ABSTRACT
In recent years, the contribution of exosomes to immunity, inflammation and host-pathogen interaction have been appreciated. Exosomes are small secreted extracellular vesicles from endosomal origin that contain a myriad of cellular molecules (protein, nucleic acids), including surface receptors. We have reported a pathogen-induced and macroautophagy/autophagy-dependent class of exosomes coined as “defensosomes”, which protect the host from membrane-targeting toxins. In a recent study, we found that defensosomes decorated with ACE2, the SARS-CoV-2 cellular receptor, are produced in the lungs of patients with COVID-19, and that increased concentration of ACE2-loaded defensosomes is associated with decreased hospitalization length. Mechanistically, SARS-CoV-2 induces the production of ACE2-coated defensosomes, a process requiring the autophagy machinery, which in turn binds and neutralizes the virus. We propose that defensosomes represent a new form of autophagy-mediated innate immunity that contributes to the host’s armamentarium against pathogens.
The autophagy pathway constitutes an important arm of host defense by directly degrading pathogens, regulating immunity, and maintaining cell homeostasis. Prior work from our lab discovered that mice deficient for the core autophagy protein ATG16L1 are more susceptible to lethal infections by Staphylococcus aureus due to heightened sensitivity to a pore-forming toxin produced by this bacterial pathogen called α-toxin, which kills target cells by binding the surface receptor ADAM10. Rather than a role for ATG16L1 in targeting autophagosome cargos to the lysosome for degradation, we found that autophagy proteins mediate release of ADAM10 on exosomes, small extracellular vesicles derived from endosomal compartments such as multivesicular bodies. These ADAM10-bearing exosomes serve as decoys that prevent α-toxin from targeting host cells by binding and inactivating free toxin subunits (). We showed that activation of the innate immune sensor TLR9 by bacterial DNA stimulates exosome production, and that other toxins that target host surface proteins such as the S. aureus bi-component toxin LukED and the diphtheria toxin are also inhibited through this mechanism. These findings suggest that exosomes dependent on autophagy proteins are part of an innate immune response to infection to counter virulence strategies that target surface cellular membrane proteins. We refer to these defensive exosomes as “defensosomes” to acknowledge the potential for distinct mechanisms of biogenesis and function from their constitutively released counterparts.
Figure 1. Defensosomes protect against bacterial toxins and viral infection. Defensosomes are produced in response to recognition of ssRNA, dsRNA, and unmethylated DNA by innate immune receptors TLR3, TLR7 and TLR9 in an ATG16L1-dependent process. ADAM10 on the surface of defensosomes binds α-toxin produced by S. aureus leading to oligomerization of this pore-forming toxin, thereby protecting the cells from lysis. SARS-CoV-2 virions are neutralized when a sufficient concentration of ACE2+ exosomes are present. Outstanding questions include how endosomal TLR signaling leads to defensosome release and what role ATG16L1 plays in this process. Created with BioRender.com.
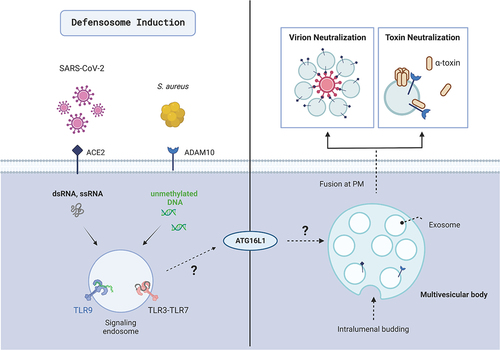
Having demonstrated a role for counteracting bacterial toxins, we next investigated whether ATG16L1-mediated release of defensosomes is involved in defense against other pathogens such as viruses. Many viruses have evolved to exploit the autophagic machinery and use host membranes for their replication. One example is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of COVID-19. SARS-CoV-2 can generate double-membraned vesicles resembling autophagosomes from ER-derived lipids to house their replication complexes, while the viral proteins Orf3a and Nsp15, block autophagic flux and limit autophagy signaling. The main receptor SARS-CoV-2 uses for cellular entry is ACE2 (angiotensin converting enzyme 2), a molecule that controls various aspects of the cardiovascular system, as well as fluid and electrolyte balance. The high affinity interaction between the SARS-CoV-2 Spike protein and ACE2 made this pathogen an attractive infection model to test the efficacy of defensosomes.
During the peak of the pandemic in New York City, many patients were intubated, creating a unique opportunity to analyze exosomes from the lungs of critically ill patients who had undergone bronchoscopies for lower airway clearance. Using flow cytometry, we found that the concentration of ACE2+ exosomes in bronchiolar lavage fluid/BALF display remarkably high variation across individuals [Citation1]. Incorporation of patient clinical data revealed that COVID patients experience shorter hospitalization times if they have an increased concentration of ACE2+ exosomes present in their lungs and higher levels of ACE2 on the surface of each exosome. These data suggested that proper loading of ACE2 onto exosomes influences the level of protection against SARS-CoV-2.
To determine whether exosomes are induced and protective against SARS-CoV-2 infection, we turned to in vitro models using tractable cell lines and an organotypic air-liquid interface culture derived from human airway epithelial cells/HAECs. SARS-CoV-2 infection robustly induces exosomes in a manner dependent on ATG16L1. To interrogate what viral or host factors trigger exosome release, we screened agonists mimicking viral ligands or immune danger signals. We found that dsRNA (TLR3), ssRNA (TLR7) and unmethylated DNA (TLR9) all elicit exosomes. In contrast, ligands for cytosolic sensors such as RIGI, IFIH1/MDA5, and CGAS-STING1 do not induce exosome release, although they have been implicated in SARS-CoV-2 pathogenesis.
To test for viral neutralization, we mixed virions with ACE2+ or ACE2− exosomes prior to infection. We observed an ACE2- and dose-dependent protective effect by the exosomes, data in line with the in vivo observation that the concentration of ACE2+ exosomes is a contributing factor to hospital length of stay in COVID patients. Notably, ATG16L1-deficient cells are not more susceptible to SARS-CoV-2 infection, indicating that loss of ATG16L1 does not have an impact on viral entry or replication in these conditions, potentially reflecting the ability of the virus to evade autophagy-mediated defense once inside cells. Finally, ultrastructural imaging reveals close associations between virions and ACE2+, but not ACE2− exosomes. 3D reconstructions suggest that there may be a possible deformation of the ACE2 receptor once bound to Spike, rendering the virus inactive if surrounded by a sufficient number of exosomes. Thus, these ACE2+ exosomes display the key hallmarks of defensosomes – inducible in response to infection, dependence on an autophagy protein, and promotes host defense by binding its target through surface protein interactions ().
Our work raises many questions surrounding defensosome biology including how these vesicles regulate immune cells upon uptake, and whether the functions of defensosomes differ based on tissue of origin. Our results also suggest that signaling localized at the endosome is a common feature important for exosome production. How the endosome is altered or differentially trafficked after these signaling events are triggered remains unclear. Additional questions include what components downstream of TLR signaling are required for exosome release, and what role ATG16L1 plays in this process. Are autophagosomes required? Finally, it will be interesting to examine whether defensosome production is impaired in other human diseases in which autophagy is suspected to be disrupted. Overall, our work highlights a new aspect of autophagy-mediated immunity. Further understanding of the cell biological mechanisms regulating defensosome biogenesis could provide new opportunities to help treat human diseases.
Disclosure statement
K.C. has received research support from Pfizer, Takeda, Pacific Biosciences, Genentech, and Abbvie. K.C. has consulted for or received honoraria from Vedanta, Genentech, and Abbvie. K.C. is an inventor on U.S. patent 10,722,600 and provisional patent 62/935,035 and 63/157,225. V.J.T. receives research support from Janssen Biotech Inc. V.J.T has consulted for Janssen Research & Development, LLC and have or received honoraria from Genentech and Medimmune. V.J.T. is an inventor on patents and patent applications filed by New York University, which are currently under commercial license to Janssen Biotech Inc. provides research funding and other payments associated with a licensing agreement.
Additional information
Funding
Reference
- Ching KL, de Vries M, Gago J, et al. ACE2-containing defensosomes serve as decoys to inhibit SARS-CoV-2 infection. PLoS Biol. 2022;20(9):e3001754.
