Abstract
Genomic imprinting disorders often exhibit delayed neurobehavioral development, suggesting this unique mechanism of epigenetic regulation plays a role in mental and neurological health. While major errors in imprinting have been linked to adverse health outcomes, there has been little research conducted on how moderate variability in imprinted gene expression within a population contributes to differences in neurobehavioral outcomes, particularly at birth. Here, we profiled the expression of 108 known and putative imprinted genes in human placenta samples from 615 infants assessed by the Neonatal Intensive Care Unit (NICU) Network Neurobehavioral Scales (NNNS). Data reduction identified 10 genes (DLX5, DHCR24, VTRNA2-1, PHLDA2, NPAP1, FAM50B, GNAS-AS1, PAX8-AS1, SHANK2, and COPG2IT1) whose expression could distinguish between newborn neurobehavioral profiles derived from the NNNS. Clustering infants based on the expression pattern of these genes identified 2 groups of infants characterized by reduced quality of movement, increased signs of asymmetrical and non-optimal reflexes, and increased odds of demonstrating increased signs of physiologic stress and abstinence. Overall, these results suggest that common variation in placental imprinted gene expression is linked to suboptimal performance on scales of neurological functioning as well as with increased signs of physiologic stress, highlighting the central importance of the control of expression of these genes in the placenta for neurobehavioral development.
Introduction
Genomic imprinting is the monoallelic expression of a subset of genes in a conserved, parent-of-origin fashion. This process is regulated by epigenetic mechanisms, including DNA methylation and histone modifications.Citation1 Complete loss of imprinting status, leading to either functional nullisomy or biallelic expression of specific imprinted genes, is implicated in a number of congenital syndromes, including Beckwith-Weidemann, Prader-Willi, and Angelman, all of which are characterized by developmental delays or impaired neurobehavioral functioning.Citation2-4 Alterations in DNA methylation at imprinted sites, suggestive of altered imprinting status, have been linked to greater risk for the development of schizophrenia in humans,Citation5 as well as with general cognitive and behavioral defects in mice.Citation6-8
The placenta is an important mediator in the communication between the developing fetus and the maternal environment. This organ regulates nutrient and waste exchange, facilitates interactions with the maternal immune system, and acts as a neuroendocrine organ by producing important hormones and growth factors. Thus, it is an appropriate tissue for the investigation of fetal pathophysiology and neurobehavioral development.Citation9,10 Placental imprinting, uniquely, plays an important role in the developing fetus and placenta, as imprinted genes are critical for maternal nutrient regulation, fetal growth control, and the development of metabolic organs in utero.Citation11,12 In addition, the pattern of expression of imprinted genes in the placenta demonstrates significant similarity to that observed in the developing brain.Citation13 Prior investigations examining placental expression of imprinted genes have demonstrated low transcriptional noise, highlighting the importance of the fine-tuned regulation of this gene subset for proper infant development.Citation14 The specific importance of placental expression of the imprinted gene IGF2 has also been demonstrated through placental specific disruption in mice, resulting in increased reactivity to stress stimuli; this phenotype was not observed in total knockouts.Citation7
A better understanding of molecular mechanisms underlying the regulation of neurodevelopment could aid in identifying individuals who may benefit from early life interventions, when such interventions may be most successful. Our group has previously explored the role of placental imprinted gene expression in neurodevelopmental outcomes.Citation15 The current study expands upon both the number of genes examined and the number of individuals involved with the measurement of 108 known and putative imprinted genes in 615 infant placental samples. We have linked the expression patterns of these genes to outcomes quantified through the Neonatal Intensive Care Unit (NICU) Network Neurobehavioral Scales (NNNS).Citation16 The NNNS provides a comprehensive evaluation of the neurobehavioral status of infants from birth to approximately one month of age and includes neurologic and behavioral measures and signs of stress.Citation16,17 This assessment has demonstrated predictive validity for medical outcomes, such as cerebral palsy, neurologic abnormalities, as well as developmental outcomes, including mental and motor functioning, behavior problems, school readiness, and IQ through 4 ½ years of age.Citation18-21 This study represents the largest, to date, to examine placental genomic imprinting and its link to newborn neurodevelopment.
Results
Characteristics of the study population are presented in , and NNNS summary score descriptive statistics are provided in . Briefly, average maternal age was 29.5 years, average infant gestational age was slightly over 39 weeks, and the newborn population was comprised of 51.5% females. Based upon the a priori design of the cohort, small for gestational age (SGA) and large for gestational age (LGA) infants were overrepresented within our study population. A model-based clustering strategyCitation22 was used to classify infants into discrete, mutually exclusive profiles based on the NNNS summary. Figure S1 provides a visualization of the pattern of normalized summary scores across the 7 profiles.
Table 1. Descriptive characteristics of the study population (n = 615)
Table 2. NICU Network Neurobehavioral Scale (NNNS) score characteristics of the study population
The Random Forest algorithm produced a ranking of the 108 imprinted or putatively imprinted genes expressed in the placenta in order of the importance each played in classifying infants into their respective NNNS profile groups. Following 20,000 iterations of training, the error rate for sample placement into the correct profile was 58% compared to an expected error rate of approximately 86% due to chance alone. Using the 10 highest ranking genes based on importance score (), k-means clustering defined 3 expression clusters, with the assignment of 319, 77, and 219 infants into clusters 1, 2, and 3, respectively (). As expected, gene expression differed by cluster (P < 0.01, ANOVA) for each of the 10 genes used for profiling (Supplemental Fig. 2).
Figure 1. Heatmap of expression (total n = 615) for the 10 genes used to assign membership to clusters 1 (n = 319), 2 (n = 77), and 3 (n = 219).
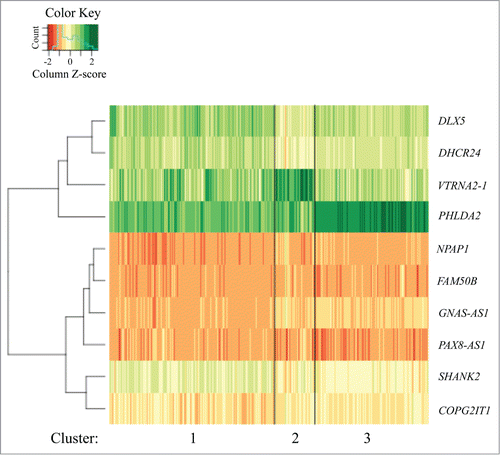
Table 3. Top 10 genes by random Forest importance score for predicting an infant NNNS profile membership
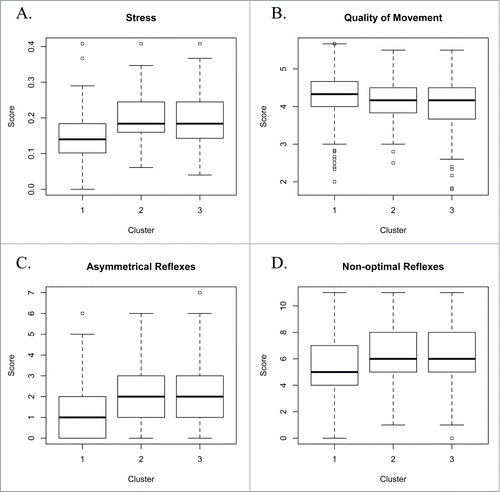
In order to more explicitly examine how gene expression impacts particular domains of neurobehavior, we examined the association between the 13 NNNS summary scores and gene expression cluster membership. Average scores of infant stress abstinence (P < 0.001), quality of movement (P < 0.001), asymmetrical reflexes (P < 0.001), and non-optimal reflexes (P < 0.001) differed across the gene expression-based clusters (). Infants in gene expression clusters 2 and 3 exhibited more signs of stress and abstinence (), reduced quality of movement (), and increased signs of asymmetrical () and non-optimal reflexes ().
Figure 2. Of the 13 NNNS summary scores analyzed, 4 yielded differential scores based upon cluster assignments, including (A) infant stress, (B) quality of movement, (C) asymmetrical reflexes, and (D) non-optimal reflexes.
To further model these relationships including covariates, we dichotomized infants based on NNNS summary scores falling outside the 10th or 90th percentile of the population, depending on which would be indicative of a poor outcome for that scale; for stress abstinence, asymmetrical reflexes, and non-optimal reflexes, scores above the 90th percentile are considered sub-optimal, while for quality of movement, scores below the 10th percentile are sub-optimal. In a logistic regression (), with membership in expression cluster 1 being the referent and including covariates, membership in expression clusters 2 and 3 was associated with an 8.36 (95% CI: 3.28–21.31) and 6.83 (95% CI: 3.07–15.18), respectively, increased odds of having stress abstinence scores above the 90th percentile. Membership in cluster 3, but not cluster 2, led to a significant 2.1 (95% CI: 1.16–3.83) fold increased odds for infants to have quality of movement scores below the 10th percentile, compared to those in cluster 1. Clusters 2 and 3 also displayed heightened odds of greater than 90th percentile asymmetrical reflex scores. Clusters 2 and 3 also displayed elevated odds of non-optimal reflex scores above the 90th percentile, though these risks were not statistically significant.
Table 4. Expression cluster membership and odds of an infant having an NNNS score within the sub-optimal 10th percentile of our population
Discussion
Germline alterations to imprinted genes are linked to congenital syndromes, nearly all of which include neurologic, mental, and behavioral deficits or delays.Citation23,24 In addition, a number of GWAS have identified genetic variation at or near imprinted genes associated with autism and other mental deficits.Citation13 Importantly, the placenta and brain are the 2 organs that have the most active imprinted genome because both organs are under tremendous selective pressure to optimize growth and development, and both are thought to have undergone co-adaptation, leading to co-expression of many of the same imprinted genes.Citation13,25 Although the expression of imprinted genes is highly regulated, our group and others have demonstrated subtle variation in the expression of these genes in the placenta of healthy newbornsCitation15,26, 27; yet, little is known about the relationships between this variation in expression and newborn and childhood outcomes. We demonstrated that infants with specific, coordinated profiles of expression of subset of imprinted genes in the placenta were at increased risk for sub-optimal performance on measures of movement quality and for exhibiting an elevated number of signs of physiologic stress of abstinence.
The NNNS is a validated assessment of infant neurobehaviorCitation16; the results of which have been shown to be predictive of later childhood medical, behavioral, and cognitive outcomes.Citation18,28 A review of the NNNS scoring system and its prediction of outcomes in comparison to other infant neurodevelopmental scoring systems may be found in portions of El-Dib et al.Citation21 Specifically, in work by Liu et al.Citation29 latent profile analysis was used to classify infants into neurobehavioral profiles based on the NNNS summary scores that were able to identify infants with neurologic and brain disease outcomes, as well as abnormal scores on measures of behavior problems, school readiness, and IQ through 4.5 y of age. Sucharew et al.Citation19 also utilized a latent profiling strategy, but among a cohort of infants with no known exposure to illicit drugs or other significant neurobehavioral risk factors, and demonstrated that infants whose NNNS profile was characterized as hypotonic demonstrated a significant 2–3 standard deviation reduction in Psychomotor Development Index scores, and 1–2 standard deviation reduction in measures of externalizing behaviors compared to infants in the other profiles. Although more work is needed to examine the predictive potential of the NNNS, as well as our derived imprinting markers, there is consistent evidence of the predictive potential of these measures.
We also created profiles using the NNNS data, and then used the random forest algorithmCitation30 to ascertain the subset of imprinted genes whose expression was most influential in differentiating infants into these neurobehavioral profiles. Subsequently, using only the 10 identified genes, we grouped individuals into 3 distinct clusters based upon their expression profile. This allowed us to explore the specific domains of neurobehavior affected by variation in the co-expression of these imprinted genes.
In our prior work, which investigated only a limited subset of 22 imprinted genes in ∼100 infants, we observed relationships between the coordinated expression of these 22 genes and sub-optimal performance in the NNNS quality of movement and handling scores.Citation15 In this study, we expanded the examination to detect the expression of 108 known and putative imprinted genes in a larger population of 615 infants. We used NanoString technology, which allows for direct detection of mRNA transcript abundance, allowing for more reproducible and robust measures of expression compared to PCR-based analyses used in prior studies.Citation31
Consistent with our prior results,Citation15 we also observed that coordinated variation of a subset of imprinted genes was associated with neurological function, defined as poor quality of movement scores, as well as increased signs of asymmetrical and non-optimal reflexes. Interestingly, we did not observe a relationship with handling in this larger cohort, but did identify an association with increased signs of overall stress. There may be several reasons for discrepancies between our previous study of gene expression and infant neurodevelopment and the current one. Among them is the larger sample size in the study presented here, the use of NanoString technology here rather than qPCR, along with the data reduction method employed here, which was not used in the prior study. Specifically, genes previously identified as having association with neurodevelopment, including homeobox A11 (HOXA11) and HOXD10, were included on the NanoString array, but not on downstream analysis due to low expression levels. These subtle differences between the studies may account for aspects of the discrepancies between them.
Among the genes to be the most discriminative, several have previously been linked to developmental disorders. This includes the paternally expressed COPG2 imprinted transcript 1 (COPG2IT1), which is located within intron 20 of COPG2,Citation32 a region identified as a candidate for the behavioral disorder Silver-Russell syndrome.Citation33 While it is unclear how this gene is specifically influencing disease phenotype, approximately 10% of Silver-Russell patients display maternal uniparental disomy (mUPD) for chromosome 7, containing this gene.Citation34 Maternal uniparental disomy for this gene would result in loss of expression, while our results suggest that increased expression of this gene was evident in individuals within expression cluster 2, who demonstrated increased risks for sub-optimal performance on the NNNS. This may suggest that within a general population, variation in both directions could be important or that overexpression of this gene in conjunction with altered expression of other genes within the cluster is contributing to phenotype.
Our analysis also identified the paternally expressed nuclear pore associated protein 1 (NPAP, previously identified as C15orf2), which has been implicated in Prader-Willi syndrome and encodes a protein associated with formation of the nuclear pore complex (NPC).Citation35,36 Several other genes associated with the formation or maintenance of the NPC have also been linked to neurodevelopment. These include nucleoporin 210 kDa (Nup210), which appears to be integral to neuronal differentiation,Citation37 and Nup133, for which a loss of function inhibited nuclear progenitor cells from terminally differentiating into neurons.Citation38 These results are interesting, as the NNNS domains that were most affected by imprinted gene expression in our study are related to neurologic functioning, and could suggest that variation to the NPC may play a role in neurodevelopmental abnormalities.
ProSAP/Shank proteins act as major scaffolding proteins at postsynaptic densities within the central nervous system. This family of proteins crosslinks receptors with cytoskeletal elements creating the necessary framework for the formation of these postsynaptic densities.Citation39 Our analysis identified SH3 and multiple ankyrin repeat domains 2 (SHANK2), a gene within this family, as a major discriminatory factor in the formation of our clusters. SHANK2 localizes within the growth cones of developing neurons,Citation40 and mice lacking all isoforms of the gene showed a marked reduction in glutamatergic transmission,Citation41 demonstrating the protein's role in neurotransmitter signaling. Mutations within this gene have been associated with autism spectrum disorder and intellectual disability in humans,Citation42,43 while loss of function in mice has led to observable autism-like behavior and hyperactivity.Citation41 The observed reduction in expression in the at-risk clusters 2 and 3 would suggest that reduced levels of this gene may play a role in the sub-optimal neurological functioning observed among infants in these expression clusters.
The results of our study are strengthened by several factors, including the robust sample size as well as the measurement of expression for the complete set of reported as well as putative imprinted genes. In addition, the placenta is a key organ in the communication between the maternal environment and the developing brain,Citation9 making it not only a valid proxy but likely a key functional feature driving infant behavioral outcomes. Yet, even with this fairly large sample size, the confidence intervals for our estimates of risk among infants within certain expression profiles are wide, due to the relatively small number of infants demonstrating the most sub-optimal performance on the NNNS evaluation. Our newborn population is relatively healthy, and at low-risk, based on classical risk factors like preterm delivery or maternal substance abuse, for significant developmental impairment, and so more importantly, these findings demonstrate that there is potentially important variation in neurobehavior even among a normative population, and suggests that this may be useful in elucidating the risk for later developmental impairment that has so far been unexplained. While this may be the case, the data was limited by our lack of information on long-term outcomes in these infants. Our use of term placentas also allows us to only examine this relationship as a cross-sectional assessment and, thus, we cannot definitively define causality or determine if there are specific periods during development where variation in imprinting may be even more relevant. In addition, while we aimed to address potential confounding factors present and assessed within the study population, it is possible that the expression of imprinted genes is a byproduct of a distinct factor outside of our analysis. Further investigation, particularly in more at-risk populations, may help to determine if imprinted gene expression is truly a mediator of some environmental factor on neurodevelopment or if this expression is just a marker of the environment, but does not play a causal role.
In summary, the study presented here is the most comprehensive look, to date, at the expression of imprinted genes within the placenta and the potential role of variation in the expression of these genes in newborn neurobehavioral outcomes. We have identified specific imprinted gene expression profiles that taken in combination with neurobehavioral profiles at birth may allow for the identification of at-risk infants so that early intervention may be implemented.
Methods
Study population
The subjects involved were part of the Rhode Island Child Health Study (RICHS), which enrolled a total of 899 mother/infant pairs at Woman and Infant's Hospital of Rhode Island (Providence, RI, USA), from Sept 1, 2009 through July 31, 2014. All subjects provided written informed consent approved by the Institutional Review Boards at Women and Infants Hospital and Dartmouth College. Mothers were between the ages of 18 and 40, free of life threatening conditions, and there were no congenital or chromosomal abnormalities in the infant. Infants were singleton births, with gestation to term (≥ 37 weeks). Term infants born small for gestational age (SGA, <10th percentile), or large for gestational age (LGA, >90th percentile), based on birth weight and gestational age calculated from the Fenton growth chart,Citation44 were selected; infants appropriate for gestational age (AGA, ≥10th percentile and ≤90th percentile) matched on gender, gestational age (±3 days), and maternal age (±2 years) were also enrolled. This design led to an overrepresentation of SGA and LGA infants within the cohort. A structured chart review was conducted to collect maternal inpatient information from the delivery. Anthropometric and clinical data was collected from the inpatient medical record from delivery, and all participants included had available data on infant birth weight, maternal age, and gestational age.
Neurodevelopmental status at birth was assessed with the NNNS, by certified psychometrists blinded to the study hypothesis. The exam was administered during the newborn inpatient stay at least 24 hours after birth but before discharge, resulting in a total of 615 infants being examined. Norms have been developed for this exam in healthy, full-term infants.Citation45
Items on the NNNS were reduced to 13 previously established summary scores.Citation16 In a prior study we utilized a latent profiling approach to classify infants into 7 distinct neurobehavioral profiles based on these summary scoresCitation22 and used these previously defined profiles to initially examine membership in these profiles with imprinted gene expression (described below). We further examined how membership in clusters defined by imprinted gene expression was associated with specific summary scores.
Placenta collection and RNA extraction
For each subject and within 2 hours of delivery, 12 samples of placenta parenchyma, 3 samples from each of 4 quadrants (totaling approximately 8–10 g of tissue) were excised. All samples were taken from the fetal side of the placenta, 2 cm from the umbilical cord insertion site, free of maternal decidua. The samples were placed immediately in RNAlater (Life Technologies, #AM7020) and stored at 4°C. At least 72 hours later, placenta samples were removed from RNAlater, blotted dry, snap-frozen in liquid nitrogen, homogenized by pulverization using a stainless steel cup and piston unit (Cellcrusher), and stored at −80°C until needed for examination. RNA was extracted by using the RNeasy kit (Qiagen, #74106) supplemented by double DNase I (Qiagen, #79254) on column digestion in order to clear any DNA contamination. Extracted RNA was finally quantified with Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., #ND-2000) and stored at −80°C.
RNA expression quantification
Placental RNA was profiled using a custom-designed code-set that included 144 known and putative imprinting genes (NanoString technologies). Briefly, RNA (100 ng) was incubated in the presence of reporter and capture probes overnight at 65°C. Following hybridization, unbound probes were removed, and the purified complexes were aligned and immobilized on imaging cartridges using an nCounter Prep Station II. Cartridges were then sealed and scanned in an nCounter Digital Analyzer for code count detection.
Code count data was first normalized against the geometric mean of spike-in controls to account for differences in hybridization and recovery. Differences in sample content were accounted for by normalizing the data against the geometric mean of the housekeeping genes GAPDH, RPL19, and RPLP0. While ACTB and B2M were also included on the array as housekeeping genes, we did not include these within the normalization due to a high level of variability between individuals, suggesting they are not appropriate for use in normalization within the placenta. Finally, the background threshold of detection was set at 2 standard deviations above the mean of the included negative controls. Expression below background threshold was set to the value of the limit of detection divided by the square root of 2 to maintain sample variability.
The subsequent analysis was restricted to the subset of expressed genes, with expression defined as surpassing the background threshold in a minimum of 60% of the samples. Based on these criteria, 108 of the 144 genes included in the code-set were determined to be expressed, and were retained for downstream analysis (Table S1). In total, expression of imprinted genes was measured for 683 unique samples, of which 6 were removed due to poor assay performance. Of the remaining 677, NNNS data was available for 615 infants.
Statistical analysis
Expression data was available for 108 genes from 615 individuals. To reduce multiple comparisons, we applied the Random Forest algorithm, through the package randomForest, a supervised classification,Citation30 to identify expression pattern similarities most related to an infant's NNNS profile. randomForest creates a ranked list of factors influential within the classification, for which we selected the top 10 influential genes to use for further analysis. We allowed the algorithm to produce 20,000 trees with 10 genes included within each for its assessment, and used the model to predict membership to the NNNS profiles 1–7.
Pearson correlations were calculated to determine the association between expression levels of the selected imprinted genes. Following the machine learning driven data reduction, individuals were binned into clusters using kmeans-clustering based upon their expression profile for the 10 selected genes. We allowed kmeans to create 3 clusters, as determined by the R package ‘nbclust’, which uses multiple tests to infer the optimal number for a specified data set.Citation46 Clustering analysis was also performed using the top 5 and 15 from the randomForest; however, 10 genes yielded optimal NNNS prediction along with differential gene expression by cluster.
Associations between cluster membership and neurobehavioral outcome were tested using a one-way ANOVA for each of the 13 NNNS summary scores. For those scores demonstrating a significant association with clustering, a logistic regression was used to model cluster membership and odds of a poor outcome score (falling within the 10th percentile associated with poor outcome) while correcting for infant gender, maternal age, infant gestational age, and birth weight. All data was analyzed using the R package version 3.1.0.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
BG drafted the manuscript and performed the statistical analyses. MK, DA, DG collected, processed, and assisted in the interpretation of expression data. BL was involved in subject recruitment, and obtaining and interpreting NNNS scores. LL, AS, JC, and CM assisted in conceiving the study and participated in its design and coordination, as well as interpretation of gene expression data. All authors read and edited the manuscript.
1073880_Supplemental_Material.zip
Download Zip (749.2 KB)Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work is supported by NIH-NIMH R01MH094609, NIH-NIEHS R01ES022223 and NIH-NIEHS P01 ES022832, US EPA grant RD83544201 in addition to NIH-R01 HD067611/R01ES022223-01A1 and the Mount Sinai Children's Environmental Health Center Pilot Fund. AJS is supported by NIH grants DA033660, HG006696, HD073731, and MH097018, and research grant 6-FY13-92 from the March of Dimes Foundation. The contents are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the publication.
References
- Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet 2014; 15:517-30; PMID:24958438; http://dx.doi.org/10.1038/nrg3766
- Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet 2010; 154C:365-76; http://dx.doi.org/10.1002/ajmg.c.30273
- Kent L, Bowdin S, Kirby GA, Cooper WN, Maher ER. Beckwith Weidemann syndrome: A behavioral phenotype-genotype study. Am J Med Genet B: Neuropsychiatric Genet 2008; 147B:1295-7; http://dx.doi.org/10.1002/ajmg.b.30729
- Cassidy SB, Dykens E, Williams CA. Prader-Willi and Angelman syndromes: Sister imprinted disorders. Am J Med Genet 2000; 97:136-46; PMID:11180221; http://dx.doi.org/10.1002/1096-8628(200022)97:2%3c136::AID-AJMG5%3e3.0.CO;2-V
- Pun FW, Zhao C, Lo WS, Ng SK, Tsang SY, Nimgaonkar V, Chung WS, Ungvari GS, Xue H. Imprinting in the schizophrenia candidate gene GABRB2 encoding GABAA receptor β-2 subunit. Mol Psychiatry 2011; 16:557-68; PMID:20404824; http://dx.doi.org/10.1038/mp.2010.47
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, et al. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet 2005; 37:625-9; PMID:15908950; http://dx.doi.org/10.1038/ng1577
- Mikaelsson MA, Constância M, Dent CL, Wilkinson LS, Humby T. Placental programming of anxiety in adulthood revealed by Igf2-null models. Nat Commun 2013; 4:2311; PMID:23921428; http://dx.doi.org/10.1038/ncomms3311
- Champagne FA, Curley JP, Swaney WT, Hasen NS, Keverne EB. Paternal influence on female behavior: The role of Peg3 in exploration, olfaction, and neuroendocrine regulation of maternal behavior of female mice. Behav Neurosci 2009; 123:469-80; PMID:19485553; http://dx.doi.org/10.1037/a0015060
- Maccani MA, Marsit CJ. Epigenetics in the Placenta. Am J Reprod Immunol 2009; 62:78-89; PMID:19614624; http://dx.doi.org/10.1111/j.1600-0897.2009.00716.x
- Novakovic B, Saffery R. The ever growing complexity of placental epigenetics - Role in adverse pregnancy outcomes and fetal programming. Placenta 2012; 33:959-70; PMID:23102655; http://dx.doi.org/10.1016/j.placenta.2012.10.003
- Gutierrez-Marcos JF, Constância M, Burton GJ. Maternal to offspring resource allocation in plants and mammals. Placenta 2012; 33, Supplement 2:e3-e10; PMID:22995735; http://dx.doi.org/10.1016/j.placenta.2012.08.006
- Frost J, Moore G. The importance of imprinting in the human placenta. PLoS Genet 2010; 6:e1001015; PMID:20617174; http://dx.doi.org/10.1371/journal.pgen.1001015
- Keverne EB. Genomic imprinting, action, and interaction of maternal and fetal genomes. Proc Natl Acad Sci U S A 2014; 112(22):6834–40; PMID:25404322.
- Diplas AI, Lambertini L, Lee M-J, Sperling R, Lee YL, Wetmur JG, Chen J. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics 2009; 4:235-40; PMID:19483473; http://dx.doi.org/10.4161/epi.9019
- Marsit CJ, Lambertini L, Maccani MA, Koestler DC, Houseman EA, Padbury JF, Lester BM, Chen J. Placenta-imprinted gene expression association of infant neurobehavior. J Pediatr 2012; 160:854-60.e2; PMID:22153677; http://dx.doi.org/10.1016/j.jpeds.2011.10.028
- Lester BM, Tronick EZ. History and description of the neonatal intensive care unit network neurobehavioral scale. Pediatrics 2004; 113:634-40; PMID:14993523
- Lester BM, Tronick EZ, in collaboration with T. Berry Brazelton M. The neonatal intensive care unit network neurobehavioral scale procedures. Pediatrics 2004; 113:641-67; PMID:14993524
- Liu J, Bann C, Lester B, Tronick E, Das A, Lagasse L, Bauer C, Shankaran S, Bada H. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics 2010; 125:e90-e8; PMID:19969621; http://dx.doi.org/10.1542/peds.2009-0204
- Sucharew H, Khoury JC, Xu Y, Succop P, Yolton K. NICU network neurobehavioral scale profiles predict developmental outcomes in a low-risk sample. Paediatr Perinatal Epidemiol 2012; 26:344-52; PMID:22686386; http://dx.doi.org/10.1111/j.1365-3016.2012.01288.x
- Stephens BE, Liu J, Lester B, Lagasse L, Shankaran S, Bada H, Bauer C, Das A, Higgins R. Neurobehavioral assessment predicts motor outcome in preterm infants. J Pediatr 2010; 156:366-71; PMID:19880137; http://dx.doi.org/10.1016/j.jpeds.2009.09.042
- El-Dib M, Massaro AN, Glass P, Aly H. Neurodevelopmental assessment of the newborn: An opportunity for prediction of outcome. Brain Dev 2011; 33:95-105; PMID:20494536; http://dx.doi.org/10.1016/j.braindev.2010.04.004
- Lesseur C, Armstrong DA, Murphy MA, Appleton AA, Koestler DC, Paquette AG, Lester BM, Marsit CJ. Sex-specific associations between placental leptin promoter DNA methylation and infant neurobehavior. Psychoneuroendocrinology 2014; 40:1-9; PMID:24485470; http://dx.doi.org/10.1016/j.psyneuen.2013.10.012
- Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol 2002; 192:245-58; PMID:12124770; http://dx.doi.org/10.1002/jcp.10129
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet 2005; 21:457-65; PMID:15990197; http://dx.doi.org/10.1016/j.tig.2005.06.008
- Kelsey G. Epigenetics and the brain: transcriptome sequencing reveals new depths to genomic imprinting. Bioessays 2011; 33:362-7; PMID:21425303; http://dx.doi.org/10.1002/bies.201100004
- Diplas AI, Lambertini L, Lee MJ, Sperling R, Lee YL, Wetmur J, Chen J. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics 2009; 4:235-40; PMID:19483473; http://dx.doi.org/10.4161/epi.9019
- Lambertini L, Marsit CJ, Sharma P, Maccani M, Ma Y, Hu J, Chen J. Imprinted gene expression in fetal growth and development. Placenta 2012; 33:480-6; PMID:22465419; http://dx.doi.org/10.1016/j.placenta.2012.03.001
- Bagner DM, Sheinkopf SJ, Miller-Loncar C, LaGasse LL, Lester BM, Liu J, Bauer CR, Shankaran S, Bada H, Das A. The effect of parenting stress on child behavior problems in high-risk children with prenatal drug exposure. Child Psychiatry Human Dev 2009; 40:73-84; PMID:18626768; http://dx.doi.org/10.1007/s10578-008-0109-6
- Liu J, Bann C, Lester B, Tronick E, Das A, Lagasse L, Bauer C, Shankaran S, Bada H. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics 2010; 125:e90-8; PMID:19969621; http://dx.doi.org/10.1542/peds.2009-0204
- Liaw A, Wiener M. Classification and Regression by randomForest. R News 2002; 2:18-22
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008; 26:317-25; PMID:18278033; http://dx.doi.org/10.1038/nbt1385
- Yamasaki K, Hayashida S, Miura K, Masuzaki H, Ishimaru T, Niikawa N, Kishino T. The Novel Gene, γ2-COP (COPG2), in the 7q32 Imprinted Domain Escapes Genomic Imprinting. Genomics 2000; 68:330-5; PMID:10995575; http://dx.doi.org/10.1006/geno.2000.6265
- Bentley L, Nakabayashi K, Monk D, Beechey C, Peters J, Birjandi Z, Khayat FE, Patel M, Preece MA, Stanier P, et al. The imprinted region on human chromosome 7q32 extends to the carboxypeptidase A gene cluster: an imprinted candidate for Silver-Russell syndrome. J Med Genet 2003; 40:249-56; PMID:12676894; http://dx.doi.org/10.1136/jmg.40.4.249
- Kotzot D, Balmer D, Baumer A, Chrzanowska K, Hamel BC, Ilyina H, Krajewska-Walasek M, Lurie IW, Otten BJ, Schoenle E, et al. Maternal uniparental disomy 7-review and further delineation of the phenotype. Eur J Pediatr 2000; 159:247-56; PMID:10789928; http://dx.doi.org/10.1007/s004310050064
- Neumann LC, Markaki Y, Mladenov E, Hoffmann D, Buiting K, Horsthemke B. The imprinted NPAP1/C15orf2 gene in the Prader-Willi syndrome region encodes a nuclear pore complex associated protein. Hum Mol Genet 2012; 21:4038-48; PMID:22694955; http://dx.doi.org/10.1093/hmg/dds228
- Wawrzik M, Unmehopa U, Swaab D, van de Nes J, Buiting K, Horsthemke B. The C15orf2 gene in the Prader-Willi syndrome region is subject to genomic imprinting and positive selection. Neurogenetics 2010; 11:153-61; PMID:20020165; http://dx.doi.org/10.1007/s10048-009-0231-z
- D'Angelo Maximiliano A, Gomez-Cavazos JS, Mei A, Lackner Daniel H, Hetzer Martin W. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell 2012; 22:446-58; PMID:22264802; http://dx.doi.org/10.1016/j.devcel.2011.11.021
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell 2008; 14:831-42; PMID:18539113; http://dx.doi.org/10.1016/j.devcel.2008.03.011
- Grabrucker AM, Schmeisser MJ, Schoen M, Boeckers TM. Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol 2011; 21:594-603; PMID:21840719; http://dx.doi.org/10.1016/j.tcb.2011.07.003
- Du Y, Weed SA, Xiong W-C, Marshall TD, Parsons JT. Identification of a novel cortactin sh3 domain-binding protein and its localization to growth cones of cultured neurons. Mol Cell Biol 1998; 18:5838-51; PMID:9742101
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, Janssen A-L, Udvardi PT, Shiban E, Spilker C, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 2012; 486:256-60; PMID:22699619
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet 2010; 42:489-91; PMID:20473310; http://dx.doi.org/10.1038/ng.589
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem 2002; 81:903-10; PMID:12065602; http://dx.doi.org/10.1046/j.1471-4159.2002.00931.x
- Fenton T, Nasser R, Eliasziw M, Kim J, Bilan D, Sauve R. Validating the weight gain of preterm infants between the reference growth curve of the fetus and the term infant. BMC Pediatr 2013; 13:92; PMID:23758808; http://dx.doi.org/10.1186/1471-2431-13-92
- Tronick EZ, Olson K, Rosenberg R, Bohne L, Lu J, Lester BM. Normative neurobehavioral performance of healthy infants on the neonatal intensive care unit network neurobehavioral scale. Pediatrics 2004; 113:676-8; PMID:14993526
- Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: an r package for determining the relevant number of clusters in a data set. J Statistical Software 2014; 61.
