ABSTRACT
Although numerous imprinted genes have been described in several lineages, the phenomenon of genomic imprinting presents a peculiar evolutionary problem. Several hypotheses have been proposed to explain gene imprinting, the most supported being Haig's kinship theory. This theory explains the observed pattern of imprinting and the resulting phenotypes as a competition for resources between related individuals, but despite its relevance it has not been independently tested. Haig's theory predicts that gene imprinting should be present in eusocial insects in many social scenarios. These lineages are therefore ideal for testing both the theory's predictions and the mechanism of gene imprinting. Here we review the behavioral evidence of genomic imprinting in eusocial insects, the evidence of a mechanism for genomic imprinting and finally we evaluate recent results showing parent of origin allele specific expression in honeybees in the light of Haig's theory.
Introduction
Genomic imprinting is the differential expression of alleles in diploid individuals, with expression being dependent upon the sex of the parent from which the allele was inherited.Citation1 Genomic imprinting is an evolutionary paradox. Natural selection is expected to favor expression of both alleles to protect against recessive mutations that render a gene ineffective.Citation2 What then is the benefit of silencing one copy of a gene, making the organism functionally haploid at that locus? Several explanations for the evolution of genomic imprinting have been proposed.Citation3
Haig's kinship theory, is the subject of this review. It is the most developed and best supported theoryCitation3 and the most applicable to social insects.Citation4 Outside the scope of this review are several non-conflict based theories to explain the evolution of genomic imprinting that have been recently reviewed by Spencer and Clark.Citation5
Haig's theory is based on the idea that maternally (matrigene) and paternally (patrigene) inherited genes in the same organism can have different selectional pressures (described in greater detail in the “Haig's kinship theory” section and ). For example, in a species with multiple paternity, a patrigene has a lower probability of being present in siblings that are progeny of the same mother than does a matrigene. As a result, a patrigene will be selected to value the survival of the individual it is in more highly, compared with the survival of siblings. This is not the case for a matrigene. In mammals and angiosperms, this conflict is played out in the provisioning of offspring with resources taken from the mother.Citation2 A patrigene will benefit by causing more maternal resources to be allocated to the individual it is in, but a matrigene will benefit from sharing resources among all the siblings. A method to differentiate matrigenes and patrigenes of these genes (i.e., imprinting) would spread in the population.
Haig's kinship theory is central to our evolutionary understanding of imprinting effects in human health and plant breeding;Citation6,7 yet, despite its importance, the theory still lacks an independent test. Social insects have been suggested as such an independent test of Haig's kinship theory for the evolution of genomic imprinting.Citation4 Here we review the behavioral evidence of genomic imprinting in social insects, evaluating recent results showing parent of origin allele specific expression in honeybees asking whether Haig's kinship theory predictions are supported. We also review the evidence for mechanisms in social insects for genomic imprinting in light of what is known in mammals. We then explore advances in technologies (e.g., next-generation sequencing, chromatin immunoprecipitation, CRISPR) that can be used in dissecting this mechanism genetically and molecularly. Finally, we propose a direction for future research suggesting a merger of genetic and molecular approaches to resolve pressing evolutionary and mechanistic questions on the nature of genomic imprinting.
Genomic imprinting
Genomic imprinting is a phylogenetically widespread phenomenon, and genes have been shown to be imprinted in mammals,Citation8 plants, and fungi,Citation9,10 as well as insects,Citation11 although see Coolon et al.Citation12 In mammals, genomic imprinting is associated with both DNA and histone methylation and these epigenetic markers are established in the germline of parents and are preserved during development (see Box 1 Genomic Imprinting: the mammalian way).Citation13
Haig's kinship theory
Haig's kinship theory is an extension of the theory of inclusive fitness that was developed by W. D. Hamilton in 1964.Citation14 The inclusive fitness theory postulates that the fitness of an individual is not solely dependent upon their own survival and reproduction, but it is also reliant on the survival and reproduction of those that share the same genes. Thus, an individual will favor actions that appear to be altruistic toward related individuals that will ensure shared genes are passed on to future generations.
Haig's kinship theory is currently the most widely accepted theory for the evolution of genomic imprinting.Citation5 Developed in the late 1990s,Citation15 Haig assumes that in a polyandrous mating system the genetic relatedness of the offspring is higher for maternally inherited alleles (matrigenes) than it is for paternally inherited alleles (patrigenes). Therefore, selection is expected to act on patrigenes to increase individual resource allocation from the mother, thus increasing paternal inclusive fitness. Whereas matrigenes should be selected to favor equal resource distribution among offspring to maximise maternal inclusive fitness.Citation16 In this context the same genes may experience different evolutionary pressures depending upon whether they are in the parents or in their progeny (F1). Genes in the mother (or father) are referred to as maternal (or paternal) genes. Genes from the mother (or the father) in the progeny are called matrigenic (or patrigenic) and a matrigene (or a patrigene) refers to an allele in the progeny derived from the mother (or the father).
This theory explains for example how the cost-benefit ratio changes during sibling competition for parental resources (i.e., time, food).Citation4 If there is no genomic imprinting and no difference in the coefficient of relatedness between patrigenes and matrigenes of offspring, an individual should weigh its own benefit (relatedness coefficient, r:1) against the cost for the sibling affected (full sibling, r:1/2; half sibling, r:1/4) and selfish or selfless behavior should not be particularly associated to patrigenes or matrigenes (). In a different situation where there is imprinting and the cost falls on siblings with a different father, the coefficient of relatedness is different between matrigenes (r:1/2) and patrigenes (r:0) (). In this situation patrigenes may be selected to be always selfish, while for matrigenes selfishness is advantageous only when the benefit is greater than 1/2 the cost to a sibling of a different father.
This theory has been used to explain why imprinting is prominent in angiosperms and mammals with extended parental care.Citation1,2 The theory also provides an explanation to why many genes expressed in embryos, placenta, and sperm are imprinted, as well as why paternally and maternally imprinted genes have opposite effects on offspring size.Citation1,4 Paternal genes are expected to favor larger offspring while maternal genes should favor smaller offspring in species with a polyandrous mating system that also provide maternal care.Citation1 Larger offspring are able to elicit more resources from the mother, which is beneficial for the individual but detrimental for the siblings. This would be advantageous to patrigenes, that may not be present in all siblings, but disadvantageous to a matrigene that has a 50% chance of being present in the siblings. For example, inactivation of the paternal allele of the Mest gene in mice (maternally imprinted) produced smaller, lighter pups,Citation17 giving weight to this theory. The function of most identified imprinted genes in mammals support this theory.Citation18,19
The theory also extends beyond systems with known imprinting; QuellerCitation4 expands on Haig's original suggestions and applies the kinship theory to social insect colonies. The haplodiploid sex determination of eusocial insects creates a potential conflict of interest between patrigenes and matrigenes in a given offspring due to different degrees of relatedness with brothers and sisters ().Citation1,4 QuellerCitation4 is able to make predictions for genomic imprinting based on various social contexts, for example queen competition and insertion of new queens, male killing, and worker reproduction. These predictions provide a solid ground with which to test the validity of the kinship theory and provide evidence for conflict within an individual genome.
Do social insects possess the machinery for genomic imprinting?
Haig's kinship theory predicts imprinting for some genes in certain conditions, but this implies that the parents are able to differentially label their genes. How do they do that? Imprints are established in gametogenesis and embryogenesis, and are present in different tissues predominantly involved in the provision of nutrients to offspring. In mammals for example, most of the identified imprinted genes are expressed in the placenta.Citation20 Moreover, the majority of imprinted genes in the angiosperm lineage are expressed in the endosperm which surrounds the plant embryo and is primarily a source of starch, but also oils and proteins.Citation21 Genes have also been reported to be imprinted in the mammalian brain,Citation22,23 and potentially in the angiosperm embryo too [see;Citation24 however also seeCitation25,26].
Although much is still not understood, many molecular components of the mechanism of genomic imprinting (e.g., DNA methylation, histone modifications) are well studies in mammals and in plants (for recent reviews of gene imprinting mechanism in plants seeCitation27,28). Below we review the evidence for the presence of these imprinting mechanism in eusocial insects [seeCitation29].
DNA methylation
Among the epigenetic markers, the methylation of the fifth carbon of cytosine (5mC) is found to be associated with control of transcription and regulation of splicingCitation30 (see Box 1, Genomic Imprinting: the mammalian way and ).
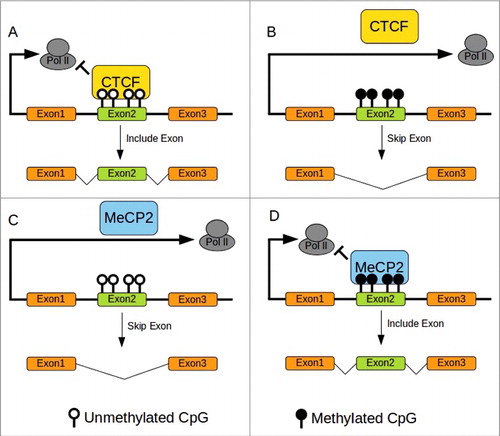
Figure 1. DNA methylation and splicing in mammals. (A)CTCF Binding to unmethylated CTCF binding sites pause Pol II elongation allowing retention of exon2. (B)CTCF cannot bind to methylated sites resulting in skipping exon2. (C)MeCP2 does not bind to unmethylated sites allowing rapid progression of Pol II resulting in skipping of exon2. (D)MeCP2 binding to methylated site pause elongation of Pol II permitting retention of exon2. Redrawn from Yan et al. 2015.Citation30 CTCF binding site with methylation sensitive CpG in bold: ATGCAGCTAGATGGCGCTC.Citation74
DNA encoding for the DNA methyltransferases (DNMTs) and 5mC have been found in many hymenopteran species.Citation31 In mammals, distinct classes of DNMTs are responsible for maintaining methylation through cell division (DNMT1) and de novo methylation (DNMT3s),Citation32 but the insect catalytic activities of these enzymes have not been formally characterized. Some species, like the silkworm B. mori, have only a single DNMT with dual DNMT1/3 functions.Citation33 The fruit fly, D. melanogaster, has lost both DNMT 1 and 3 and encodes for a DNMT2 enzyme that is involved in RNA methylation.Citation33 Hymenoptera maintain all 3 classes of enzymes (DNMT1, 2, and 3) in single or multiple copies.Citation31,33-36 The hymenoptera methylation toolkit also includes 10–11 translocation enzymes (TET) that catalyze the oxidation of 5mC to 5-hydroxymethyl cytosine (5hmC).Citation37 This enzyme and 5hmC are thought to be part of the process of demethylation in mammalsCitation38 and may also have similar roles in insects.
The distribution of DNA methylation in hymenoptera shows similarities as well as differences with that of mammals. In mammals more than 70% of CpGs are methylated and methylation can be found in promoters as well as gene bodies,Citation39 while in hymenoptera less than 2% of CpGs are methylated and are primarily localized in gene bodies.Citation31,35,40
In mammals, whole genome assays uncovered a substantial difference between isolated CpG (usually heavily methylated) and hypomethylated grouped CpGs (CpG islands or CGIs: region of 500–2000 bp).Citation41 A long held view associates DNA methylation with transcriptional repression, but deep analysis of CGIs and intragenic methylation began to erode this classic view. CGIs tend to correlate with promoter regions and can be classified as High, Intermediate or Low CpG density promoters (HCPs, ICPs, and LCPs respectively). HCPs are linked to transcriptionally active genes and increases in methylation indeed result in gene silencing. ICPs are also inactive when methylated, although the level of methylation changes significantly during differentiation.Citation42,43 In contrast, LCPs are usually hypermethylated and transcriptionally active.Citation42,44 Moreover, active genes in the inactive imprinted X chromosome, show a high level of gene body methylation. This could simply be a consequence of the transcription availability since in this case the genes would also be accessible to the DNA methylases,Citation45 or the higher level of methylation may be related to RNA transcription and/or splicing given that the pattern of methylation marks intron-exon boundaries.Citation46
In eusocial insects, the caste system imposes another layer of organization to the reprogramming and cell lineage specification that may be controlled by epigenetic mechanisms. It is possible that like different cell lineages acquire a particular epigenome during development, also different castes are epigenetically defined during development.Citation47,48 There may be programming and re-programming windows during development and later in life, as for example suggested by a study in honeybee (A. mellifera) where phenotypic plasticity between nurses and foragers correlated with variation in DNA methylation in the brain.Citation49 However, re-programming outside these coding windows may be difficult and result in less than fully penetrating phenotypes.Citation50,51
Larvae, during development, may be considered dual potent because they retain the ability to develop into different castes (e.g., queens vs. workers) and many epigenetic tools (DNA methylation, miRNA, piRNA, etc.) as well as alternative splicing were implicated in this plasticity in the honeybee A. mellifera and to a lesser extent in the ants Camponotus floridanus and Harpegnathos saltator.Citation33-35,52 Interestingly, DNMT1a is maternally transmitted to the progeny in the wasp N. vitripennis and its downregulation results in a block of development 10–12 h after egg laying, while DNMT3 is not essential for early embryo development.Citation53 Similarly in mice, deletion of DNMT1 is lethal at gastrulation resulting in a global loss of DNA methylation.Citation54 Differences in behavior in adult castes may be also associated with a difference in epigenetic programming. The role of DNA methylation in this context was recently tested with contrasting results in the honeybee A. mellifera, the ants C. floridanus, H. saltator, Solenopsis invicta, and Ooceraea biroi.Citation31,35,49,55,56 Moreover, the function of DNA methylation is reported to be of less importance in the simple eusocial societies of the wasp Polistes canadensis and the ant Dinoponera quadriceps where these species are more reliant on transcriptional network re-organization to determine phenotype differences, in comparison with highly eusocial insects.Citation57
Libbrecht et al. 2016, questioned the results of previous studies reporting caste dependent differences in DNA methylation, suggesting that they used a false positive-prone method in their next-generation sequencing data analyses.Citation55 However, using methylation-sensitive AFLP, Amarasinghe and colleaguesCitation58 also found differences in methylation in the heads of reproductive and non-reproductive workers in queenless B. terrestris colonies. Drug manipulation of the methylation level clearly correlated DNA methylation with changes in phenotype/behavior of the workers. Importantly, this data support Haig's kingship theory predictions that genomic imprinting should be important in worker reproductive behavior. The theory predicts in fact that there should be conflict between maternally and paternally derived alleles in loci involved with workers reproduction resulting in genomic imprintingCitation4 (see section: Parent of origin allele specific expression).
Whether differences in DNA methylation are hallmarks of different caste specific behavior is still to be resolved. It is worth noting here that DNA methylation plays an important role in learning and memory formation in both mammals (rats) and honeybees and is dynamically regulated in mammalian adult brains.Citation59,60
Gene body methylation and splicing
CpG methylation in gene bodies seem to have a conserved function between mammals and hymenoptera. CpG methylation was found mostly in exons marking intron-exon boundaries and was associated with controlling splicing in the honeybee A. mellifera,Citation34,49,61,62 in the wasp N. vitripennisCitation63 (but see alsoCitation40) and in both the ants C. floridanus and H. saltator.Citation35 Highly methylated genes were found to be uniformly and transcriptionally active in different conditions and representing housekeeping genes expressed in most cell types.Citation34,35,40,64,65 This also holds true in a social insect outside of hymenoptera, the termite Zootermopsis nevadensis (isoptera) shows the same intragenic methylation associated with highly expressed genes and alternative splicing.Citation66 In contrast, sparsely methylated genes are associated with caste-specific gene expression in germ lines in honeybees.Citation67 How can DNA methylation mediate splicing and even participate in the process of alternative splicing? Recent data from mammals may provide some clues.
DNA methylation can change the availability of a locus to a protein or a protein complex. This is the case of ZFP57, a protein important to maintaining genomic imprinting during development in mammals (see Box 1: Genomic Imprinting: the mammalian way). ZFP57 affinity for its binding site increases with methylation.Citation68
Another example is the allele-specific expression of H19/IGF2 locus. The allele-specific level of methylation of the imprinting control region (ICR) in this locus mediates the binding of CCCTC-binding factor (CTCF) that in turn controls the expression of H19 (maternal allele) or IGF2 (paternal allele).Citation69 The binding of CTCF is a pausing signal for RNA polymerase II (Pol II). DNA methylation inhibits CTCF binding, therefore enabling Pol II elongation resulting in skipping of an exon ().Citation70 This mechanism could explain why hypomethylation was associated with alternative gene splicing in the gene alk in the honeybee A. mellifera and lipoprotein receptor 2 in the ant C. floridanus.Citation71,72
Conversely, the mammalian methyl-CpG-binding protein 2 (MeCP2) is involved in exon retention, but mirrors CTCF in its affinity to DNA methylation ().Citation73 MeCP2 has greater affinity for its methylated binding site inducing a pause in Pol II elongation and retention of the target exon (). High levels of methylation were associated with retention of exons in the ants C. floridanus and H. saltatorCitation35 that could be achieved via a MeCP2 like mechanism. These 2 mechanisms (MeCP2 like, CTCF like) could be operating in concord, resulting in a complex relationship between DNA methylation and control of alternative splicing.
It is important here to note that MeCP2 was implicated in neuronal function that in mammals relies heavily on alternative splicing.Citation75 Similarly in A. mellifera the transition from nursing to foraging associates with alternative splicing events.Citation49 In the plant Arabidopsis thaliana and in the green spotted puffer-fish Tetraodon nigroviridis a particular histone variant (H2A.Z) is enriched in hypomethylated loci and could serve as a marker for alternative splicingCitation76,77 while in mammals the methylation of H3K9 by the Histone H3K9 methyltransferase G9a recruits DNMTs to unmethylated loci and changes chromatin status.Citation78 Although little evidence is at the moment available, it has been shown that histone deacetylases (HDACs) in the ant C. floridanus are involved in the transition to foraging/scouting suggesting a possible role for histone modifications in determining caste-specific patterns of behavior.Citation79 It is probable that DNA methylation is therefore acting in concert with other epigenetic modifiers (e.g., ncRNAs, histone modifications) to regulate alternative splicing in hymenopteraCitation71,80 in a cell specific manner.
Further investigation is needed to reveal the mechanistic role of DNA methylation and other epigenetic modifications in regulating alternative splicing in hymenoptera.
Other epigenetic mechanisms
Regardless of the role of DNA methylation, other epigenetic markers could be important in defining a specific caste related behavior. Long non-coding RNA (lncRNA) and microRNA (miRNA) are potential additions to the epigenetic tool kit in defining behaviors. miRNA may have a role in determining caste-specific expression profiles in A. mellifera, targeting unmethylated genes during development.Citation81
Changes in miRNA expression profiles are also associated with the nurse-forager transition.Citation82 Another class of small RNA (piRNA, PIWI interacting RNA) are associated to genomic imprinting of the paternally methylated Rasgrf1 in mammals.Citation83 In addition, some lncRNA have being implicated in mediating aspects of brain development, functional diversification (kakusei,Citation84), and the transition from nursing to foraging (Nb-1,Citation85) in A. mellifera. In mammals, lncRNA are also related to genomic imprinting (for recent reviews seeCitation86 and Citation87). All clusters of imprinted genes in mammals include one or more lncRNACitation88 and the genes and the lncRNA in these clusters are often expressed from the opposite parental chromosomes.Citation89 lncRNA use a variety of molecular strategies to epigenetically regulate transcription of imprinted clusters.Citation86,87
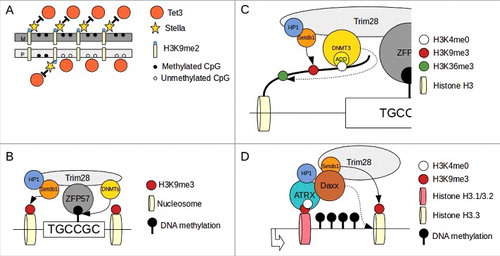
Figure 2. Mechanism of gene imprinting in mammals. (A)Stella binding to H3K9me2 prevent TET3 dependent de-methylation. Maternal (M) DNA is enriched of H3K9me2 compared with paternal (P) and therefore maternal imprinted regions are protected from active de-methylation. (B)After zygote formation and during the firsts cell divisions, level of methylation at imprinted loci is maintained by ZFP57/TRIM28 complex binding to methylated consensus and recruitment of DNMTs. (C)DNMT3a/b interact with H3 tail via ADD domain and their activity is permitted only when H3K4 is unmethylated (H3K4me0). This modification is enriched in the DNA methylated allele of imprinted loci (see text). DNMT3s interact also via a PWWP domain (dotted arrow) to H3K36me3 (enriched in gene bodies of active genes). H3K9me3 methylation is maitained by Setdb1 (continuous arrow). (D)At transcriptionally active imprinted loci the S-phase specific H3.1/H3.2 histones are exchange for the cell cycle independent H3.3 histone. The ATRX/Daxx complex is responsible for the exchange of H3.1/3.2 with H3.3 (dotted arrow). ATRX bind H3K4me0 via an ADD domain and interact also with to H3K9me3. Daxx recruit Setdb1 that maintain H3K9me3 methylation (continuous arrow). Re-draw from Messerschmidt et al. 2014 and Voon and Gibbons 20168,91
More work is required to reveal how the different epigenetic markers are intertwined together to generate the different pattern of behavior and expression, genomic imprinting during gameto- and embryogenesis resulting in phenotypic plasticity in hymenoptera.
It seems that even the variant of histone allocated to imprinted loci contributes to maintaining imprinting. During development the H3.3 histone variant was found to be enriched in heterochromatic regions including the DNA methylated allele in imprinted loci.Citation125,126 The chaperone responsible for the allocation of H3.3 in this region is the ATRX/Daxx complex that preferentially targets the methylated allele.Citation125,126 ATRX has an ADD domain to interact with H3K4me0 and can also directly bind H3K9me3 and HP1.Citation127 It has been shown in addition that Daxx interacts with TRIM28 and Setdb1.Citation128 So the ATRX/Daxx complex could represent another way (alternatively or in combination with ZFP57) to recruit TRIM28/Setdb1 to imprinted regions and maintain DNA methylation during development ().
Whether such an exquisite epigenetic reprogramming with successive waves of de-methylation and re-methylation during gametogenesis and embryogenesis exists in hymenoptera, is not currently known. The role of histone modification is also unknown. In honeybee at the present time there is no evidence for a comprehensive de-methylation program during embryogenesis.Citation129 Moreover, methylation levels are retained through early stages of development, as they also are in other animals (e.g., Danio rerio, Caenorhabditis elegans) although the specific mechanism is still to be determined.Citation130,131
Box 1. Genomic Imprinting: The mammalian way
The expression of imprinted genes in mammals is coordinated by allele-specific (maternal or paternal) DNA methylation in imprinting control regions (ICRs).Citation90 Allele-specific methylation or parent-of-origin-specific methylation is introduced during gametogenesis and it is preserved throughout the entire life. During gametogenesis in fact, the somatic epigenetic code is removed in the primordial germ cells (PGCs). A new sex specific and germ specific epigenetic code and transcription profile is established and finally a post-fertilization removal of this epigenetic code is activated to allow further embryonic development.Citation91 The ICRs allele specific methylation therefore must be protected from the post-fertilization wave of de-methylation.
The first wave of de-methylation in the PGCs erases the somatic epigenetic code and establishes totipotency. The global level of DNA methylation at this stage is dramatically reduced and parental imprinting and X-chromosome silencing are removed in mouse embryos.Citation92 This reprogramming is achieved trough a combination of passive and active de-methylation. During the PGCs proliferation the de-novo DNMT3s are repressed and Np95 (an essential cofactor for DNMT1) is either downregulated or excluded from the nucleus.Citation92,93 This passive de-methylation affects the genome globally, but some regions (including imprinted genes, CGIs of inactive X-chromosomes) are hypomethylated only after an active de-methylation involving TET1/2 and 5hmC.Citation92,94
Following this PGC re-programming, a new sex-specific epigenetic code and ICR methylation is established. The exact mechanism of this precise re-methylation is still to be uncovered, but it appears to be related more to the chromatin structure, histone modifications and transcription factors, as opposed to sequence specificity.Citation95,96
The second wave of epigenetic reprogramming takes place post fertilization during early embryogenesis and shows important differences to the quasi total de-methylation occurring in PGCs. The kinetics of de-methylation of the zygote genome for example is very different compared with PGCs, and imprinted regions tend to be protected during this de-methylation process, allowing parent-of-origin-specific gene expression.Citation97-100 The level of methylation and the kinetics of de-methylation of the paternal and maternal DNA is very different. The sperm DNA has double the level of methylation (80–90% CpGs) than DNA of the egg (40% CpGs), and is quickly de-methylated soon after zygote formation.Citation97-99 On the contrary, the maternal DNA undergoes a delayed, replication-dependent de-methylation.Citation97-99 This implies that an efficient active de-methylation mechanism must be in place soon after fertilization and that the maternal DNA must be shadowed to it (). This differential de-methylation required an active TET3 that specifically localizes to the paternal pronucleous inducing a conversion from 5mC to 5hmC as a first step in the de-methylation program.Citation101 Since the affinity of DNMT1 for oxidized 5mC (5hmC, 5fC and 5caC see below: A molecular future) is extremely poor, it is also possible that methylation is reduced passively during cell division.Citation102-104
The maternal genome in the zygote is protected from the TET3 activity by a protein called STELLA (also PGC7 or Dppa3). STELLA has a strong affinity for the maternally enriched dimethylated histone H3 Lys9-marked chromatin (H3K9me2) and this interaction results in a change of chromatin structure that prevents TET3 binding.Citation105 The protection from the rapid de-methylation driven by differential paternal-maternal H3K9me2/STELLA interaction could also be a mechanism to maintain genomic imprinting. In fact, imprinted gene loci undergo a complete loss of methylation if STELLA is not present and some paternally imprinted genes are found to retain H3K9me2-marked chromatin, preserving them from rapid de-methylation post fertilization ().Citation105
The maternal DNA sees a passive loss of methylation achieved by nuclear exclusion of DNMT1. The level of methylation of maternally imprinted genes is however preserved and a low level of nuclear DNMT1 is essential to maintain genomic imprinting.Citation106 DNMT1 in this context is part of a complex of proteins that has at its center ZFP57 (Krueppel-associated box (KRAB) domain zinc finger protein) and TRIM28 (also KAP1).Citation107 Other components of this complex are nucleosome remodelling and histone deacetylation (NuRD), H3K9me3-catalyzing histone methyltransferase (Setdb1), heterochromatin protein 1 (HP1), and DNMT3a/bCitation108-110 (). This complex localizes to ICRs, possibly thanks to a recognition site for ZFP57 (TGCCGC).Citation111 ZFP57 affinity for this binding site is methylation dependent and increases when the site is methylated.Citation68,108 So during early cell divisions the ZFP57/TRIM28 complex, brings DNMT1 to ICRs in a ZFP57 target-dependent manner, enabling the maintenance of DNA methylation during this early stage.Citation112 TRIM28 also interacts with HP1 and Setdb1.Citation113 Since H3K9me3 associates strongly with DNA methylation, the TRIM28/HP1/Setdb1 complex may participate (via histone modifications) in the preservation of DNA methylation during this early stage of development.
This mechanism may be robust enough to permit even some initial loss of methylation in imprinted regions (as it was observed) as long as it does not affect the ZFP57 target sites.Citation114,115 The ZFP57/TRIM28 complex recruit DNMT3a/b that may be able to compensate for the initial loss of methylation.Citation114,115 ZFP57 is expressed during embryogenesis, but is restricted to ovaries and testes in the adultsCitation116 suggesting that ZFP57 is necessary to maintain methylation at imprinted loci during development.
DNA methylation is often associated with histone modifications. We have already encountered 2 of these modifications (H3K9me2–3), but also H3K4me3 and H3K36me3 play fundamental roles in the preservation of DNA methylation in imprinted loci. H3K4me3 for example specifically inhibits DNA methylation and is relegated to the unmethylated allele in imprinted regions.Citation117 DNMT3a/b and their co-factor DNMT3L interact to the histone 3 (H3) tail via a ATRX-DNMT3-DNMT3L (ADD) domain. Methylation of H3K4 (H3K4me3) either prevents this interaction (with DNMT3L) or inhibits DNMT3 activity (DNMT3a/b).Citation118 The PWWP domain of DNMT3a/b is important for the interaction with H3K36me3, which intriguingly is found to be enriched in gene bodies of active genes.Citation119-121 So DNMT3a/b localization to a specific imprinted region depends on the absence of H3K4me3 (H3K4me0) and the presence of H3K9me3 (see above).Citation122-124
Therefore, methylation at imprinted loci is maintained thanks to the ZFP57/TRIM28 localization to these regions that results in the preservation of H3K9me3 via Setdb1 and DNMT3a/b recruitment ().
Behavioral evidence of genomic imprinting in social insects
Differences in worker ovary size and stinging behavior in A. mellifera show strong parent-of-origin effectsCitation132,133 suggesting that an imprinting mechanism may be in place.
It has been extensively demonstrated that honeybee defensive behaviors are heritable in a parent-of-origin dependent manner. Africanised honeybees (Apis mellifera scutellata) are known to exhibit a more rapid stinging response to alarm stimuli (visual or pheromonal), that is more sensitive, with a lower threshold before initiating a sting response, compared with the European subspecies.Citation132 Through backcrossing and reciprocal cross studies this defensive behavior has been correlated with paternal inheritance. Drones from F1 queens of a cross of Africanised drones and European queens were used for backcrosses in Stort and Goncalves' study which found increased defensive behaviors in backcross colonies.Citation134 Guzman-Novoa and Page report workers of a cross with an Africanised paternity have a sting response equal to that of those with full Africanised inheritance.Citation135 Backcrossing of F1 gynes twice with European drones resulted in the same worker stinging response as full European control workers. Similarly, DeGrandi-Hoffman and colleagues found that colonies with an Africanised paternity showed a higher level of defensive behaviors regardless of whether the colony was founded by an Africanized or European queen.Citation136 Reciprocal crosses by Guzman-Novoa et al. also showed F1 workers with Africanised paternity to have a greater sting response compared with F1 workers with European paternity. F1 colonies with European paternity exhibited a response intermediate to that of control European and Africanised colonies.Citation137
If imprinting is present, one would expect these asymmetries in behavioral phenotypes between F1 colonies of a reciprocal cross. These examples of parent-specific behaviors are consistent with the theory of imprinting via increased patrigenic expression or silencing of matrigenic expression. High expression of patrigenic alleles involved in increased aggressive behaviors with conspecifics is likely to promote the lineage of those individuals.Citation137 Whereas matrigenic alleles of the same genes are benefited when silenced as it will reduce the costs of increased colony defensive behaviors.
It is also worth noting a non-social, parasitic wasp (Nasonia vitripennis) of the same lineage (hymenoptera), also has been reported to show a “grandfather effect” in which F1 females of reciprocal crosses of closely related species show mating behaviors similar to their paternal heritage.Citation138
Although the heritability of defensive behaviors has been well recorded, it has been questioned whether there is a sufficient range of empirical studies regarding the effect of imprinting on conflict-resolution behaviors.Citation139 Though more recently, other examples of potential behavioral effects of imprinting in social insects have been identified in the reproductive behavior of honey beesCitation133 and caste allocation and behavior effects in ants.Citation140,141
A reciprocal cross between 2 honey bee subspecies (Apis mellifera capensis and A. m. scutellata) found F1 workers with an A. m. capensis father to produce on average 30% more ovarioles than F1 workers with an A. m. scutellata father.Citation133 In the wild, A. m. capensis workers are unique in the ability to produce reproductive female offspring through thelytokous parthenogenesis.Citation142 Hence male A. m. capensis individuals may experiencing an increased “motivation” for their worker daughters to produce a greater number of ovarioles and monopolise gyne reproduction. Whereas founding queens are neither benefited or at a disadvantage from worker gyne production.
Parent-of-origin specific effects on caste and behavior have also been reported in ants.Citation140,141 Libbrecht et al.Citation141 conducted Linepithema humile crosses to reveal the proportion of queens and workers produced in a colony as determined by the paternal lineage. The proportions of queens and males, and all females and males were affected by the interaction between parental lineages. L. humile ants from the same crosses were also used to demonstrate that the maternal lineage affected the behaviors of efficiency to collect pupae, foraging propensity, and the distance between non-brood-tenders and brood.Citation140 Similarly, the paternal lineage influenced 2 other ant behaviors: the efficiency to feed larvae and the distance between brood-tenders and brood. Therefore, differences in parental selection pressures exist, and this observed behavior is highly consistent with the traits of genomic imprinting.
Parent of origin allele specific expression
Haig's theory predicts workers reproduction genes should be imprinted
Haig's kinship theory predicts different outcomes in terms of imprinted genes and selfish/selfless behavior depending on the specific social structure considered. Here we outline the theory in relation to 2 important species: Apis mellifera (European honeybee), a single diploid queen mated by multiple haploid males and Bombus terrestris (Buff-tailed bumblebee), a single diploid queen mated by a single haploid male. We also discuss how the theories predictions change for each species depending on the presence or absence of the queen, referred to as a queenright colony or queenless colony respectively ().
Figure 3. Genetic basis for intragenomic conflict in A. mellifera (left) and B. terrestris (right). In honey bee colonies a single queen (diploid) mates with multiple haploid males. In bumblebee colonies the queen mates only with a single male. Re-drawn from Queller 2003 and Galbraith et al. 2016.Citation4,143
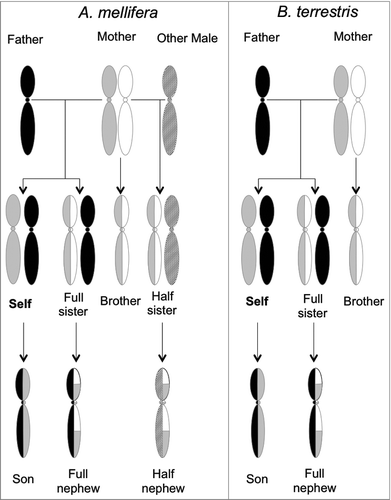
Why should a worker (self; ) take care of her sisters instead of reproducing? In haplodiploid species females hatch from fertilized (diploid) eggs, while males hatch from unfertilized (haploid) eggs, so the degree of relatedness between offspring is different for matrigenes and patrigenes. In the case of bumblebees, the coefficient of relatedness between self ( right) and other sisters is r:3/4 (they share 3/4 of their genome on average), which is higher than their relatedness with their own offspring (r:1/2), so this may explain their selfless behavior.
These coefficients of relatedness between bumblebee sisters or toward offspring change when calculated independently from the perspective of a patrigene and a matrigene. For a patrigene r:1 since all sisters have the same father, while r:1/2 for a matrigene. In addition, for patrigenes in self, r:0 with any males produced by the queen. So in a queenright bumblebee colony, the only chance for the self patrigene to be passed to the next generation is to favor self reproduction or reproduction by her sisters (either workers or new queens) at the cost of rearing fewer brothers. Matrigenes however should be selected to regulate self reproduction when it is associated with high costs to brothers as these genes are less likely to occur in nephews than they are in brothers.
However, should the singly mated bumblebee queen die and the self begin to reproduce, a patrigene would have the same probability to be passed to the next generation either by self or self's sisters (r:1/2). Patrigenes should now not experience selective pressure for reproductive behavior, meaning sterility and nursing behavior are just as beneficial as reproductive behavior. For a matrigene the situation is different since this probability is reduced (r:1/4) in the self's sisters. So under queenless conditions with self reproducing, matrigenes may be imprinted to promote reproduction and avoid sterility.
In honeybee colonies queens mate with multiple males, so a worker (self; left) has both full sisters and half sisters. The coefficient of relatedness between self and sisters varies between r:3/4 for full sisters and r:1/4 for half sisters. From a patrigene perspective r:1 with full sisters but r:0 with half sisters because the latter have a different father. To a matrigene half sisters and full sisters looks the same having both r:1/2. The matrigene in a worker has an equal (50%) chance of being in a brother as well as in self's offspring. Therefore, the matrigene tries to ensure an equal distribution of her resources among both her sons and daughters. In a queenright situation once again patrigene may promote self reproduction because this is the only way to be passed to the next generation. In a queenless situation self should be selected to begin to reproduce, since the patrigene (with no relatedness to half sisters and their progeny) will only increase its fitness if self, or full sisters, reproduce. On the contrary, matrigenes having the same degree of relatedness with both full and half nephews (r:1/4) should be selected to moderate reproduction if this comes at a high cost to half sisters. So in the case of honey bees when a queen dies and self begin to reproduce, patrigenes may been imprinted to promote reproduction and avoid sterility.
Haig's kinship theory provides the theoretical basis to predict genomic imprinting in hymenopteran eusocial insects (e.g., A. mellifera, B. terrestris, C. floridanus). These insects have full epigenetic toolkits including DNMTs and TETs, they display complex social structures with caste specific behaviors, and differences in matrigene-patrigene relatedness due to the haplo-diploid sex determination, which are all elements that reinforce the expectation for genomic imprinting. So, is there any evidence for genomic imprinting in hymenoptera?
Evidence for parent of origin allele specific expression
Testing candidate genes using allele-specific (parent-specific) amplifications, Amarasinghe and colleaguesCitation145 identified 2 genes (ecdysone 20-monooxygenase-like and IMP-L2-like) with parent-of-origin specific expression in B. terrestris. The pattern of expression was consistent with the prediction of Haig's kinship theory that genes associated with initiation of worker reproduction should be paternally expressed in queenright B. terrestris workers, and genes that inhibit reproduction should be maternally expressed. Indeed, Ecdysone 20-monooxygenase-like, a gene involved in ovary activation,Citation146 was paternally expressed and IMP-L2-like, that was implicated in inhibition of worker reproduction,Citation147 was maternally expressed. This expression profile was evocative of a possible genomic imprinting mechanism at work.
Recently a few studies have looked for parent-specific gene expression (PSGE), possibly due to genomic imprinting, with interesting results in the honeybee A. mellifera.Citation143,144 These studies took advantage of reciprocal crosses to investigate the prediction that PGSE should affect reproductive and cooperative behaviors of non- reproductive workers (). This approach uncouples parent-of-origin effects from lineage-specific effects, generating single drone insemination between 2 different lineages. In Kocher et al. 2015 the authors used 2 different lineages from Europe (Apis mellifera carnica) and Africa (A. mellifera scutellata) while in Galbraith et al. 2016 they used A. mellifera ligustica from both continents. Crucially for the identification of any PSGE, the lines used showed behavioral differences and most importantly they had many single nucleotide polymorphisms (SNPs) that allowed identification of maternal and paternal alleles.Citation148 In Kocher et al. 2015, the authors identified only a small group of significant PSGE, mostly maternally biased, while in Galbraith et al. 2016, the authors reported an association between patrigene expression and worker reproduction (). In Kocher et al. 2015 the parental allele expression was compared in queen right guard worker bees (full body), larvae (full body), and individual forager brains. They found 12 (larvae), 17 (guards), and 23 (brains) maternally biased genes and 3–4 (guard brains) paternally biased genes. In this environment (queenright) one may expect to find imprinted genes that repress worker reproduction and mediate social behavior (perhaps reducing aggression). The adult tissues where it is more likely that genomic imprinting would result in PGSE could reasonably be the ovary (being directly affected by changes in reproduction) and the brain (mediating reproduction related behavior). Indeed one of the genes in the brain with PGSE was huntingtin that has been associated with stinging behavior,Citation137 and a maternally expressed gene in larvae (Neural Lazarillo) that downregulates insulin signaling, a pathway important for queen-worker differentiation.Citation149
Figure 4. Reciprocal crosses. Females from different lineages (A and B) are crossed reciprocally with single drones of the opposite lineages (B and A). Because of SNPs between the lineages maternal and paternal alleles are recognizable in F1. The global level of expression of maternal (Mat) and paternal (Pat) alleles in a tissue can be tested by RNA-seq. Parental Bias results in an overexpression of the Parental allele (Mat or Pat) in both crosses. Lineage Bias results in an overexpression of the lineage specific allele (A or B). Redraw from Kocher et al 2015.Citation144
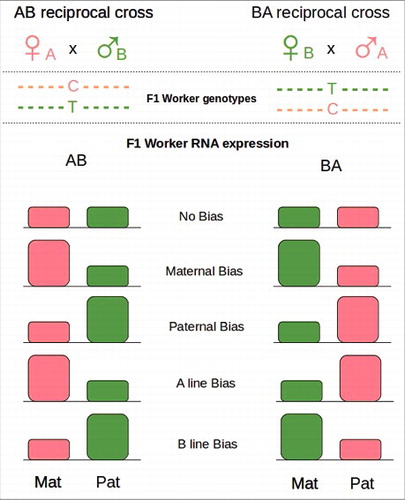
Table 1. Comparison of PSGE in 2 Apis mellifera studies, Kocher et al. 2015 and Galbraith et al. 2016.
Table 2. Useful Technologies for addressing questions of mechanism of GI.
Most likely a combination of different reasons explained the small number of maternally biased genes identified in Kocher et al. 2015. For instance no particular phenotypic or behavioral difference was imposed on the bees tested. Moreover, full bodies were used for larvae and guards, masking any possible tissue specific effects (even though they used forager brains with similar results)(). But most importantly they suffer from a strong lineage-of-origin bias that would cloud any other allele-specific bias. In fact, they suggest that mito-nuclear incompatibilities in the hybrids were affecting the results, and more recently they expand on that observation confirming it.Citation150
In the paper by Galbraith and colleagues, the authors took advantage of a clear phenotypic and behavioral difference between African and European bees where African workers tend to have larger ovaries and become more readily reproductively activeCitation151 (). These differences allow the kinship theory prediction that patrigenes should promote reproduction to be tested. In the reciprocal crosses a strong patrigenic effect was evident at the phenotypic level. In fact workers from the reciprocal crosses with African fathers showed an increased number of ovarioles and were more likely to become reproductive compared with workers with European fathers. To test for PSGE, workers from the reciprocal crosses were maintained in an environment (queenless and broodless) that was inducive to reproduction, and difference in expression was measured in ovaries and fat bodies of reproductive and non-reproductive individuals. In this condition one would expect that regardless of the lineage-of-origin of the father, patrigenes should be upregulated in both non-reproductive and reproductive workers but even more so in the latter. Of the 303 identified transcripts with an allele bias, the number of patrigenes over represented in sterile and reproductive workers was 143 and 181 respectively with an additional 12 and 13 matrigenes. These results broadly confirmed the initial predictions. Genes of the ecdysone-vitellogenin pathway, controlling egg production, including vitellogenin (that encode for a yolk precursor protein), showed paternal expression. Some genes that were paternally biased only in reproductive workers included yolkless, ecdysone receptor (ECR) and ecdysone-induced protein 75 (E75).
Comparing transcription of ovaries between reproductively active and inactive individuals could include an intrinsic source of noise. In fact, although oocytes are not transcriptionally active and the on-set of zygotic transcription is usually placed after egg deposition (5 h after egg laying in the wasp N. vitripennisCitation152), the oocytes include several maternally transferred mRNA (for a review seeCitation153) that may not be represented in tissue from non reproductive individuals.
Both papers provide evidence for parent-of-origin effects in the progeny, but they offer very different results. This may suggest that PSGE is conditional to the specific context in which the difference in expression is tested, implying that different genes may be parentally imprinted in different cell lineages. PSGE may therefore be revealed only in specific environmental contexts. For example, an imprinted gene promoting ovary development, may result in a difference in expression solely when a worker becomes reproductive in a queenless, broodless situation and in a specific tissue (e.g., ovary). In the same animal a gene may be imprinted in one tissue and not in another, and a difference in expression becomes visible only in a tissue-specific, condition-specific manner.
How parental allele bias is achieved is still unknown but some interesting observations can be drawn from these papers. In mammals, genomic imprinting is often associated with DNA methylation but Kocher et al. 2015 did not find any association between their parentally biased genes and known methylated genes (). We compared Galbraith et al. 2016 list of PSGE with known methylated genes (taken fromCitation67,34,31 andCitation49), not only did we not find a correlation between the 2, in fact there were zero genes in common between the 2 lists. These findings have also been mirrored in,Citation154 where only a small list of genes were found to overlap between Galbraith et al. 2016 PSGE list and gamete/embryo methylated genes from.Citation155 It is entirely possible little overlap is seen because the samples between these studies are too varied, PSGE and methylation may differ depending on tissue and developmental stage. When investigating the role of methylation in PSGE it is worth noting epialleles, where the sequence itself determines methylation status, can potentially be confused for parent-specific methylation.Citation156 It is vital that future studies, exploring methylation as a regulatory mechanisms for genomic imprinting in hymenoptera, take this potential confusion into consideration. However, in hymenoptera, methylation is often found in gene bodies marking intron-exon junctions and is associated with splicing more than gene silencing. Chromatin remodelling in the fruit fly Drosophila melanogaster was associated with parent-of-origin expressionCitation11 and both Kocher et al. 2015 and Galbraith et al. 2016 identified genes involved in this process. So it is quite possible that other epigenetic mechanisms (e.g., histone modifications) are generating the genomic imprinting that provoke the PSGE observed in these papers.
A molecular future: Prospects
Next generation sequencing technologies have permitted an extraordinary advance in our understanding of the genetics of many hymenoptera species in a relatively short time. RNA-seq and sequencing of bisulfite converted DNA (BS-seq) are powerful tools enabling researchers to ask and answer new and important questions about the distribution and functions of DNA methylation in hymenoptera as well as to test the prediction of Haig's kinship theory. As noted above careful analyses of these data are required, epiallelesCitation156 and poor statistical analysisCitation64 could lead to genomic imprinting being incorrectly labeled. In fact Wedd et al. 2016 describe a differentially methylated epiallele (of the gene AmLAM ) in the honeybee, where an increase in methylation correlated with an increase in gene expression, but only in certain developmental stages and tissue types.Citation157 This shows how important it is to take into account epialleles, splice forms, tissue type and developmental stage when exploring methylation as a mechanism for genomic imprinting in insects.
Recently other modifications on cytosines have been described in many tissues in mammals including 5-hydroxymethylationcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). TET enzymes can convert 5mC to 5hmC and further oxidise it to 5fC and 5caC in a possible demethylation pathway back to unmodified C.Citation38,158,159 5hmC in addition may be an epigenetic mark on its ownCitation160 for example changing the DNA interaction dynamics of 5mC binding proteins and functioning as an off-switch.Citation161 The identification of intermediate C modifications may therefore be important in the context of rapid re-programming associated with response to environmental changes, tissue specific epigenomes, caste specification, and genomic imprinting. BS-sequencing cannot discriminate between 5mC and 5hmC because both are resistant to deamination by sodium bisulfite treatments so until recently whole genome quantification of 5hmC has been very difficult. New technologies however are now available to map both 5hmC and 5fC at a genome level with single base resolution.Citation162-164 Tet- assisted bisulfite sequencing (TAB-Seq) and oxidative bisulfite sequencing (oxBS-Seq) combined with BS-seq has allowed mapping of full genome 5hmC at single-nucleotide resolution,Citation162,163 while reduced bisulfite sequencing (redBS-Seq) permits identification of 5fC with the same resolution.Citation164 To increase their statistical and biologic power, future experimental design should consider carefully both number of independent biologic replicates, tissue specificity as well as developmental and social context.
Although DNA methylation seems to have some conserved functions between mammals and hymenoptera (e.g., control of splicing) it is not clear whether its role in genomic imprinting is shared by the 2 groups. Genomic imprinting is widely found in mammalsCitation165 and plants,Citation166 and imprinted chromosomal regions have also been identified in insects,Citation167 fishCitation168 and nematodes.Citation169 The epigenetic markers that are most frequently associated with genomic imprinting are histone modifications. Methylation of some of histone 3 (H3K9, H3K27, H3K4) and 4 (H4K20) lysines are among the most common modifications associated with genomic imprinting.Citation170 In particular H3K9me is a shared feature of imprinting in mammals (see Box 1, Genomic Imprinting: the mammalian way), plants (A. thaliana)Citation171 and insects (Planococcus citri ).Citation172 Even in Drosophila, where only a small fraction of the genome shows any methylation at all (0.5% in early embryoCitation173), both H3K9 and H3K4 methylation are associated with genomic imprintingCitation11,174 suggesting that these modifications may be part of an ancestral common mechanism. Chromatin immunoprecipitation sequencing (ChIP-seq) is a versatile technology that identifies genome wide DNA-protein interactionsCitation175 and has been successfully used to investigate histone modificationsCitation176 and chromatin remodelling complexesCitation177 as well as RNA polymerase and transcription factor binding regions.Citation178,179 This approach used in the context of hymenoptera species could provide key answers regarding changes in chromatin structure and histone modifications related to different development stages, caste definition, and possible mechanisms of genomic imprinting. A limitation of this approach is that it requires a large sample size (1–10 million cells) that could be problematic to achieve especially in the context of environment-specific/tissue-specific epigenetic re-programming.Citation175,180 Nevertheless, both ChIP and ChIP-seq have been successfully used in D. melanogaster,181182 and recent advances in this technologies such as indexing-first ChIP (iChIP)Citation183 and high- throughput ChIP (HT-ChIP)Citation184 dramatically reduce the amount of cells required and could be adapted to work in insects.
CRISPR gene editing is one of the most exciting and far reaching tools available today to the molecular biologist for manipulation of target genes. CRISPR has been successfully used in a variety of insect species including Diptera (Drosophila melanogaster, Anopheles gambiae, Aedes aegypti),Citation185-187 Coleoptera (Tribolium castaneum),Citation188 Lepidoptera (Bombyx mori ),Citation189 and lately hymenoptera (Nasonia vitripennis, Apis mellifera, Ooceraea biroi ).Citation190-192 Due to differences in life cycle, social structure, and ease of laboratory manipulation, some hymenoptera species may be more suitable than others to support CRISPR approaches. At the very least this technology could be used to induce knockout or mutations of genes suspected to be involved in genomic imprinting (e.g., DNMTs, TETs, Histone modification proteins), but it has also the potential to facilitate the functional investigation of protein domains and control target expression. Mimicking an approach that has been already tested for mammalian DNMTs,Citation193 CRISPR could facilitate the generation of transgenic flies (D. melanogaster) expressing hymenoptera genes (e.g., DNMTs, TETs, Histone modification proteins) in null backgrounds, therefore enabling their functional characterization, contributing greatly to our understanding of the mechanism of genomic imprinting. The functional characterization of DNMTs could also help to understand why hymenoptera and mammals differ so dramatically in their level of CpG methylation. A possibility is that the “common” status of hymenoptera DNMTs is to be functionally/molecularly inactive or in a “closed” conformation and require a cofactor(s) to be “open” and/or active. Alternatively, an unknown cofactor(s) may strongly repress DNMTs limiting their activity to specific sites. In addition, other epigenetic mechanisms (e.g., histone modifications) may contribute to the observed CpG methylation difference between mammals and hymenoptera, making DNA more or less accessible to DNMTs, implying in fact that the majority of hymenoptera DNA is unavailable at any given time/tissue. CRISPR may also permit the implementation of DNA adenine methyltransferase (Dam) identification (DamID)Citation194 a powerful technology that has been used for the identification of chromatin interacting proteins and other DNA-protein interactions (see Pindyurin et al., 2016195 for a recent DamID application in D. melanogaster). Recently a very exciting application of CRISPR-Cas9 technology has been used to manipulate the level of methylation of specific cytosines in a target specific manner. Mammalian cells were transformed with a catalytic inactive form of Cas9 fused with either TET1 or DNMT3a along with gRNAs that direct enzymatic activity in a sequence specific manner.Citation196,197 This technology, adapted to be used in vivo in hymenoptera, will be a powerful tool to study the function of specific CpG methylation and the contribution of DNA methylation to genomic imprinting, splicing, caste definition, tissue specificity, and much more.
Using a combination of these technologies in a hypothesis driven context (e.g., Haig's kinship theory predictions) will allow us to explore the mechanism of genomic imprinting in hymenoptera. Moreover, it will be possible to explore commonalities as well as differences between hymenoptera and other insects, mammals, and plants in the mechanism of genomic imprinting.
Closing remarks
The quest for understanding genomic imprinting and the mechanisms that generate and maintain it in hymenoptera is only beginning. Although many questions are still unanswered, some relevant observations are already possible. Genomic imprinting may be not a common characteristic to all hymenoptera since evidence are lacking for some species (e.g N. vitripennis) and may be more common in eusocial insects as initial investigation in A. mellifera suggests. Genomic imprinting may not have a binary status but be very much a variable of time (developmental time), social context (e.g., caste definition, queenright or queenless), tissue, and physiologic status. By its very nature, the epigenetic code (including DNA methylation, C intermediate status, histone modifications, and more) is plastic and responsive to environmental changes, tissue specific, and probably variable during development. So it is not far fetched to suppose that also genomic imprinting, relying on epigenetic modification for its generation and maintenance, could be as plastic. The definition of a specific context (e.g., tissue specificity, social context, age) is particularly important for studies that apply an expression only approach. A poor choice of tissue and or social context may produce results of difficult interpretation. Future work therefore will be facing the challenge of defying with particular care the context of genomic imprinting investigation and should combine multiple technologies to investigate differences in expression as well as changes in epigenetic markers.
Whether DNA methylation is associated with genomic imprinting in hymenoptera is still an open question; however, its role in controlling splicing has been supported in different species in many studies. Alternative splicing could be a possible target of genomic imprinting where the expression of a splicing variant is parent-of-origin specific. Examples of sex specific alternative splicing are found both in the fruit fly D. melanogasterCitation205 and mammalsCitation206 and DNMT1 alternative exon splicing results in maternal/paternal differential expression in mammalian germ cells.Citation207 In hymenoptera, DNA methylation could play a crucial role mediating the inclusion/exclusion of introns/exons in a parent-of-origin specific manner determining the parent-of-origin variant expression. The investigation of other epigenetic markers including intermediate C modifications and euchromatin signals (e.g., histone modifications) is becoming crucial for the understanding of the mechanism of genomic imprinting. It is now fundamental to identify the proteins mediating genomic imprinting as well as defining their molecular functions. This task as challenging as it is, may be facilitated by the availability of technologies such as CRISPR and Chomatin IP that allow us to explore the role of target genes as well as whole genomes. The vast amount of knowledge from mammals, plants, and Drosophila regarding the genes and histone modifications involved in genomic imprinting as well as the resources already available for these species, could serve as a vantage starting point for the challenges ahead.
Is it possible to generate and maintain gene imprinting without DNA methylation in hymenoptera, and what is the role of H3K9 and H3K4 methylation and other histone modifications? What is the relation if any between 5mC, 5hmC and 5fC and gene imprinting/alternative splicing? Is alternative splicing a target of genomic imprinting? What are the proteins mediating genomic imprinting? These are but few of the questions that need to be answered to understand the mechanism of genomic imprinting in hymenoptera.
The studies today availableCitation143,144 suggest that genomic imprinting is present in the honeybee A. mellifera but in a very complex interaction between several variables. Questioning the prediction of Haig's kin theory in other species has become now essential to reveal whether it is a common strategy in eusocial insects. Will genomic imprinting be pervasive in other eusocial insects (e.g., B. terrestris, C. floridanus)?
We think that we are now at the dawn of a very exciting time having the extraordinary opportunity to dissect the epigenetic code and genomic imprinting with unprecedented detail and from both mechanistic and evolutionary points of view using population genetics and molecular biology approaches. This will provide answers to many of today's questions and hopefully allow us to ask many more.
Disclosure of potential conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Haig D. The kinship theory of genomic imprinting. Annual review of ecology and systematics 2000; 9-32 doi:10.1146/annurev.ecolsys.31.1.9
- Hurst LD. Evolutionary theories of genomic imprinting. In W. Reik and A. Surani, editors, Genomic imprinting. IRL Press, Oxford, 1997.
- Patten MM, Ross L, Curley JP, Queller DC, Bonduriansky R, Wolf JB. The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity 2014; 113(2):119-28; PMID:24755983; https://doi.org/10.1038/hdy.2014.29
- Queller DC. Theory of genomic imprinting conflict in social insects. BMC Evol Biol 2003; 3(1):1; PMID:12515583; https://doi.org/10.1186/1471-2148-3-15
- Spencer HG, Clark AG. Non-conflict theories for the evolution of genomic imprinting. Heredity 2014; 113(2):112-8; PMID:24398886; https://doi.org/10.1038/hdy.2013.129
- Ubeda F. Evolution of genomic imprinting with biparental care: Implications for Prader-Willi and Angelman syndromes. PLoS Biol 2008; 6(8):e208; https://doi.org/10.1371/journal.pbio.0060208
- George C, Hui KYH, Scorza R, Nip W-K. Transgenic Plants and Crops. CRC Press, March 2002. ISBN 978-0-203-91097-9.
- Voon HPJ, Gibbons RJ. Maintaining memory of silencing at imprinted differentially methylated regions. Cell Mol Life Sci 2016; 73(9):1871-79; PMID:26883803; https://doi.org/10.1007/s00018-016-2157-6
- Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 2001; 293(5532):1070-4; PMID:11498574; https://doi.org/10.1126/science.293.5532.1070
- Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 2007; 23(4):192-9; PMID:17316885; https://doi.org/10.1016/j.tig.2007.02.004
- Joanis V, Lloyd V. Genomic imprinting in Drosophila is maintained by the products of suppressor of variegation and trithorax group, but not polycomb group, genes. Mol Genet Genomics 268(1):103-12, 2002; PMID:12242505; https://doi.org/10.1007/s00438-002-0731-0
- Coolon JD, Stevenson KR, McManus CJ, Graveley BR, Wittkopp PJ. Genomic Imprinting absent in Drosophila melanogaster adult females. Cell Rep 2012; 2(1):69-75. ISSN 22111247. URL http://linkinghub.elsevier.com/retrieve/pii/S2211124712001738; https://doi.org/10.1016/j.celrep.2012.06.013
- Wood AJ, Oakey RJ. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet 2006; 2(11):e147; PMID:17121465; https://doi.org/10.1371/journal.pgen.0020147
- Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol 1964; 7:1-16. URL https://doi.org/10.1016/0022-5193(64)90038-4
- Haig D. Placental hormones, genomic imprinting, and maternal—fetal communication. J Evolutionary Biology 1996; 9(3):357-380; https://doi.org/10.1046/j.1420-9101.1996.9030357.x
- Wilkins JF, Haig D. Inbreeding, maternal care and genomic imprinting. J Theor Biol 2003; 221(4):559-64; PMID:12713940; https://doi.org/10.1006/jtbi.2003.3206
- Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene mest. Nat Genet 1998; 20(2):163-9; PMID:9771709; https://doi.org/10.1038/2464
- Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol 2002; 192(3):245-58. ISSN 0021-9541, 1097-4652. https://doi.org/10.1002/jcp.10129. URL http://doi.wiley.com/10.1002/jcp.10129
- Ogunwuyi O, Upadhyay A, Adesina SK, Puri R, Foreman TM, Hauser BR, Cox J, Afoakwah E, Porter A, Annan E, et al. Genetic imprinting: Comparative analysis between plants and mammals. Plant Tissue Culture and Biotechnology 2016; 26(2):267-84; https://doi.org/10.3329/ptcb.v26i2.30576
- Renfree MB, Suzuki S, Kaneko-Ishino T. The origin and evolution of genomic imprinting and viviparity in mammals. Philosophical Transactions of the Royal Society B: Biological Sciences 2012; 368(1609):20120151-20120151. ISSN 0962-8436, 1471-2970; URL http://rstb.royalsocietypublishing.org/cgi/doi/10.1098/rstb.2012.0151; https://doi.org/10.1098/rstb.2012.0151
- Gehring M. Genomic imprinting: insights from plants. Genetics 2013; 47(1):187; https://doi.org/10.1146/annurev-genet-110711-155527
- Keverne EB. Importance of the matriline for genomic imprinting, brain development and behaviour. Philosophical Transactions of the Royal Society B: Biological Sciences 2012; 368(1609):20110327-20110327. ISSN 0962-8436, 1471-2970. https://doi.org/10.1098/rstb.2011.0327. URL http://rstb.royalsocietypublishing.org/cgi/doi/10.1098/rstb.2011.0327
- Swaney WT, Curley JP, Champagne FA, Keverne EB. Genomic imprinting mediates sexual experience-dependent olfactory learning in male mice. Proc Natl Acad Sci U S A 2007; 104(14):6084-9. ISSN 0027-8424, 1091-6490. https://doi.org/10.1073/pnas.0609471104. URL http://www.pnas.org/cgi/doi/10.1073/pnas.0609471104
- Jahnke S, Scholten S. Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol 2009; 19(19):1677-81. ISSN 09609822; PMID:19781944; https://doi.org/10.1016/j.cub.2009.08.053. URL http://linkinghub.elsevier.com/retrieve/pii/S0960982209016303
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh C-T, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM. Parent-of-Origin effects on gene expression and DNA Methylation in the maize endosperm. Plant Cell 2011; 23(12):4221-33. ISSN 1040-4651, 1532-298X. URL http://www.plantcell.org/cgi/doi/10.1105/tpc.111.092668; https://doi.org/10.1105/tpc.111.092668
- Gehring M, Missirian V, Henikoff S. Genomic analysis of Parent-of-Origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 2011; 6(8):e23687. ISSN 1932-6203. URL http://dx.plos.org/10.1371/journal.pone.0023687; https://doi.org/10.1371/journal.pone.0023687
- Rodrigues JA, Zilberman D. Evolution and function of genomic imprinting in plants. Genes Dev 2015; 29(24):2517-2531
- Satyaki PRV, Gehring M. DNA methylation and imprinting in plants: machinery and mechanisms. Crit Rev Biochem Mol Biol 2017; 52(2):163-75; PMID:28118754; https://doi.org/10.1080/10409238.2017.1279119
- Galbraith DA, Yi SV, Grozinger CM. Evaluation of possible proximate mechanisms underlying the kinship theory of intragenomic conflict in social insects. Integr Comp Biol 2016; 56(6):1206-14; PMID:27940613; https://doi.org/10.1093/icb/icw111
- Yan H, Bonasio R, Simola DF, Liebig J, Berger SL, Reinberg D. DNA methylation in social insects: how epigenetics can control behavior and longevity. Annu Rev Entomol 2015; 60:435-52; PMID:25341091; https://doi.org/10.1146/annurev-ento-010814-020803
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol 2010; 8(11):e1000506
- Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci 2014; 39(7):310-8; PMID:24947342; https://doi.org/10.1016/j.tibs.2014.05.002
- Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet 2011; 27(4):127-31; PMID:21288591; https://doi.org/10.1016/j.tig.2011.01.003
- Foret S, Kucharski R, Pellegrini M, Feng S, Jacobsen SE, Robinson GE, Maleszka R. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci U S A 2012; 109(13):4968-73; https://doi.org/10.1073/pnas.1202392109
- Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol 2012; 22(19):1755-64; PMID:22885060; https://doi.org/10.1016/j.cub.2012.07.042
- Sadd BM, Barribeau SM, Bloch G, de Graaf DC, Dearden P, Elsik CG, Gadau J, Grimmelikhuijzen CJ, Hasselmann M, Lozier JD, et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol 2015; 16(1):1
- Wojciechowski M, Rafalski D, Kucharski R, Misztal K, Maleszka J, Bochtler M, Maleszka R. Insights into DNA hydroxymethylation in the honeybee from in-depth analyses of tet dioxygenase. Open Biol 2014; 4(8):140110
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333(6047):1300-3; PMID:21778364.
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 2010; 107(19):8689-94; https://doi.org/10.1073/pnas.1002720107
- Wang X, Wheeler D, Avery A, Rago A, Choi JH, Colbourne JK, Clark AG, Werren JH. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet 2013; 9(10):e1003872; PMID:24130511
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011; 25(10):1010-22; https://doi.org/10.1101/gad.2037511
- Meissner A1, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008; 454(7205):766-70; PMID:18600261
- Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nature Genet 2010; 42(12):1093-100; PMID:21057502
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genet 2007; 39(4):457-66; PMID:17334365; https://doi.org/10.1038/ng1990
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science 2007; 315(5815):1141-43; PMID:17322062; https://doi.org/10.1126/science.1136352
- Anastasiadou C, Malousi A, Maglaveras N, Kouidou S. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol 2011; 30(5):267-75; PMID:21545276; https://doi.org/10.1089/dna.2010.1094
- Patalano S, Hore TA, Reik W, Sumner S. Shifting behaviour: epigenetic reprogramming in eusocial insects. Curr Opin Cell Biol 2012; 24(3):367-73; PMID:22429916; https://doi.org/10.1016/j.ceb.2012.02.005
- Chittka A, Wurm Y, Chittka L. Epigenetics: the making of ant castes. Curr Biol 2012; 22(19):R835-38; PMID:23058801; https://doi.org/10.1016/j.cub.2012.07.045
- Herb BR, Wolschin F, Hansen KD, Aryee MJ, Langmead B, Irizarry R, Amdam GV, Feinberg AP. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat Neurosci 2012; 15(10):1371-73; PMID:22983211
- Shi YY, Huang ZY, Zeng ZJ, Wang ZL, Wu XB, Yan WY. Diet and cell size both affect queen-worker differentiation through DNA methylation in honey bees (Apis mellifera, apidae). PloS One 2011; 6(4): e18808
- Sumner S, Kelstrup H, Fanelli D. Reproductive constraints, direct fitness and indirect fitness benefits explain helping behaviour in the primitively eusocial wasp, Polistes canadensis. Proc Biol Sci 2010; 277(1688):1721-8. page rspb20092289
- Glastad KM, Hunt BG, Yi SV, Goodisman MA. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol 2011; 20(5):553-65; PMID:21699596; https://doi.org/10.1111/j.1365-2583.2011.01092.x
- Zwier MV, Verhulst EC, Zwahlen RD, Beukeboom LW, van de Zande L. DNA methylation plays a crucial role during early Nasonia development. Insect Mol Biol 2012; 21(1):129-38; PMID:22122805; https://doi.org/10.1111/j.1365-2583.2011.01121.x
- Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyl- transferases. PLoS Genet 2012; 8(6):e1002750
- Libbrecht R, Oxley PR, Keller L, Kronauer DJ. Robust DNA methylation in the clonal raider ant brain. Curr Biol 2016; 26(3):391-5; PMID:26804553
- Glastad KM, Hunt BG, Yi SV, Goodisman MA. Epigenetic inheritance and genome regulation: is DNA methylation linked to ploidy in haplodiploid insects? Proceedings of the Royal Society of London B: Biological Sciences 2014; 281(1785):20140411; https://doi.org/10.1098/rspb.2014.0411
- Patalano S, Vlasova A, Wyatt C, Ewels P, Camara F, Ferreira PG, Asher CL, Jurkowski TP, Segonds-Pichon A, Bachman M, et al. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc Natl Acad Sci U S A 2015; 112(45):13970-5; https://doi.org/10.1073/pnas.1515937112
- Amarasinghe HE, Clayton CI, Mallon EB. Methylation and worker reproduction in the bumble-bee (Bombus terrestris). In Proc Biol Sci 2014; 281:20132502
- Lockett GA, Wilkes F, Maleszka R. Brain plasticity, memory and neurological disorders: an epigenetic perspective. Neuroreport 2010; 21(14): 909-13; PMID:20717061; https://doi.org/10.1097/WNR.0b013e32833e9288
- Biergans SD, Jones JC, Treiber N, Galizia CG, Szyszka P. DNA methylation mediates the discriminatory power of associative long-term memory in honeybees. PloS One 2012; 7(6):e39349
- Li-Byarlay H, Li Y, Stroud H, Feng S, Newman TC, Kaneda M, Hou KK, Worley KC, Elsik CG, Wickline SA, et al. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc Natl Acad Sci U S A 2013; 110(31):12750-55; https://doi.org/10.1073/pnas.1310735110
- Lockett GA, Kucharski R, Maleszka R. DNA methylation changes elicited by social stimuli in the brains of worker honey bees. Genes, Brain and Behavior 2012; 11(2):235-42. ISSN 16011848; https://doi.org/10.1111/j.1601-183X.2011.00751.x
- Park J, Peng Z, Zeng J, Elango N, Park T, Wheeler D, Werren JH, Yi SV. Comparative analyses of DNA methylation and sequence evolution using Nasonia genomes. Mol Biol Evol 2011; 28(12):3345-54; PMID:21693438
- Libbrecht R, Oxley PR, Keller L, Kronauer DJ. Robust DNA methylation in the clonal raider ant brain. Curr Biol 2016; 26(3):391-5. ISSN 09609822; https://doi.org/10.1016/j.cub.2015.12.040. URL http://linkinghub.elsevier.com/retrieve/pii/S0960982215015717.
- Hunt BG, Brisson JA, Yi SV, Goodisman MA. Functional conservation of DNA methylation in the pea aphid and the honeybee. Genome Biol Evol 2010; 2:719-28; PMID:20855427
- Glastad KM, Gokhale K, Liebig J, Goodisman MA. The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci Rep 2016; 6:37110. ISSN 2045-2322; https://doi.org/10.1038/srep37110. URL http://www.nature.com/articles/srep37110.
- Elango N, Hunt BG, Goodisman MA, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A 2009; 106(27):11206-11; https://doi.org/10.1073/pnas.0900301106
- Liu Y1, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of ZFP57 recognition of CpG methylation within a specific DNA sequence. Genes Dev 2012; 26(21):2374-79; https://doi.org/10.1101/gad.202200.112
- Min HY, Lee SC, Woo JK, Jung HJ, Park KH, Jeong HM, Hyun SY, Cho J, Lee W, Park JE, et al. Essential role of DNA methyltransferase 1-mediated transcription of Insulin-like growth factor 2 in resistance to histone deacetylase inhibitors. Clin Cancer Res 2016; 23(5):1299-311. pages clincanres–0534.
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011; 479(7371):74-9; PMID:21964334
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell 2011; 144(1):16-26; PMID:21215366
- Perales R, Bentley D. “Co-transcriptionality:” the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 2009; 36(2):178-91; PMID:19854129; https://doi.org/10.1016/j.molcel.2009.09.018
- Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MECP2 to promote exon recognition. Cell Res 2013; 23(11):1256-69; PMID:23938295; https://doi.org/10.1038/cr.2013.110
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016; 529(7584):110-4; PMID:26700815
- Guy J, Cheval H, Selfridge J, Bird A. The role of MECP2 in the brain. Annu Rev Cell Dev Biol 2011; 27:631-52; PMID:21721946
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010; 328(5980):916-9; PMID:20395474; https://doi.org/10.1126/science.1186366
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 2008; 456(7218):125-29; PMID:18815594; https://doi.org/10.1038/nature07324
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009; 10(5):295-304; PMID:19308066; https://doi.org/10.1038/nrg2540
- Simola DF, Graham RJ, Brady CM, Enzmann BL, Desplan C, Ray A, Zwiebel LJ, Bonasio R, Reinberg D, Liebig J, et al. Epigenetic (re) programming of caste-specific behavior in the ant Camponotus floridanus. Science 2016; 351(6268):aac6633; PMID:26722000; https://doi.org/10.1126/science.aac6633
- Bonasio R. Emerging topics in epigenetics: ants, brains, and noncoding RNAs. Ann N Y Acad Sci 2012; 1260(1):14-23; PMID:22239229; https://doi.org/10.1111/j.1749-6632.2011.06363.x
- Ashby R, Forêt S, Searle I, Maleszka R. microRNAs in honey bee caste determination. Sci Rep 2016; 6:18794
- Greenberg JK, Xia J, Zhou X, Thatcher SR, Gu X, Ament SA, Newman TC, Green PJ, Zhang W, Robinson GE, et al. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav 2012; 11(6):660-670; https://doi.org/10.1111/j.1601-183X.2012.00782.x
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse RASGRF1 locus. Science 2011; 332 (6031):848-52; PMID:21566194; https://doi.org/10.1126/science.1203919
- Kiya T, Ugajin A, Kunieda T, Kubo T. Identification of kakusei, a nuclear non-coding RNA, as an immediate early gene from the honeybee, and its application for neuroethological study. Int J Mol Sci 2012; 13(12):15496-509; PMID:23443077; https://doi.org/10.3390/ijms131215496
- Tadano H, Yamazaki Y, Takeuchi H, Kubo T. Age and division-of-labour dependent differential expression of a novel non-coding RNA, Nb-1, in the brain of worker honeybees, Apis mellifera. Insect Mol Biol 2009; 18(6):715-26; PMID:19817910; https://doi.org/10.1111/j.1365-2583.2009.00911.x
- Wu R, Su Y, Wu H, Dai Y, Zhao M, Lu Q. Characters, functions and clinical perspectives of long non-coding RNAs. Mol Genet Genomics 2016; 291(3):1013-33; PMID:26885843; https://doi.org/10.1007/s00438-016-1179-y
- Kanduri C. Long noncoding RNAs: Lessons from genomic imprinting. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2016; 1859(1):102-11; https://doi.org/10.1016/j.bbagrm.2015.05.006
- Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long non-coding RNAs. Epigenetics 2009; 4(5):277-86; https://doi.org/10.4161/epi.4.5.9242
- Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 2014; 6(2):a018382; PMID:24492710; https://doi.org/10.1101/cshperspect.a018382
- Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 2011; 12(8):565-75; PMID:21765458; https://doi.org/10.1038/nrg3032
- Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 2014; 28(8):812-28; https://doi.org/10.1101/gad.234294.113
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 2012; 48(6):849-62; PMID:23219530
- Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J 2013; 32(3):340-53; PMID:23241950
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of TET1 and TET2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 2013; 24(3):310-23; PMID:23352810; https://doi.org/10.1016/j.devcel.2012.12.015
- Strogantsev R, Ferguson-Smith AC. Proteins involved in establishment and maintenance of imprinted methylation marks. Brief Funct Genomics 2012; 11(3):227-39; PMID:22760206; https://doi.org/10.1093/bfgp/els018
- Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Phil. Trans. R. Soc. B 2013; 368(1609):20110336
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Embryogenesis: demethylation of the zygotic paternal genome. Nature 2000; 403(6769): 501-2; PMID:10676950
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 2000; 10(8):475-8; PMID:10801417; https://doi.org/10.1016/S0960-9822(00)00448-6
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241(1):172-82; PMID:11784103; https://doi.org/10.1006/dbio.2001.0501
- Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction 2004; 127(6):643-51; PMID:15175501; https://doi.org/10.1530/rep.1.00221
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of TET3 DNA Dioxygenase in epigenetic reprogramming by oocytes. Nature 2011; 477(7366):606-10; PMID:21892189; https://doi.org/10.1038/nature10443
- Inoue A, Zhang Y. Replication-dependent loss of 5- hydroxymethylcytosine in mouse preimplantation embryos. Science 2011; 334 (6053):194-194; PMID:21940858; https://doi.org/10.1126/science.1212483
- Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 2011; 21(12):1670-76; PMID:22124233; https://doi.org/10.1038/cr.2011.189
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 2012; 40(11):4841-9, page gks155
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 2012; 486(7403):415-9; PMID:22722204
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic DNMT1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 2008; 22(12):1607-16; https://doi.org/10.1101/gad.1667008
- Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Kru¨ppel-associated Box Zinc finger protein family. Mob DNA 2015; 6(1):1
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 2011; 44(3):361-72; PMID:22055183; https://doi.org/10.1016/j.molcel.2011.08.032
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 2011; 286(30):26267-76; PMID:21652716; https://doi.org/10.1074/jbc.R111.252569
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. Zinc Finger Protein ZFP57 requires its co-factor to recruit DNA Methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem 2012; 287(3):2107-18; PMID:22144682; https://doi.org/10.1074/jbc.M111.322644
- Strogantsev R, Krueger F, Yamazawa K, Shi H, Gould P, Goldman-Roberts M, McEwen K, Sun B, Pedersen R, Ferguson-Smith AC. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol 2015; 16 (1):1; PMID:26025256
- Messerschmidt D. Should I stay or should I go: protection and maintenance of DNA methylation at imprinted genes. Epigenetics 2012; 7(9):969-75; PMID:22869105
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. Setdb1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to Hp1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 2002; 16(8):919-32
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 2011; 138 (5):811-20; PMID:21247965
- Kobayashi H1, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 2012; 8(1):e1002440
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, ZFP57, maintains both maternal and paternal imprints. Dev Cell 2008; 15(4):547-57; PMID:18854139
- Singh P, Wu X, Lee DH, Li AX, Rauch TA, Pfeifer GP, Mann JR, Szabó PE. Chromosome-wide analysis of parental allele-specific chromatin and DNA methylation. Mol Cell Biol 2011; 31(8):1757-70; PMID:21321082
- Li BZ, Huang Z, Cui QY, Song XH, Du L, Jeltsch A, Chen P, Li G, Li E, Xu GL. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res 2011; 21(8):1172-81; PMID:21606950
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schübeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3b in genic methylation. Nature 2015; 520(7546):243-47; PMID:25607372
- Morselli M, Pastor WA, Montanini B, Nee K, Ferrari R, Fu K, Bonora G, Rubbi L, Clark AT, Ottonello S, et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. Elife 2015; 4:e06205; PMID:25848745; https://doi.org/10.7554/eLife.06205
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 2012; 489(7416):452-55; PMID:22914091; https://doi.org/10.1038/nature11326
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with Hp1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 2003; 31(9):2305-12; PMID:12711675; https://doi.org/10.1093/nar/gkg332
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the Hp1 chromo domain. Nature 2001; 410(6824):120-24; PMID:11242054; https://doi.org/10.1038/35065138
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for Hp1 proteins. Nature 2001; 410(6824):116-20; PMID:11242053; https://doi.org/10.1038/35065132
- Voon HP, Hughes JR, Rode C, De La Rosa-Velázquez IA, Jenuwein T, Feil R, Higgs DR, Gibbons RJ. ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep 2015; 11(3):405-18; PMID:25865896; https://doi.org/10.1016/j.celrep.2015.03.036
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010; 140(5):678-91; PMID:20211137; https://doi.org/10.1016/j.cell.2010.01.003
- Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, Ragozin S, Jeltsch A. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of H4 and K9. Hum Mol Genet 2011; 20(11):2195-203; PMID:21421568; https://doi.org/10.1093/hmg/ddr107
- Elsässer SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 2015, 522(7555):240-44; PMID:25938714; https://doi.org/10.1038/nature14345
- Kucharski R, Foret S, Maleszka R. EGFR gene methylation is not involved in Royalactin controlled phenotypic polymorphism in honey bees. Sci Rep 2015; 5
- Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nat Rev Genet 2006; 7(5):395-401; PMID:16534512; https://doi.org/10.1038/nrg1834
- Remy JJ. Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr Biol 2010; 20(20):R877-78; PMID:20971427; https://doi.org/10.1016/j.cub.2010.08.013
- JShorter JR, Arechavaleta-Velasco M, Robles-Rios C, Hunt GJ. A genetic analysis of the stinging and guarding behaviors of the honey bee. Behav Genet 2012; 42(4):663-74; PMID:22327626; https://doi.org/10.1007/s10519-012-9530-5
- Oldroyd BP, Allsopp MH, Roth KM, Remnant EJ, Drewell RA, Beekman M. A parent-of-origin effect on honey- bee worker ovary size. Proceedings of the Royal Society of London B: Biological Sciences 2014; 281(1775):20132388
- Stort AC, Goncalves LS. Genetics of defensive behavior. In The “African” honey bee, pages 329-56. Westview Press, Boulder, CO, spivak m, fletcher djc, and breed md, eds edition, 1991.
- Guzman-Novoa E, Page RE. Backcrossing Africanized honey bee queens to European drones reduces colony defensive behavior. Annals of the Entomological Society of America 1993; 86(3):352-55. ISSN 0013-8746, 1938-2901. URL https://academic.oup.com/aesa/article-lookup/doi/10.1093/aesa/86.3.352; https://doi.org/10.1093/aesa/86.3.352
- DeGrandi-Hoffman G, Collins A, Martin JH, Schmidt JO, Spangler H. Nest defense behavior in colonies from crosses between Africanized and European Honey Bees (Apis mellifera L.) (Hymenoptera: Apidae). Journal of Insect Behaviour 1998; 11(1):37-45; https://doi.org/10.1023/A:1020862432087
- Guzman-Novoa E, Hunt GJ, Page RE, Uribe-Rubio JL, Prieto-Merlos D, Becerra-Guzman F. Paternal effects on the defensive behavior of honeybees. J Hered 2005; 96(4):376-80
- Beukeboom LW, van den Assem J. Courtship and mating behaviour of interspecific Nasonia hybrids (Hymenoptera, Pteromalidae): a grandfather effect. Behaviour Genetics 2001; 31(2):167-77; https://doi.org/10.1023/A:1010201427204
- Keller L. Adaptation and the genetics of social behaviour. Philos Trans R Soc Lond B Biol Sci 2009; 364(1533):3209-16, ISSN 0962-8436, 1471-2970. URL http://rstb.royalsocietypublishing.org/cgi/doi/10.1098/rstb.2009.0108; https://doi.org/10.1098/rstb.2009.0108
- Libbrecht R, Keller L. Genetic Compatibility affects division of labor in the argentine ant Linepithema humile. Evolution 2013; 67(2):517-24. ISSN 00143820. URL http://doi.wiley.com/10.1111/j.1558-5646.2012.01792.x; https://doi.org/10.1111/j.1558-5646.2012.01792.x
- Libbrecht R, Schwander T, Keller L. Genetic components to caste allocation in a multiple-queen ant species. Evolution 2011; 65(10):2907-15, ISSN 00143820. URL http://doi.wiley.com/10.1111/j.1558-5646.2011.01348.x; PMID:22023572; https://doi.org/10.1111/j.1558-5646.2011.01348.x
- Anderson RH. The laying worker in the cape honeybee, Apis mellifera capensis. Journal of Apicultural Research 1963; 2(2):85-92. ISSN 0021-8839, 2078-6913. https://doi.org/10.1080/00218839.1963.11100065. URL http://www.tandfonline.com/doi/full/10.1080/00218839.1963.11100065
- Galbraith DA, Kocher SD, Glenn T, Albert I, Hunt GJ, Strassmann JE, Queller DC, Grozinger CM. Testing the kinship theory of intragenomic conflict in honey bees (Apis mellifera). Proc Natl Acad Sci U S A 2016; 113(4):1020-25
- Kocher SD, Tsuruda JM, Gibson JD, Emore CM, Arechavaleta-Velasco ME, Queller DC, Strassmann JE, Grozinger CM, Gribskov MR, San Miguel P, et al. A search for parent-of-origin effects on honey bee gene expression. G3: Genes—Genomes—Genetics 2015; 5(8):1657-62; https://doi.org/10.1534/g3.115.017814
- Amarasinghe HE, Toghill BJ, Nathanael D, Mallon EB. Allele specific expression in worker reproduction genes in the bumblebee Bombus terrestris. PeerJ 2015; 3:e1079; https://doi.org/10.7717/peerj.1079
- Geva S, Hartfelder K, Bloch G. Reproductive division of labor, dominance, and ecdysteroid levels in hemolymph and ovary of the bumble bee Bombus terrestris. J Insect Physiol 2005; 51(7):811-23; PMID:15885700; https://doi.org/10.1016/j.jinsphys.2005.03.009
- Cardoen D, Wenseleers T, Ernst UR, Danneels EL, Laget D, DE Graaf DC, Schoofs L, Verleyen P. Genome-wide analysis of alternative reproductive phenotypes in honeybee workers. Molecular Ecology 2011; 20(19):4070-84; PMID:21902748
- Page RE Jr, Amdam GV. The making of a social insect: developmental architectures of social design. Bioessays 2007; 29(4):334-43; PMID:17373656
- Mullen EK, Daley M, Backx AG, Thompson GJ. Gene co-citation networks associated with worker sterility in honey bees. BMC Syst Biol 2014; 8(1):1
- Gibson JD, Arechavaleta-Velasco ME, Tsuruda JM, Hunt GJ. Biased allele expression and aggression in hybrid honeybees may be influ- enced by inappropriate nuclear-cytoplasmic signaling. Frontiers Genet 2015; 6
- Linksvayer TA, Rueppell O, Siegel A, Kaftanoglu O, Page RE Jr, Amdam GV. The genetic basis of transgressive ovary size in honeybee workers. Genetics 2009; 183(2):693-707; PMID:19620393
- Verhulst EC, Beukeboom LW, van de Zande L. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 2010; 328(5978):620-23; PMID:20431014
- Kumano G. Polarizing animal cells via mRNA localization in oogenesis and early development. Dev Growth Differ 2012; 54(1):1-18
- Galbraith DA, Yi SV, Grozinger CM. Evaluation of possible proximate mechanisms underlying the kinship theory of intragenomic conflict in social insects. Integr Comp Biol 2016; 56(6):1206-14. ISSN 1540-7063, 1557-7023. https://doi.org/10.1093/icb/icw111. URL https://academic.oup.com/icb/article-lookup/doi/10.1093/icb/icw111
- Drewell RA, Bush EC, Remnant EJ, Wong GT, Beeler SM, Stringham JL, Lim J, Oldroyd BP. The dynamic DNA methylation cycle from egg to sperm in the honey bee Apis mellifera. Development 2014; 141(13):2702-11; PMID:24924193
- Remnant EJ, Ashe A, Young PE, Buchmann G, Beekman M, Allsopp MH, Suter CM, Drewell RA, Oldroyd BP. Parent-of-origin effects on genome-wide DNA methylation in the cape honey bee (Apis mellifera capensis) may be confounded by allele-specific methylation. BMC Genomics 2016; 17(1):1
- Wedd L, Kucharski R, Maleszka R. Differentially methylated obligatory epialleles modulate context-dependent LAM gene expression in the honeybee Apis mellifera. Epigenetics 2016; 11(1):1-10. ISSN 1559-2294, 1559-2308. https://doi.org/10.1080/15592294.2015.1107695. URL http://www.tandfonline.com/doi/full/10.1080/15592294.2015.1107695
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011; 145(3):423-34; PMID:21496894; https://doi.org/10.1016/j.cell.2011.03.022
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA Glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011; 146(1):67-79; PMID:21722948; https://doi.org/10.1016/j.cell.2011.06.020
- Branco MR, Ficz G, Reik W. Uncovering the role of 5- hydroxymethylcytosine in the epigenome. Nat Rev Genet 2012; 13(1):7-13
- Valinluck V, Sowers LC. Endogenous cytosine damage prod-ucts alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 2007; 67(3):946-50; PMID:17283125; https://doi.org/10.1158/0008-5472.CAN-06-3123
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 2012; 149(6): 1368-80; PMID:22608086; https://doi.org/10.1016/j.cell.2012.04.027
- Booth MJ1, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5- methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 2012; 336(6083):934-7; PMID:22539555; https://doi.org/10.1126/science.1220671
- Booth MJ, Marsico G, Bachman M, Beraldi D, Balasubramanian S. Quantitative sequencing of 5-formylcytosine in DNA at single- base resolution. Nat Chem 2014; 6(5):435; PMID:24755596; https://doi.org/10.1038/nchem.1893
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet 2005; 21(8):457-65; PMID:15990197; https://doi.org/10.1016/j.tig.2005.06.008
- Raissig MT, Baroux C, Grossniklaus U. Regulation and flexibility of genomic imprinting during seed development. The Plant Cell 2011; 23(1):16-26; PMID:21278124; https://doi.org/10.1105/tpc.110.081018
- Lloyd V. Parental imprinting in Drosophila. Genetica 2000; 109(1-2):35-44; PMID:11293793; https://doi.org/10.1023/A:1026592318341
- McGowan RA, Martin CC. DNA methylation and genome imprinting in the zebrafish, Danio rerio: some evolutionary ramifications. Biochem Cell Biol 75(5):499-506, 1997; PMID:9551175; https://doi.org/10.1139/o97-070
- Bean CJ, Schaner CE, Kelly WG. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nature genetics 2004; 36(1):100-5; PMID:14702046; https://doi.org/10.1038/ng1283
- Cheung P, Lau P. Epigenetic regulation by histone methylation and histone variants. Molecular Endocrinology 2005; 19(3):563-73; PMID:15677708; https://doi.org/10.1210/me.2004-0496
- Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature 2004 431(7006):364-70; PMID:15372044; https://doi.org/10.1038/nature02875
- Bongiorni S, Pugnali M, Volpi S, Bizzaro D, Singh PB, Prantera G. Epigenetic marks for chromosome imprinting during spermatogenesis in coccids. Chromosoma 2009; 118(4):501-12; PMID:19458957; https://doi.org/10.1007/s00412-009-0214-8
- Takayama S, Dhahbi J, Roberts A, Mao G, Heo SJ, Pachter L, Martin DI, Boffelli D. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res 2014; 24(5):821-30; PMID:24558263; https://doi.org/10.1101/gr.162412.113
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schäfer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 2007; 26(1):103-15; PMID:17434130; https://doi.org/10.1016/j.molcel.2007.02.025
- Kidder BL, Hu G, Zhao K. ChIP-Seq: technical considerations for obtaining high-quality data. Nature Immunology 2011; 12(10):918-22; PMID:21934668; https://doi.org/10.1038/ni.2117
- ENCODE-Project-Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489(7414):57-74; PMID:22955616; https://doi.org/10.1038/nature11247
- Shimbo T, Du Y, Grimm SA, Dhasarathy A, Mav D, Shah RR, Shi H, Wade PA. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet 2013; 9(12):e1004028
- Zentner GE, Henikoff S. High-resolution digital profiling of the epigenome. Nat Rev Genet 2014; 15(12):814-27; PMID:25297728; https://doi.org/10.1038/nrg3798
- Pepke S, Wold B, Mortazavi A. Computation for ChIP-Seq and RNA-Seq studies. Nat Methods 2009; 6:S22-32; PMID:19844228; https://doi.org/10.1038/nmeth.1371
- Furey TS. ChIP–Seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat Rev Genet 2012; 13(12):840-52; PMID:23090257; https://doi.org/10.1038/nrg3306
- Shlyueva D, Meireles-Filho ACA, Pagani M, Stark A. Genome-wide Ultrabithorax binding analysis reveals highly targeted genomic loci at developmental regulators and a potential connection to polycomb- mediated regulation. PloS one 2016; 11(8):e0161997.
- Löser E, Latreille D, Iovino N. Chromatin preparation and chromatin immuno-precipitation from Drosophila embryos. Methods Mol Biol (Clifton, NJ) 2016; 1480:23
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. Chromatin state dynamics during blood formation. Science 2014; 345(6199):943-9; PMID:25103404; https://doi.org/10.1126/science.1256271
- Blecher-Gonen R, Barnett-Itzhaki Z, Jaitin D, Amann-Zalcenstein D, Lara-Astiaso D, Amit I. High-throughput Chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc 2013; 8(3):539-54; PMID:23429716; https://doi.org/10.1038/nprot.2013.023
- Bassett AR, Liu JL. CRISPR/Cas9 and genome editing in Drosophila. J Genet Genomics 2014; 41(1):7-19; PMID:24480743; https://doi.org/10.1016/j.jgg.2013.12.004
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol 2016; 34(1):78-83; PMID:26641531; https://doi.org/10.1038/nbt.3439
- Dong S, Lin J, Held NL, Clem RJ, Passarelli AL, Franz AW. Heritable CRISPR/Cas9-mediated genome editing in the yellow fever mosquito, Aedes aegypti. PloS One 2015; 10(3):e0122353
- Gilles AF, Schinko JB, Averof M. Efficient CRISPR-mediated gene targeting and transgene replacement in the beetle Tribolium castaneum. Development 2015; 142(16):2832-39; PMID:26160901; https://doi.org/10.1242/dev.125054
- Liu Y, Ma S, Wang X, Chang J, Gao J, Shi R, Zhang J, Lu W, Liu Y, Zhao P, et al. Highly efficient multiplex targeted mutagenesis and genomic structure variation in Bombyx mori cells using CRISPR/Cas9. Insect Biochem Mol Biol 2014; 49:35-42; PMID:24698835; https://doi.org/10.1016/j.ibmb.2014.03.010
- Li M, Cook LY, Douglah D, Chong A, White BJ, Ferree P, Akbari OS. Generation of heritable germline mutations in the jewel wasp Nasonia vitripennis using CRISPR/Cas9. Scientific Reports 2017; 7(1):901.
- Kohno H, Suenami S, Takeuchi H, Sasaki T, Kubo T. Production of knockout mutants by CRISPR/Cas9 in the European honeybee, Apis mellifera. Zoolog Sci 2016; 33(5):505-12; PMID:27715425; https://doi.org/10.2108/zs160043
- Trible W, Chang N-C, Matthews BJ, McKenzie SK, Olivos-Cisneros L, Oxley PR, Saragosti J, Kronauer DJC. orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 2017; 170(4):727-735.
- Mund C, Musch T, Strödicke M, Assmann B, Li E, Lyko F. Comparative analysis of DNA methylation patterns in transgenic Drosophila overexpressing mouse DNA methyltransferases. Biochem J 2004; 378(3):763-8; PMID:14636159; https://doi.org/10.1042/bj20031567
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA Adenine methyltransferase. Nat Genet 2001; 27(3):304-8; PMID:11242113; https://doi.org/10.1038/85871
- Pindyurin AV, Pagie L, Kozhevnikova EN, van Arensbergen J, van Steensel B. Inducible DamID systems for genomic mapping of chromatin proteins in Drosophila. Nucleic Acids Res 2016; 44(12):5646-57 page gkw176.
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, Jaenisch R. Editing DNA methylation in the mammalian genome. Cell 2016; 167(1):233-47; PMID:27662091; https://doi.org/10.1016/j.cell.2016.08.056
- Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, Klasić M, Zoldoš V. Repurposing the CRISPR/Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016; 44(12):5615-28 page gkw159.
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. A survey of best practices for RNA-Seq data analysis. Genome Biol 2016; 17(1):13
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5(7):621-8; PMID:18516045; https://doi.org/10.1038/nmeth.1226
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013; 500(7463):477-81; PMID:23925113; https://doi.org/10.1038/nature12433
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 2010; 28 (10):1097-105; PMID:20852635; https://doi.org/10.1038/nbt.1682
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129(4):823-37; PMID:17512414; https://doi.org/10.1016/j.cell.2007.05.009
- Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, Yen CA, Lin S, Lin Y, Qiu Y, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 2015; 518(7539):350-4; PMID:25693566; https://doi.org/10.1038/nature14217
- Yan H, Tian S, Slager SL, Sun Z, Ordog T. Genome-wide epigenetic studies in human disease: A primer on-omic technologies. Am J Epidemiol 2016; 183(2):96-109; PMID:26721890
- Sosnowski BA, Belote JM, McKeown M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 1989; 58(3):449-59; PMID:2503251; https://doi.org/10.1016/0092-8674(89)90426-1
- Blekhman R, Marioni JC, Zumbo P, Stephens M, Gilad Y. Sex-specific and lineage-specific alternative splicing in primates. Genome Res 2010; 20(2):180-9; PMID:20009012; https://doi.org/10.1101/gr.099226.109
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development 1998; 125(5):889-97; PMID:9449671
