candidates only Ede1, the initiator of CME in yeast, directly binds Atg8. Bioinformatic analysis showed that Ede1 contains several putative Atg8-interacting motifs (AIMs), and deletion or mutation of the three most C-terminal AIMs (Ede1[AIMR]) abrogates the interaction with Atg8. A high-resolution crystal structure further supports AIM-dependent interaction between Ede1 and Atg8. In addition, immunoprecipitation experiments showed that Ede1 binds also to the scaffold protein Atg11, which associates with most known autophagy receptors and is required for phagophore formation.
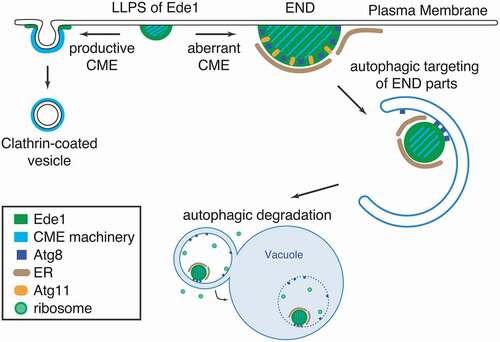
To test whether Ede1 serves as an autophagy receptor for other CME proteins, we compared the Atg8 interactome of cells expressing wild-type Ede1 or the Ede1[AIMR] mutant. These experiments revealed that AIM-dependent interaction of Ede1 and Atg8 is required for the co-enrichment of other CME machinery proteins with Atg8. To visualize the sites of interaction between Atg8 and Ede1 in vivo, we performed live-cell imaging of cells in which both proteins were fluorescently labeled. We found that Atg8 colocalizes with Ede1 in an AIM-dependent manner at distinct PM sites where Ede1 strongly accumulates in approximately 5% of proliferating wild-type cells. Compared to regular sites of endocytosis, these atypical Ede1-Atg8 clusters show strong enrichment of Ede1, they lack normal endocytic mobility, and also strongly accumulate other early CME proteins. We therefore named these aberrant clusters Ede1-dependent eNdocytic protein Deposits (ENDs). Importantly, formation of ENDs is enhanced in mutants known to disturb the early phase of CME either by overexpression of Ede1 or by simultaneous deletion of three endocytic adaptors (yap1801∆, yap1802∆, and apl3∆), hinting at a function for autophagy in quality control of CME assembly.
Next, we investigated if END is a target of autophagy. Indeed, guided by the Atg8-Ede1 interaction, we found that unproductive END assemblies are cleared by the core autophagy machinery in an Atg11-dependent manner even without external autophagy induction. Genetic perturbation of the early phase of CME increases not only END formation but also Ede1-dependent autophagic clearance under the same conditions. Time-lapse microscopy experiments showed that an initial burst of Atg8 at sites of ENDs results in pinching off of small pieces of Atg8 and Ede1 double-positive structures from the ENDs, which are later found within autophagic bodies in atg15∆ mutant cells as revealed by correlative cryo-ET. Collectively, these data support the function of Ede1 as an autophagy receptor for clearing aberrant CME assemblies.
To understand the physical properties of the END, we performed fluorescence recovery after photobleaching (FRAP) experiments on eGFP-Ede1-labeled cells. We found that Ede1 moves dynamically within this compartment and is constantly exchanged with the environment, which is in contrast to non-soluble protein aggregates in which the constituents are static. Dynamic protein compartments can be generated by local enrichment of proteins through multivalent interactions in a process called liquid-liquid phase separation (LLPS). Accordingly, we observed the disappearance and reappearance of ENDs after hexanediol addition and washout, an aliphatic alcohol that reversibly dissolves liquid-like assemblies. Ede1 encompasses an intrinsically disordered low complexity region (LCR), which we found is, together with the coiled-coil domain, necessary and sufficient to drive END formation. Autophagic degradation of CME machinery proteins is strictly dependent on the ability of Ede1 to phase separate, highlighting the role of cargo-receptor concentration in establishing high avidity between the receptor and Atg8.
In summary, our study unveiled a role for selective autophagy in the removal of aberrant CME protein assemblies in S. cerevisiae (). Interestingly, Ede1 has a dual function as an initiator of CME and an autophagy receptor for the removal of aberrant assemblies. We proposed to call this subtype of autophagy receptors “intrinsic autophagy receptors”. They are integral and functionally important subunits within normal complexes but can, if needed, direct them to phagophores for degradation. Consequently, intrinsic autophagy receptors may provide a built-in quality control for functionality or assembly of the complex itself.
Figure 1. Schematic depiction of the autophagic END pathway. In the early phase of CME, Ede1 and other CME machinery proteins assemble at the plasma membrane by liquid-liquid phase separation (LLPS). If the early phase of CME assembly is disturbed, Ede1-dependent endocytic protein deposits (ENDs) accumulate. ENDs contain, besides the intrinsic autophagy receptor Ede1, other CME proteins as well as the autophagy proteins Atg8 and Atg11. This results in an engulfment of END pieces into phagophores, which are subsequently delivered to the vacuole for degradation
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Reference
- Wilfling F, Lee CW, Erdmann PS, et al. A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits. Mol Cell. 2020;80:764–778.
