ABSTRACT
Animal and plant somatic cells have the capacity to switch states or reprogram into stem cells to adapt during stress and injury. This ability to deal with stochastic changes or reprogramming of somatic cells also needs macroautophagy/autophagy. Here, we expand on this notion and provide a primary example of how overexpression of ATG8/LC3 in the moss Physcomitrium patens enhances the ability to reprogram somatic cells into stem cells when subjected to severe wounding. This observation suggests that autophagy is not only required for cells to dedifferentiate but also makes cells more competent to do so.
Abbreviations: ATG: autophagy related; atg5: AUTOPHAGY 5; ATG8/LC3: AUTOPHAGY 8/microtubule associated protein 1 light chain 3; GFP: green fluorescent protein
Introduction
Autophagy is a widely conserved cytosolic recycling process essential for cellular homeostasis and stress adaptation [Citation1,Citation2]. The process involves ATG (autophagy related) genes and knockout of ATG genes is often detrimental or severely affect lifespan and adaptive responses [Citation3,Citation4], while overexpression of ATG genes gives both animals and plants increased fitness [Citation5–7].
In multicellular organisms, cells adjacent to wounded tissue need to reenter the cell cycle to replace dead cells [Citation8–10]. Autophagy has also been implicated in cellular regeneration and induced pluripotent stem cell formation in animals [Citation11–13]. We recently showed that pluripotent cells of autophagy deficient Arabidopsis mutant plants exhibit de-regulated cell growth and organogenesis, while somatic cells of autophagy deficient plants struggle to dedifferentiate and produce pluripotent cells. This was also observed in the bryophyte Physcomitrium patens when we investigated wound-induced reprogramming and stem cell differentiation of somatic gametophore cells [Citation10,Citation14]. For Physcomitrium to activate this type of reprogramming, the gametophore leaves are isolated and wounded by cutting them in half. The detached gametophore cells are then monitored overtime to detect filament protrusion in the cut site. These filaments indicate that the gametophore cells have reprogrammed into a new cell type [Citation10,Citation15].
However, while autophagy plays a major role during somatic reprogramming, it is currently unknown if autophagic activity can improve reprogramming efficiency. We therefore investigated if overexpression of the limiting gene ATG8/LC3 (AUTOPHAGY 8/microtubule associated protein 1 light chain 3) would affect wound induced reprogramming in Physcomitrium.
Results
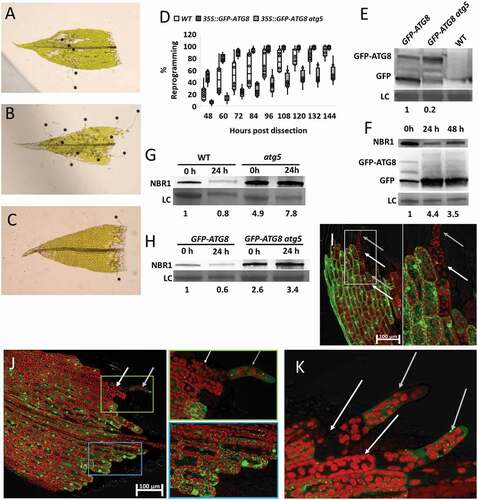
First, we selected moss plants expressing GFP-ATG8/LC3 under the control of the constitutive 35S promoter. Next, we compared the development of tip growth, as a proxy for reprogramming, after wounding the gametophore leaves of 35S::GFP-ATG8/LC3 plants and the parental wild-type control Gransden. Tip growth from cells adjacent to the wound site from both genotypes became visible 48 h after wounding, however, the percentage of leaves with tip growth was much higher for 35S::GFP-ATG8/LC3 plants. Moreover, 96 h after cutting, 100% of 35S::GFP-ATG8/LC3 leaves exhibited tip growth while this could only be observed in approximately 60% of the leaves from WT Gransden. (). To examine if the positive effect we see on reprogramming in moss plants overexpressing ATG8/LC3 depended on functional autophagy, we introduced 35S::GFP-ATG8/LC3 in an atg5 (AUTOPHAGY 5) background and analyzed tip growth development. Interestingly, 35S::GFP-ATG8/LC3 atg5 plants had a very low reprogramming efficiency: Only 35% of this line’s leaves had initiated reprogramming 96 h after wounding and increasing time to 144 h only resulted in 50% of the leaves reprogramming (). This is consistent with our earlier observations for atg5 mutants exhibiting impaired wound-induced reprogramming [Citation14]. Our data indicates that overexpression of ATG8/LC3 augment reprogramming efficiency via functional autophagy. We corroborated this by examining autophagic flux in 35S::GFP-ATG8/LC3 plants through GFP-ATG8/LC3 cleavage assay and NBR1 (NEXT TO BRCA1 gene 1) turnover [Citation16]. Free GFP was massively present in protein extracts of 35S::GFP-ATG8/LC3 plants but almost absent from protein extracts of 35S::GFP-ATG8/LC3 atg5 plants (). In addition, there is a 4-fold increase in the free GFP:GFP-ATG8/LC3 ratio already 24 h after wounding and NBR1 levels declined at the same timepoint indicating increased autophagic activity and autophagy induction before visible signs of reprogramming become apparent (). To verify that the decline in NBR1 levels 24 h after wounding was because of autophagic-dependent turnover, we repeated the experiment in the WT and the 35S::GFP-ATG8/LC3 and compared them to atg5 and 35S::GFP-ATG8/LC3 atg5 (–H). While NBR1 decreases in WT (20%) and 35S::GFP-ATG8/LC3 (40%) 24 h after wounding, in atg5 backgrounds NBR1 accumulates, indicating NBR1 turnover through functional autophagy. Because of the increased autophagic flux observed 24 h after wounding, we decided to examine how the GFP-ATG8/LC3 signal would develop during reprogramming in the hours leading up to the protrusion event. In the 35S::GFP-ATG8/LC3 line, a strong GFP signal was visible in most of the leaf cells (). However, in cells (white arrows) with a visible tip protrusion (gray arrows), GFP signal was mostly absent (, white box). Because we observed GFP signals in most cells and subtle changes may thus be masked in the 35S::GFP-ATG8/LC3 line, we investigated a line expressing GFP-ATG8/LC3 from its native promoter [Citation17]. When expressed from the ATG8/LC3 native promotor, GFP-ATG8/LC3-positive foci accumulated in the periphery of the cut site, where the cells destined for reprogramming are located (, see blue box). Here we also observed less signal in cells (white arrow, green box) which already exhibited tip growth (gray arrow). In addition, cells adjacent to cells with tip growth showed almost no sign of GFP-ATG8/LC3 (). This indicates that autophagy is mainly active in the cells during the initial reprogramming phase but terminates once the reprogramming is finished. This observation aligns with previous observations that cells which successfully reprogram blocks adjacent cells from doing so [Citation18].
Figure 1. Overexpression of ATG8/LC3 enhances wound-induced reprogramming in Physcomitrium patens. (A–C) Representative gametophores with tip growth after 96 h, for WT (A) 35S::GFP-ATG8/LC3 (B) and 35S::GFP-ATG8/LC3 atg5 (C). Stars mark tip protrusions. (D) Boxplot showing the percentage of gametophores with at least one cell showing tip growth, for each of the genotypes tested. Box limits indicate the 25th and 75th percentiles and whiskers extend to the minimum and maximum. Data derived from 8 biological replicates. All samples at the individual timepoints show significant differences according to one-way ANOVA (E) GFP-ATG8/LC3 western blot in the different moss lines used. (F) 35S::GFP-ATG8/LC3 and NBR1 western blots after wounding. Pre-wounded (0 h); 24 h post wounding (24 h) and 48 h post wounding (48 h). Values given represent the ratio of free GFP:GFP-ATG8/LC3, relative to non-treated control (NT, set to 1). (G–H) Western blot showing NBR1 level in (G) WT and atg5 plants or (H) 35S::GFP-ATG8/LC3 and 35S::GFP-ATG8/LC3 atg5, before (0 h) and 24 h after wounding induced reprogramming (24 h). Values given represent the intensity of the NBR1 bands normalized to the loading control and relative to WT 0 h (G) or 35S::GFP-ATG8/LC3 0 h (H). (I–K) GFP-ATG8/LC3 foci in 35S::GFP-ATG8/LC3 (I) or pATG8/LC3::GFP-ATG8/LC3 (J,K) gametophores, 48 h after wounding. White arrow indicates the cell which has undergone differentiation and the gray arrow indicate the new protonema cell. In figures (I,J) regions within colored boxes were magnified and presented at the rightmost panels.
Discussion
Overexpression of key autophagy bottleneck genes is linked to increasing fitness and delayed aging [Citation5–7]. We previously showed that autophagy deficient Physcomitrium lines showed reduced wound induced reprogramming of somatic gametophore cells to protonema stem cells [Citation14]. Here we expand on this notion by showing that overexpression of ATG8/LC3 can enhance wound induced reprogramming in an autophagy dependent manner. Autophagy seems to be particularly engaged prior to the tip growth i.e. during the dedifferentiation of somatic cells into stem cells; after which autophagic activity seems to be reduced. This is the first example of how a boosted ATG gene can help facilitate reprogramming of somatic cells into stem cells.
Methods
Growth conditions
Physcomitrium patens (Gransden 2004 strain; Christina Lunde, Plant Biochemistry Laboratory, Department of Plant Biology, Faculty of Life Sciences, University of Copenhagen, DK–1871 Frederiksberg C, Copenhagen, Denmark) was grown as described previously [Citation19]. In short they were cultivated on BCD-AT media (MgSO4.7H2O 250 mg/l, KH2PO4 250 mg/l, KNO3 1010 mg/l, ammonium tartrate 920 mg/l, FeSO4.7H2O 12.5 mg/l, CaCl2.H2O 147 mg/l, trace elements 1 ml/l [H3BO3 61 mg/l, {NH4}6Mo7O24 · 4H2O 38 mg/l, CuSO4 · 5H20 6 mg/l, CoCl2 · 6H20 5.1 mg/l, ZnCl2 4.1 mg/l, MnCl2 · 4H2O 4.1 mg/l; use miliQ-water], pH 6.5 adjusted with KOH, agar 8 g/l) overlaid with cellophane discs (AA Packaging Ltd special order) when needed. P. patens was grown at 22°C 55 µEm−2s−1, with a 16 h light-8 h dark cycle. For gametophore production P. patens was grown without cellophane for 4 weeks.
Transformation
WT and atg5 moss lines were transformed using the polyethylene glycol (Sigma, P7181) transformation [Citation19] with the PTM13 vector (a gift from Mattias Thelander and Eva Sundberg, Department of Plant Biology, Swedish University of Agricultural Sciences, The Linnean Center for Plant Biology in Uppsala, Sweden) which contains a GFP-ATG8/LC3 fusion driven by two 35S promoters and a hygromycin resistance gene also driven by a 35S promoter. It was integrated into the neutral locus 108 [Citation17]. Transformed protoplasts were selected on hygromycin and genotyped.
Reprogramming assay
To investigate the rate of reprogramming initiation in Physcomitrium, the top part of 4-week-old gametophores were isolated and the tips were dissected with a surgical knife and placed on a new plate overlaid with cellophane. The gametophore tips were checked for reprogramming activation every 12 h using a Leica MZ16 F Fluorescence Stereomicroscope. For imaging, gametophore tips were dissected as described above and the individual tips were placed in small dots of 2% methyl cellulose 15cP (Sigma, M7140-100G) in an empty Petri dish, overlaid with cellophane and cooled BCD-AT media on top. Pictures were taken every 24 h using a Sony α6000 camera mounted on a Leica MZ16 F Fluorescence Stereomicroscope.
Western blot
Protein was extracted in Lacus buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 15 mM EGTA, 100 mM NaCl, 2 mM DTT, 30 mM β-glycero-phosphate, 0.1% Nonidet P-40 [Sigma, 492016]) plus phosphatase inhibitor (PhosSTOP [Roche, 04906837001]) and protease inhibitor cocktails (Complete [Roche, 11697498001]). Samples were cleared by centrifugation and boiled for 5 min in SDS-loading buffer. Total proteins were separated by SDS-PAGE and electro-blotted. Immunoblots were blocked for 1 h in TBS-Tween (0.1% Tween 20 [Merck, 8221841000] v:v) and 5% BSA (Serva, 1193402). GFP was detected by incubating with primary anti-GFP antibody (1:5000, GFP, [Acris, TP401]; or 1:5000, NBR1, [Agrisera, AS14 2805]). This was followed by incubation with anti-rabbit-IgG-HRP (1:5000; Promega, W4011). The horseradish peroxidase-conjugated antibody was visualized with ECL substrate (2 mM 41BPA, 500 µM luminol, 100 mM Tris, pH 8.8 and 1.7•10−2 H2O2) and pictures taken with a Sony A7S camera. The bands were analyzed using ImageJ. To obtain a loading control the whole membrane was stained with amido black and appropriate bands was used as control.
Confocal microscopy
All images were taken with LSM700 Zeiss confocal microscope. The images of the gametophores were taken with a 10X and 20X air objective. Image analysis was performed with Zen 2012 (Zeiss) and ImageJ.
Statistics
To test for significant differences () we compared the 35S::GFP-ATG8/LC3 to the WT, the WT to the 35S::GFP-ATG8/LC3 atg5, and the 35S::GFP-ATG8/LC3 to the 35S::GFP-ATG8/LC3 atg5 using oneway Anova. This was repeated for each timepoint.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873.
- Kim JH, Jung H, Chung T. Birth, growth, maturation, and demise of plant autophagic vesicles. J Plant Biol. 2020;63(3):155–164.
- Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13(10):1619–1628.
- Hanaoka H, Noda T, Shirano Y, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129(3):1181–1193.
- Minina EA, Moschou PN, Vetukuri RR, et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J Exp Bot. 2018;69(6):1415–1432.
- Pyo JO, Yoo S-M, Ahn H-H, et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:1–9.
- Chen Q, Soulay F, Saudemont B, et al. Overexpression of ATG8/LC3 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 2019;60(2):343–352.
- Willet SG, Lewis MA, Miao Z-F, et al. Regenerative proliferation of differentiated cells by mTORC 1‐dependent paligenosis. EMBO J. 2018;37(7):1–17.
- Marhava P, Hoermayer L, Yoshida S, et al. Re-activation of stem cell pathways for pattern restoration in plant wound healing. Cell. 2019 May;177(4):957–969.e13.
- Ishikawa M, Murata T, Sato Y, et al. Physcomitrella cyclin-dependent kinase A links cell cycle reactivation to other cellular changes during reprogramming of leaf cells. Plant Cell. 2011;23(8):2924–2938.
- Miao ZF, Lewis MA, Cho CJ, et al. A dedicated evolutionarily conserved molecular network licenses differentiated cells to return to the cell cycle. Dev Cell. 2020;55(2):178–194.e7.
- Wang S, Xia P, Ye B, et al. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13(5):617–625.
- Saera-Vila A, Kish PE, Louie KW, et al. Autophagy regulates cytoplasmic remodeling during cell reprogramming in a zebrafish model of muscle regeneration. Autophagy. 2016;12(10):1864–1875.
- Rodriguez E, Chevalier J, Olsen J, et al. Autophagy mediates temporary reprogramming and dedifferentiation in plant somatic cells. EMBO J. 2020;39(4):1–11.
- Kofuji R, Hasebe M. Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr Opin Plant Biol. 2014;17:13–21.
- Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222.
- Sanchez-Vera V, Kenchappa CS, Landberg K, et al. Autophagy is required for gamete differentiation in the moss Physcomitrella patens. Autophagy. 2017;13(11):1939–1951.
- Sato Y, Sugimoto N, Hirai T, et al. Cells reprogramming to stem cells inhibit the reprogramming of adjacent cells in the moss Physcomitrella patens. Sci Rep. 2017;7(1):1–12.
- Bressendorff S, Azevedo R, Kenchappa CS, et al. An innate immunity pathway in the moss Physcomitrella patens. Plant Cell. 2016;28(6):1328–1342.
