ABSTRACT
A multifunctional role of Atg8-family proteins (Atg8 from yeast and human LC3 and GABARAP subfamilies, all referred to here as ATG8s) in macroautophagy/autophagy is carried out by two protein domains, the N-terminal helical domain (NHD) and ubiquitin-like (UBL) domain. Previous studies showed that the NHD of PE-conjugated ATG8s mediates membrane hemifusion via a direct interaction with lipids in trans-membrane association, which would require the NHD in lipidated ATG8s to adopt a solvent-oriented, “open”, conformation that unmasks a UBL domain surface needed for membrane tethering. A purpose of the “closed” conformation of the NHD masking the tethering surface on the UBL domain, a conformation seen in the most structures of non-lipidated ATG8s, remained elusive. A recent study by Zhang et al. discussed in this article, showed that the N terminus of lipidated human ATG8s adopts the “closed” conformation when it interacts with the membrane in cis-membrane association, i.e. with the same membrane ATG8 is anchored to. This finding suggests functions for two distinct conformations of the NHD in lipidated ATG8s and raises questions about determinants controlling cis- or trans-membrane associations of the NHD in ATG8−PE.
Abbreviations: AIM, Atg8-family interacting motif; 3D-CLEM, three-dimensional correlative light and electron microscopy; FRET, Förster/fluorescence resonance energy transfer; LIR, LC3-interacting motif; MD, molecular dynamics; NHD, N-terminal helical domain; UBL, ubiquitin-like.
The architecture of ATG8 proteins operating in autophagy is comprised of two distinct domains, the N-terminal helical domain and ubiquitin-like domain. The UBL domain adopts a well-defined globular structure, whereas the NHD is less rigid and foldable into two separate α-helices. The α1-helix (approximately the first ten amino acid residues) often adopts an extended conformation, based on the NMR structures of ATG8s in solution [Citation1–3]. Each domain in ATG8s fulfills a specific function. The UBL carries two hydrophobic pockets where the Atg8-family interacting motif (AIM)/LC3-interacting region (LIR) motif of autophagy receptors or adaptors docks in order to sequester the cargo by phagophores or to connect with the autophagy machinery. The C-terminal glycine in the UBL domain is essential for conjugation of ATG8s to phosphatidylethanolamine (PE) in a process that is indispensable for autophagy. Phe77 and Phe79 in Atg8 from yeast increase the interaction of Atg8−PE with the membrane and allow for membrane deformations inducing a positive membrane curvature [Citation2]. The UBL domain also mediates membrane tethering [Citation4] and Phe104 and Tyr106 in Atg8 are needed for this process [Citation5].
The first ten amino acids in the NHD of lipidated human ATG8s carries amino acid residues that interact with lipid head groups to establish ionic interactions or they penetrate into the membrane to engage in hydrophobic interactions. These interactions are required for membrane fusion mediated by ATG8s but not for membrane tethering [Citation4]. Atg8 from yeast lacking nine N-terminal residues fails in membrane hemifusion [Citation5]. Together these findings suggest that the very N terminus of lipidated ATG8s has an evolutionarily common purpose, which is binding to the membrane. The membrane tethering-hemifusion activity of lipidated ATG8s implies that ATG8−PE anchored on one membrane attaches on an opposite membrane and brings them together. This mechanism established a model for trans-membrane association of ATG8−PE. An unsolved issue for this model was the conformation of the NHD that, according to many available structures of non-lipidated ATG8s, masks the residues (Phe104 and Tyr106 in Atg8) comprising the tethering surface in the UBL domain. Therefore, the trans-membrane association model assumes that the NHD in Atg8−PE adopts, via an unknown trigger, a solvent-oriented conformation that opens up and unmasks the tethering surface in the UBL domain in order to mediate membrane tethering and hemifusion [Citation5]. This very plausible mechanism raises unsolved questions. For example, can the NHD in the “closed” conformation, seen in non-lipidated ATG8s, exist also in ATG8−PE? Can the N-terminal peptide in ATG8−PE be in cis-membrane association, in which the N terminus binds to the same membrane where the protein is PE-conjugated? A recent study by Zhang et al. [Citation6], described here, addressed these questions by probing the N terminus of lipidated LC3B and GABARAP.
Zhang et al. labeled the N termini of LC3B and GABARAP with 7-nitrobenz-2-oxa-1,3-diazol-4-yl (NBD), a fluorescent probe that enhances fluorescence emission at approximately 535 nm in a hydrophobic environment established by protein-protein or protein-membrane interactions. NBD-labeled LC3B and GABARAP were mixed with ATG7, ATG3, the ATG12−ATG5-ATG16L1 complex, and liposomes to probe the fluorescence signal in an in vitro lipidation assay. The NBD fluorescence increases partially upon formation of complexes between ATG8s and ATG7 or ATG3 but increases significantly in the presence of all reaction components, suggesting that the N termini of lipidated ATG8s are in a hydrophobic interface created between the protein and liposomal membrane.
To investigate the arrangements of LC3B–PE and GABARAP–PE on the membrane the authors applied all-atomistic MD simulations and calculated the percentage of time the ATG8 residues contact the 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC)-containing membrane during 1 µs of the trajectory. This approach revealed four protein-membrane interfaces: the structurally flexible N- terminal region, loop 3, loop 6, and the dynamic C terminus (). The basic residues in the arginine-rich motif are not in the contact with the membrane, in contrast to CG-MD simulations applied in previous studies. The aromatic residues in LC3B and GABARAP homologous to F77 and F79 in Atg8 from yeast are also distant from the membrane. Importantly, MD simulations by Zhang et al. show the cis-membrane association of lipidated ATG8s. The results of MD simulations were validated by the NBD spectra assay where His-tagged LC3B or GABARAP are attached to Ni-NTA on large unilamellar vesicles containing various compositions of lipids. The NBD fluorescence signal is highest with 50% 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine/DOPE, 45% POPC, and 5% Ni-NTA, suggesting a facilitating effect of lipid-packing defects. Further validation of all-atomistic MD simulations was obtained using a FRET assay utilizing 2% rhodamine-PE. A strong FRET signal was detected between NBD-labeled LC3B-His or GABARAP-His and rhodamine liposomes. The FRET signal is sensitive to imidazole and to a triple mutation (M1E K2E E7A) in the N terminus of GABARAP. Neither the triple mutant nor the GABARAP ∆9 mutant showed a defect in liposome clustering relative to wild type, suggesting that the N terminus of GABARAP is not involved in membrane tethering, in agreement with a previous study [Citation4]. To explore how alterations in the UBL domain affect the interaction between the N termini of ATG8s and the membrane, the N-terminally NBD-labeled LC3B and GABARAP were mutated in the loop 3 or R-rich motif. Both mutations disrupt the hydrophobic environment in the protein-lipid interacting interface. GABARAP mutated in the R-rich motif cannot even efficiently discharge from ATG3.
Figure 1. The N terminus is engaged in cis- and trans-membrane association of lipidated ATG8s. (A) the multiple-states NMR structure of LC3C (PDB ID: 2NCN) representing the ATG8 proteins. The NHD (dark gray) precedes the UBL domain (green-cyan). Highly flexible N-terminal region (extended conformational ensemble), loop 3 (purple), loop 6 (chartreuse), and the flexible C terminus conjugated to PE have a high occupancy time on the membrane. Alterations in the loop 3 and R-rich motif (blue) interfere with the cis-membrane insertion of the ATG8 N terminus. (B) The N terminus occupies the membrane in cis-membrane association of ATG8−PE. Amino acid residues involved in this association of the N terminus deform the membrane and/or change local membrane elasticity, and, thereby, promote expansion of the phagophore. (C) the N terminus occupies the membrane in trans-membrane association of ATG8−PE. The “open” conformation of the N terminus allows for solvent exposure of aromatic amino acid residues (brown) in the UBL domain that are important for membrane tethering. This aromatic patch is masked by the N terminus in the “closed” conformation (see B). The N terminus is indispensable for membrane fusion, but not tethering, which ultimately leads to phagophore expansion.
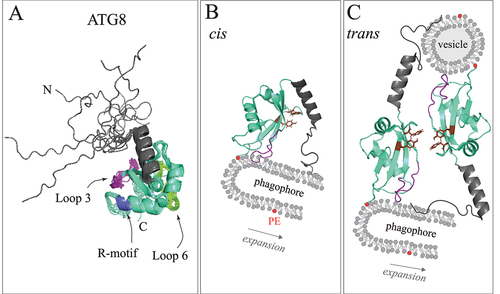
To test functional impacts of the impaired interaction between the N termini of human ATG8s and the membrane, the authors used Hexa KO cells (lacking 6 of the human ATG8 genes) expressing GABARAP mutated in the N terminus, loop 3, or the LIR motif and monitored S349-phosphorylated gel-like SQSTM1/p62 structures after starvation. Western blot analysis as well as microscopy images show a blocked SQSTM1 clearance with the LIR mutant (Y49A L50A) but not with the variants of GABARAP disrupted in membrane association of their N termini. The authors concluded that the membrane insertion of the GABARAP N terminus is not required for SQSTM1 body degradation. In contrast, a 3D-CLEM approach shows that only the LIR mutant of GABARAP can yield a similar diameter and volume of autophagosomes as wild type in starved Hexa KO cells. The diameter and volume of autophagosomes are significantly reduced with the mutants carrying the altered N terminus or loop3 in GABARAP. Restoration of autophagosome size is completely abolished with the GABARAP ∆9 mutant or in the absence of GABARAP. The authors concluded that cis-membrane association of ATG8s mediates phagophore membrane growth, possibly by promoting membrane deformations or changes in local membrane elasticity.
It is difficult to exclude a possibility that GABARAP in trans-membrane association is also present in the Hexa KO cells and that mutations targeting cis-membrane insertion affect trans-membrane association of GABARAP as well. Nevertheless, the study by Zhang et al. establishes a possibility that the N terminus of ATG8−PE can bind to lipids in two different conformations depending on the cis- or trans-membrane association (). The determinants for these two conformations remain unknown. Zhang et al. propose factors such as composition of the membrane lipids, the membrane curvature, the stage of phagophore growth, and/or cellular localization. Future research will hopefully explore these determinants, and also answer a structural question of spatiotemporal distribution of the two conformations of the ATG8 molecule. Specifically, it would be intriguing to explore whether the cis- and trans-membrane associations are spatially separated on distinct ATG8 molecules, where each molecule upon PE conjugation is structurally modified in the cell to play, for the rest of its time on the membrane, the predetermined role of either a membrane deformer (in “closed” conformation) or membrane fuser (in “open” conformation). Alternatively, the cis- and trans-membrane associations of ATG8s may arise from the two alternating conformations of the same switching ATG8 molecule that is promptly regulated in a temporal manner during phagophore growth. Finding whether the conformation of a given ATG8−PE on the membrane is constant or switching will lead to the discovery of structural modifiers or regulators and their mechanisms of action, opening a vast field of research in autophagy.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Krichel C, Mockel C, Schillinger O, et al. Solution structure of the autophagy-related protein LC3C reveals a polyproline II motif on a mobile tether with phosphorylation site. Sci Rep. 2019 Oct 2;9(1): 14167. PubMed PMID: 31578424. doi: 10.1038/s41598-019-48155-8
- Maruyama T, Alam JM, Fukuda T, et al. Membrane perturbation by lipidated Atg8 underlies autophagosome biogenesis. Nat Struct Mol Biol. 2021 Jul;28(7):583–593. PubMed PMID: 34239122. doi: 10.1038/s41594-021-00614-5
- Schwarten M, Stoldt M, Mohrluder J, et al. Solution structure of Atg8 reveals conformational polymorphism of the N-terminal domain. Biochem Biophys Res Commun. 2010 May 7;395(3): 426–431. PubMed PMID: 20382112. doi: 10.1016/j.bbrc.2010.04.043
- Weidberg H, Shpilka T, Shvets E, et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011 Apr 19;20(4): 444–454. PubMed PMID: 21497758. doi: 10.1016/j.devcel.2011.02.006
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007 Jul 13;130(1): 165–178. PubMed PMID: 17632063. doi: 10.1016/j.cell.2007.05.021
- Zhang W, Nishimura T, Gahlot D, et al. Autophagosome membrane expansion is mediated by the N-terminus and cis-membrane association of human ATG8s. Elife. 2023 Jun 8;12. PubMed PMID: 37288820. doi: 10.7554/eLife.89185
