ABSTRACT
Faroe islanders consume marine foods contaminated with methylmercury (MeHg), polychlorinated biphenyls (PCBs), and other toxicants associated with chronic disease risks. Differential DNA methylation at specific CpG sites in cord blood may serve as a surrogate biomarker of health impacts from chemical exposures. We aimed to identify key environmental chemicals in cord blood associated with DNA methylation changes in a population with elevated exposure to chemical mixtures. We studied 72 participants of a Faroese birth cohort recruited between 1986 and 1987 and followed until adulthood. The cord blood DNA methylome was profiled using Infinium HumanMethylation450 BeadChips. We determined the associations of CpG site changes with concentrations of MeHg, major PCBs, other organochlorine compounds [hexachlorobenzene (HCB), p,p’-dichlorodiphenyldichloroethylene (p,p’-DDE) and p,p’-dichlorodiphenyltrichloroethane], and perfluoroalkyl substances. In a combined sex analysis, among the 16 chemicals studied, PCB congener 105 (CB-105) exposure was associated with the majority of differentially methylated CpG sites (214 out of a total of 250). In female-only analysis, only 73 CB-105 associated CpG sites were detected, 44 of which were mapped to genes in the ELAV1-associated cancer network. In males-only, methylation changes were seen for perfluorooctane sulfonate, HCB, and p,p’-DDE in 10,598, 1,238, and 1,473 CpG sites, respectively, 15% of which were enriched in cytobands of the X-chromosome associated with neurological disorders. In this multiple-pollutant and genome-wide study, we identified key epigenetic toxicants. The significant enrichment of specific X-chromosome sites in males implies potential sex-specific epigenome responses to prenatal chemical exposures.
Introduction
DNA methylation may be affected by early-life chemical exposures with the potential to persist even after the exposure has been removed [Citation1]. Because of this enduring behavior, DNA methylation has the power to store the effect of environmental chemical exposures occurring during early development [Citation2,Citation3]. Earlier studies on DNA methylation investigated exposure-associated “global” methylation changes in several common repetitive elements, such as long interspersed nuclear elements (LINE) or Alu elements, because their sequences are widespread in the human genome and represent about 45% of methylation sites in the whole genome [Citation4,Citation5]. However, such “global” approaches are uninformative in terms of understanding specific exposure-related changes in humans, as they do not reveal the specific genes, biological pathways, or chromosomal loci involved. Genome-wide methylation analysis with single CpG site resolution overcomes this hurdle and is now the preferred platform for environmental epigenetic studies [Citation6,Citation7].
Methylation changes associated with exposure and/or later-life diseases can be detected as early as the prenatal period [Citation8,Citation9]. Differential DNA methylation in umbilical cord white blood cell (UCWBC) has been associated with maternal smoking [Citation10–Citation12], air-pollutants [Citation8,Citation9], and arsenic exposure in utero [Citation13]. Specific DNA methylation changes in UCWBC predict childhood diseases, such as asthma [Citation8]. These data suggest that environmental stressors during pregnancy may trigger specific DNA methylation changes in susceptible genes in UCWBCs. These cells may be disease-related target cells or serve as surrogates for the affected tissues/organs. DNA methylation changes in UCWBCs have been reported to be associated with earlier onset or increased prevalence of diseases via reprogramming of gene expression networks crucial to tissues/organs development, in accordance with the Developmental Origins of Health and Disease (DoHAD) concept [Citation14].
Up to now, almost all studies on environmental epigenetics focused on the effects of a single-toxicant exposure (e.g., lead [Citation15]) or a single representative from a mixture (e.g., cotinine in smoking [Citation10]). To the best of our knowledge, no studies so far were designed to identify chemicals among an exposure mixture that are associated with either the largest degree of changes (number of differentially methylated CpG sites) or the most specific (e.g., concentration in specific cytobands) DNA methylation changes in the UCWBC. Here, we refer to these toxicants as key epigenetic toxicants. They could be the most potent drivers of epigenetic changes or the key indicators of such changes within an exposure mixture. Furthermore, the potential sex differences of such changes are understudied, as most investigators remove affected CpG sites in the sex chromosomes before analyses. Therefore, significant data voids exist in the identification of the key epigenetic toxicants complex exposures, as well as any sex-specific DNA methylation changes associated with the same exposure setting.
To address these challenging questions, we took advantage of a birth cohort in the Faroe Islands born in 1986–1987 [Citation16,Citation17]. We initially reported elevated exposures to methylmercury (MeHg) and halogenated substances in this population due to consumption of pilot whale meat and blubber [Citation18]. In this birth cohort, neurodevelopmental deficits have been shown to be associated primarily with elevated prenatal exposures to methylmercury [Citation19]. The goal of this pilot study was to conduct a genome-wide analysis of DNA methylation distribution in 72 UCWBC DNA samples isolated from cord blood of Faroese cohort members and examine associations with exposure to 16 environmental chemicals measured in the cord blood. We applied Illumina HumanMethylation450 BeadChip (450K) assay to detect unbiasedly genome-wide methylation levels at more than 480,000 CpG sites in UCWBC DNA and utilized several newly developed bioinformatics methods for data-mining, uncovering key epigenetic toxicants and sex-differences in exposure-associated methylation changes.
Materials and Methods
Study design and subjects
A cohort of singleton term births was recruited at the Faroese hospitals assembled during an 18-month period in 1986–1987 [Citation20]. Children born before the 37th week of gestation or who had a severe congenital disease were excluded. At delivery, samples of umbilical cord whole blood and scalp hair from the mothers were collected for analysis of environmental chemicals. The study protocol was approved by the Ethical Review Committee for the Faroe Islands and the Institutional Review Board in Boston. We selected 72 samples of cord blood, all with no maternal smoking exposure, with sufficient volume, and that reflected the largest possible range of exposures, including samples with high MeHg and low total polychlorinated biphenyls (PCB) level or vice versa for the purpose of minimizing mutual correlations between the pollutants.
The Faroese population is of primarily Nordic and Irish origin and comparable in many aspects to other Western populations [Citation21] but is uniquely exposed to environmental chemicals due to traditional dietary habits (including pilot whale meat and blubber). Depending on the diet, the range of chemical exposures is wide, and average exposures to persistent chlorinated compounds and MeHg at the time were much higher than in most Western populations [Citation22]. The Faroese conduct their occasional subsistence whaling when pilot whale pods approach the coasts, and the whale meat and blubber are shared locally [Citation23]. Dietary habits are affected mainly by availability and, to a lesser extent, the fairly small differences in socioeconomic factors. Average concentrations of polychlorinated biphenyls (PCBs) in blubber are about 20 μg/g [Citation24], MeHg concentrations in lean whale muscle are much higher than in most fish [Citation25], and perfluoroalkyl substances (PFASs) are also at higher levels than other food items generally consumed [Citation26]. Thus, this population provides a unique natural experiment to ascertain adverse effects of elevated exposures to these marine contaminants.
Chemical exposure assessment
Cord whole blood and maternal hair were analyzed for total mercury (as indicators of prenatal MeHg exposure) [Citation20], and whole blood was also analyzed for organochlorine compounds [Citation27] and PFASs [Citation28]. The PCB analysis comprised major congeners, including the dioxin-like CB-118 and CB-105 and non-dioxin like congeners. PFAS analysis provided results on the five most common substances, including perfluorooctane sulfonic acid (PFOS), perfluorooctanoic acid (PFOA), perfluorohexane sulfonic acid (PFHxS), perfluorodecanoic acid (PFDA), and perfluorononanoic acid (PFNA). For PFOS, we determined both the straight-chain (n-PFOS) and branched isomers. As the chlorinated compounds are lipophilic and the lipid content of whole blood is fairly constant, we expressed the concentrations on a blood volume basis, as were mercury and the PFASs.
DNA isolation and genome-wide methylation analyses
Genomic DNA was isolated from total cord blood by use of a Maxwell 16 System DNA Purification Kit (Promega, Madison, WI). Each DNA sample was quantified with Nanodrop spectrometer and its quality was examined with gel electrophoresis to make sure that the DNA was not degraded.
The DNA samples were bisulfite-treated according to the instructions of the EZ DNA Methylation Kit (Zymo Research, Irvine, CA). Genome-wide methylation analyses were performed using Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA) in the Genomics, Epigenomics, and Sequencing Core at University of Cincinnati.
Data analyses
The raw methylation data were loaded using minfi package in R statistics program [Citation29]. Quality control analyses on probes and samples based on the default setting of beadcount, detection P value, and data distribution were performed using the “ENmix” package [Citation30]. Outlier probes and/or samples based on the tested criteria were excluded before further analysis. The background signals and the dye bias were corrected using a model-based background correction method (ENmix) and the regression on the logarithm of internal control probes (RELIC) method [Citation31], respectively, both embedded in the “preprocessENmix” function. Inter-array variations were quantile-normalized and the probe bias was corrected by regression on the correlated probes (RCP) method [Citation32]. Outlier probes and/or samples were further removed as recommended in the ENmix package using aforementioned criteria. The dataset supporting the results of this study is available at the National Center for Biotechnology Information Gene Expression Omnibus repository, GSE104778.
Surrogate variable analysis (SVA) [Citation33] was applied to the dataset to determine surrogate variables related to batch effects, cell type heterogeneity, and unobserved sources of variations in the cord blood samples. The unobserved sources of variation were used as covariates in linear regression models for differential methylation site analysis associated with each of the exposures (as a continuous variable) using limma package [Citation34]. SVA has been shown to be more stable than existing reference-based or other reference-free cell type mixture adjustment methods [Citation35]. CpG sites on the sex chromosomes were excluded in the mixed-sex analyses, but those CpG sites residing on the sex chromosomes were included in the respective sex-stratified analyses. Co-variates including surrogate variables, alcohol consumption status, maternal age, gestational age, sex, and slide number were included and adjusted for in the analyses.
Bioinformatics analyses
The CpG sites with methylation changes significantly associated with an exposure were selected for further analyses. To enhance the biological relevance of the analyses, only genes for which at least two CpG sites showed significant methylation changes were chosen for pathway analysis. Significant differences in the methylation level were defined as at least 10% change (increase or decrease) in the beta value per unit change in the exposure. In the case of hexachlorobenzene (HCB) analyses, the cutoff value was set at 100% change or higher in the beta value for each unit increase of HCB exposure, as all individuals in this cohort had exposure less than one unit. One-tenth unit of HCB exposure was therefore required to cause at least a 10% change in methylation level of a particular CpG site. Ingenuity Pathway analysis (Qiagen, Redwood City, CA) was applied to determine any potential gene networks or pathways enriched in each exposure evaluation. ToppFun, as a part of ToppGene Suite (https://toppgene.cchmc.org/enrichment.jsp), was used to determine whether the genes were enriched in specific cytogenetic bands of the chromosome.
Results
Stringent data preprocessing provides high-quality data for linear regression analyses
Seventy-two UCWBC DNA samples with wide-ranging exposure data were selected from the Faroese Island birth cohort 1986–1987 [Citation20] and used for genome-methylome analyses. Demographic and exposure data of the selected cohort subjects are listed in . We used the latest data analysis pipeline with stringent criteria to determine the exposure-related methylation change in these samples. Twenty-one samples and 77,967 CpG sites were removed due to poor data quality as reflected from low bead counts, poor detection P values, and abnormal signal distribution as determined by the default setting of the ENmix package [Citation30]. Thus, here we reported data from 51 high-quality samples (19 males and 32 females) ().
Table 1. Descriptive characteristics of study population (n = 72) and their exposure.
Table 2. Number of CpG sites with methylation change significantly associated with the exposure (FDR<0.05).
Identification of CpG sites whose methylation levels correlated with prenatal CB105 exposure in mixed-sex or female-only analysis, revealing concentration of sites in the ELAVL1 pathway
In the complete sample analysis with both males and females, we found 214 CpG sites with significant methylation changes associated with an increase in CB-105 exposure () (See Supplementary Tables for details). Additionally, we found 18 and 10 differentially methylated CpG sites associated with pp’-DDT and CB-101, respectively. We did not find any significant DNA methylation changes associated with the other chemical exposures. In sum, CB-105 was the only toxicant associated with the largest number of significant methylation sites among all other exposures in the mixed-sex analysis (; column “mixed”).
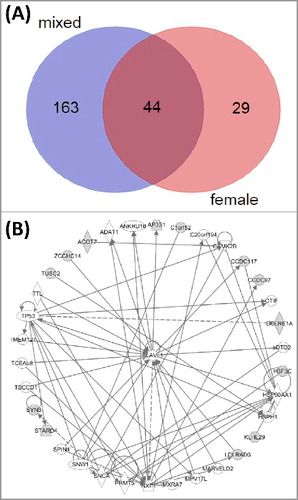
In female newborns only, we found 73 CpG sites with significant methylation changes associated with CB-105 (; column “female”), of which 44 were significant also in the complete data set (A). These 44 sites were mapped to 33 genes. Ingenuity pathway analysis (IPA®) revealed that 13 of those genes are involved in cancer, organismal injury, and abnormalities, as well as reproductive system disease, which form part of a gene network connected by Embryonic Lethal, Abnormal Vision, Drosophila, Homolog-Like 1 (ELAVL1) (B). This gene is known to play an important role in cancer progression [Citation36].
Figure 1. Common significant CpG sites found in mixed-sex and female-only analyses of PCB-105 exposure are related to cancer. (A) Venn diagram showing 44 CpG sites that were found in both analyses. (B) A fraction (30%) of 44 CpG sites was mapped to genes connected with an ELAV1-associated cancer network.
Identification of CpG sites with methylation levels correlating with prenatal exposure to nPFOS, HCB, or p,p’-DDE only in male-specific analyses (these sites are predominantly found on the chromosome X of males)
Surprisingly, we found several CpG sites with methylation changes significantly associated with exposures mainly to three other toxicants in the male-only analyses: 10,598 CpG sites associated with nPFOS, 1,238 CpG sites associated with HCB, and 1,473 CpG sites associated with p,p’-DDE (; column male)(See Supplementary Tables for details). When we compared the CpG sites associated with each of the three exposures, we found that less than 50% of the CpG sites were common to all three exposures for the joint analyses of both sexes (Supplementary Figure S1), thus suggesting that each toxicant has a unique epigenetic footprint in UCWBC DNA.
We then determined whether DNA methylation changes associated with one of the three toxicants was dependent on the exposures to the two other toxicants. We tested a multivariate model in males adjusted for all three chemicals associated with DNA methylation and found that associations of nPFOS, p,p’-DDE, and HCB were all significantly attenuated in the multiple-exposure adjusted model.
To ascertain if differences exist in the neighborhood context of the significant CpG sites associated with the three toxicants in male/female newborns compared with the combined-sexes sites, we mapped their positions in relation to the consensus CpG Islands (Island) upstream of the transcription sites of their purported genes (such as N_shelves, N_shores, or Open_Sea [Citation37]). The distribution of CpG sites was similar for CB105 in the mixed-sex analysis (PCB-105_all), for HCB in the males (HCB_m), for nPFOS in the males (nPFOS_m), and for CB105 in the females (CB105_f). However, we observed a higher percentage of island-associated CpG sites and fewer Open_Sea-associated sites for p,p’-DDE in males (p,p’-DDE_m) (Supplementary Figure S2). We further mapped these sites to their respective chromosomal loci. A higher percentage (15-25%) of the affected sites were found to the X-chromosome in the male-specific analyses (i.e., HCB_m, p,p’-DDE_m, nPFOS_m) () when compared with the average percentages (4.2 to 4.6%) mapped to somatic chromosomes. Consistent with this observation, we identified several cytogenetic bands on the X-chromosome that were statistically enriched (FDR<0.05) with affected CpG sites in the analyses ().
Figure 2. Chromosomal locations of the significant CpG sites. Percentage of the number of CpG sites with significant methylation change against each exposure (PCB-105, HCB, p,p’-DDE, and nPFOS) per chromosome was calculated. PCB-105 represents the percentage obtained from the analyses of mixed-sex with PCB-105 exposure; HCB_m represents the percentage obtained from the analyses of male with HCB exposure; p,p’-DDE_m represents the percentage obtained from the analyses of male with p,p’-DDE exposure; nPFOS_m represents the percentage obtained from the analyses of male with nPFOS exposure; PCB-105_f represents the percentage obtained from the analyses of female with PCB-105 exposure.
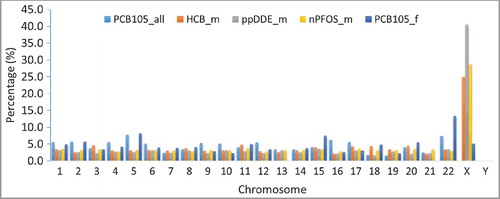
Table 3. Cytogenetic band analyses of the significant CpG sites found in HCB_m, ppDDE_m and nPFOS_m studies.
It is noteworthy that Xp11.23-p11.22 was the top cytoband identified with the highest enrichment of CpG sites associated with both HCB and p,p’-DDE in males (). According to the Human Phenotype Ontology Project (HPO Browser, http://compbio.charite.de/hpoweb/showterm?id=HP:0000118), duplication of this cytoband is associated with intellectual disability, speech delay, and a peculiar electroencephalographic pattern in childhood [Citation38]. Other diseases linked to this region include Wiskott–Aldrich syndrome, several forms of retinal degeneration and intellectual developmental disorders (http://genome.cshlp.org/content/6/11/1056.short), autism spectrum disorders (CACNA1F [Citation39]), bipolar disorders (monoamine oxidase A [Citation40]), X-linked autoimmunity-allergic dysregulation syndrome [Citation41] (JM2), immune dysregulation, polyendocrinopathy and enteropathy, X-linked (IPEX) syndrome, a rare form of X-linked immune dysfunction involving regulatory T cells (FoxP3 [Citation42]). A major susceptibility locus for sex reversal/gonadal dysgenesis also exists in an extended locus (Xp11.21-11.23) encompassing this region [Citation43].
Exposure to HCB, p,p’-DDE, or nPFOS were associated with methylation changes in CpG sites associated with genes in pathways of key physiological functions and diseases
In males only, IPA® pathway analysis was performed to determine any relationships between key physiological functions/diseases and methylation changes associated with exposures to HCB, p,p’-DDE, or nPFOS. In terms of physiological functions, all three chemical exposures were associated with pathways of “embryonic development” (). The top physiological function related to HCB and p,p’-DDE exposures was “reproductive system development and function,” whereas the top common function affected by exposure to nPFOS or p,p’-DDE was “nervous system development and function” (). HCB exposure also was associated with “behavior” and “hematological/cardiovascular system development and function” (). With respect to disease prediction, significant CpG sites were associated with genes related to “cancer” for exposure to p,p’-DDE or to nPFOS in males (). Consistent with the physiological function associations, CpG sites are associated with genes related to “hematological disorders” for HCB exposure and to “neurological disease” for p,p’-DDE exposure in males (). “Metabolic disease” also was predicted to be one of the disease pathways associated with male HCB exposure ().
Table 4. Predicted physiological functions of the genes involved in HCB, ppDDE and nPFOS analyses of male subjects.
Table 5. Predicted disease outcomes of the genes involved in HCB, ppDDE and nPFOS analyses of male subjects.
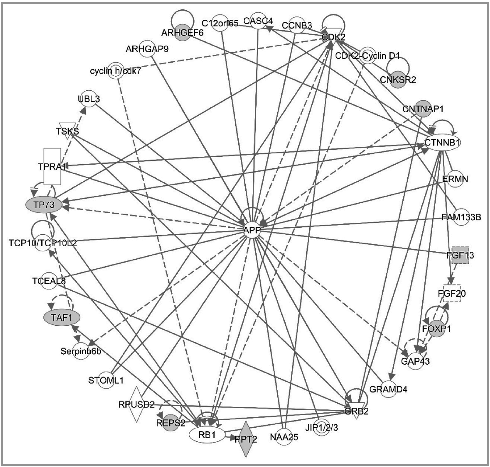
At the molecular level, gene network analyses suggested that extracellular signal-regulated kinase 1/2 (ERK1/2) was a key mediator affecting “reproductive system development and function” and “developmental disorder” in both HCB_m () and p,p’-DDE_m-related DNA methylation changes (). Of interest, the amyloid precursor protein gene (APP) was the central node linking the methylation changes associated with p,p’-DDE_m exposure (), affecting “carbohydrate metabolism and organismal development, nucleic acid metabolism”. APP also was shown to be the key connector for methylation changes affecting the top gene network related to nPFOS_m exposure, disrupting “cell cycle, nervous system, development and function, and cardiac proliferation” ().
Figure 3. Gene networks involved in the analyses of males with HCB exposure. The genes neighboring the significant CpG sites (with greater than 100% change of beta value per unit of HCB change and with more than 1 significant CpG site per gene) were subjected to pathway analyses. Left panel shows the top network involved: gene expression, reproductive system development and function, cell-to-cell signaling and interaction. Right panel shows the next top network involving cellular function and maintenance, cancer, organismal injury and abnormalities.
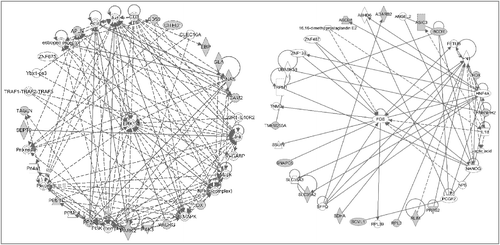
Figure 4. Gene networks involved in the analyses of males with p,p’-DDE exposure. The genes neighboring the significant CpG sites (with greater than 10% change of beta value per unit of p,p’-DDE change and with more than 1 significant CpG site per gene) were subjected to pathway analyses. The left panel shows the top network involved: developmental disorder, endocrine system disorders, and hereditary disorder. The right panel shows the next top network involving carbohydrate metabolism, organismal development, and nucleic acid metabolism.
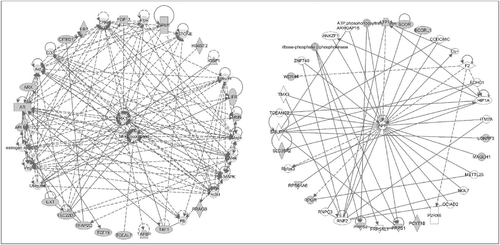
Figure 5. Networks involved in the analyses involving males with nPFOS exposure. The genes neighboring the significant CpG sites (with greater than 10% change in beta value per unit of nPFOS change and with more than 1 significant CpG site per gene) were subjected to pathway analyses. One top network involving cell cycle, nervous system, development and function, and cardiac proliferation was identified.
Discussion
Previous genome-wide methylome studies [Citation44–Citation46] reported associations of methylation changes with prenatal exposures to single toxicants/pollutants. The present study aimed at identifying “driver” epigenetic toxicants in a complex mixture of 15 marine pollutants found at elevated concentrations in cord blood from a Faroe Islands birth cohort. Of significance, we found notable sex differences in chemical-associated DNA methylation changes. In combined-sex analyses, the majority of differential methylation changes (214 sites) was associated with CB-105 but not with MeHg or other persistent organic pollutants (POPs). Further, in sex-specific analyses, we observed significant sex-dependent associations. Female-specific sites (75 sites) were found to associate primarily with exposure to CB-105 (73 sites) as a “driver” epigenetic toxicant, and a great number of them (44 sites) are concentrated in genes (33) in the ELAV1-associated cancer network. In contrast, considerably more differential methylation changes were found in males, with nPFOS (10,598 sites), HCB (1,238 sites), and p,p’-DDE (1,473 sites) emerging as “driver” epigenetic toxicants. Of interest, these male-specific sites were linked to genes regulating embryonic development, the development of or diseases in the reproductive, nervous, hematological, and cardiovascular systems, as well as carbohydrate and nucleic acid metabolism. Significantly, over 15% of the male-specific, differentially methylated CpG sites are located in unique cytobands of the X-chromosome. These include Xp11.23-p11.22, which are associated with intellectual disabilities and other neurological disorders. It was also noted that the association of these three exposures with methylation changes may not be specific, as the associations in terms of significance were weakened after mutual adjustments. Taken together, these findings suggest that prenatal exposure to a complex environmental chemical mixture engendered toxicant-/sex-/disease-specific epigenetic modifications that may be in part interdependent in cord blood DNA.
We identified nPFOS, HCB, and p,p’-DDE as potentially the most important contributors of DNA methylation changes in males and CB-105 as the most epigenetically active toxicant in females. Most previous studies focused only on establishing correlations between methylome changes with exposures to a single chemical. This approach may not be able to provide a full picture of the effects of human exposures because, unlike toxicological studies in animal models, human exposures rarely consist of single chemicals at a time. Using an unbiased, genome-wide methylome profiling approach and a newly established robust bioinformatics pipeline [Citation30–Citation32], we were able to identify chemicals that were associated with the highest numbers of CpG sites with differential methylation status in a sex-specific manner. Furthermore, mapping those CpG sites to various regions of known genes has uncovered some putative disease-/disorder-related pathways, hence offering a glimpse of probable linkages to disease risks in this population. In this regard, a noteworthy finding is that the most “epigenetically active” pollutant for female newborns was CB-105, while, for males, nPFOS, HCB, and p,p’-DDE were the most active. Of further interest is that the sex-dimorphic gene pathways have little overlap in biological or pathological functioning, suggesting toxicant-specific epigenetic reprogramming of gene expression and thereby divergent disease risks between males and females. This sexual difference could be due to differences between endocrine disrupting properties of chemicals on androgen or estrogen signaling. Our findings may also be relevant to the growing literature on sexual dimorphism in biological accumulation and degradation of these chemicals [Citation47,Citation48], thus warranting future investigation.
Although sex dimorphic effects have been frequently reported in epidemiological studies [Citation49], reports on sex-specific DNA methylation changes and the enrichment of these sites in specific X-chromosome cytobands have attracted less attention. In this study, sex-specific analyses showed CB-105 exposure as the main driver of differential DNA methylation changes in females. In contrast, the principal exposures associated with differential methylome changes in the males were found to be nPFOS, HCB, and p,p’-DDE. Moreover, the gene networks where the differentially methylated CpG sites were located seem to have minimal overlaps. The female-associated changes appear to affect the ELAV1-associated cancer network, whereas the male-associated changes have a much broader potential influence on embryonic, reproductive, neural, cardiovascular and digestive systems, and diseases associated with dysregulation of X-chromosome inactivation [Citation50,Citation51]. X-chromosome inactivation serves to balance the dosage of X-encoded genes between males and females and it occurs during female embryonic development [Citation50,Citation51]. Of note, the male-specific sites particularly exhibited enrichments in X-chromosome cytobands, thus suggesting that these pollutants did not affect X-chromosome inactivation. Since male cells have only one X-chromosome, the activation or inactivation of disease-causing genes by DNA methylation would be different when these genes are carried on X-chromosomes rather than on autosomes. In addition, changes in gene expression patterns due to escape from X-chromosome inactivation, from mosaicism, or skewing are very complex events [Citation51–Citation54] that cannot be fully elucidated within this pilot study.
We identified associations between the degree of DNA methylation changes of a large number of CpG sites and the exposures to nPFOS, HCB, and p,p’-DDE were modulated by the presence of another exposure. In other words, if an association was evaluated by itself, an association may be significant but, if adjusted for another exposure, the association became weaker. Hence, our findings support the concept that exposure-exposure interaction could affect genome-wide methylation levels. However, such observation could be also due to reduced precision (or correlation/over-adjustment bias) when more than one exposure variable is inserted into the model. Therefore, because of the small size of this study, additional advanced interaction analyses are not feasible.
In this study, we found CpG sites associated with three POPs in male newborns. The sites associated with HCB_m are on genes related to reproductive system development and function. This chemical was introduced as a fungicide in the 1940s [Citation55]. In one younger Faroese birth cohort (born in 2007–2009) maternal serum HCB concentrations during pregnancy were associated with increased body mass index in childhood with no clear evidence of sex-dimorphic associations [Citation56], but sex-specific associations with childhood growth have been reported in other birth cohorts [Citation57]. Prenatal exposure to HCB-contaminated grain has been associated with the lower proportion of male births in other populations [Citation58]. p,p’-DDE is a breakdown product of the insecticide dichlorodiphenyltrichloroethane (DDT), well-known for its anti-androgenic activities [Citation59]. Population-based studies have shown that men with adult [Citation60] or prenatal [Citation61] DDE exposure were sub-fertile [Citation62] and were also associated with impairment of cognitive functions, deficits in psychosocial developments in childhood [Citation63–Citation67], and increased risk of neural degenerative diseases at old age [Citation68]. nPFOS belongs to a group of substances that are widely used in a variety of products including surface treatments for stain resistance, surface treatments on metals, as well as firefighting foams. In this study, nPFOS-associated methylome changes were predicted to dysregulate genes involved in embryonic and nervous system development. Our findings are in agreement with published studies, including a previous study in a Faroese birth cohort [Citation69], showing inverse associations between cord blood PFOS, and head circumference, ponderal index [Citation70,Citation71], and/or birth weight [Citation71,Citation72]. In the Faroese study [Citation69], inverse associations between PFOS exposure and birth size measures were more pronounced in boys, while mainly null or positive in girls, which is in line with the sex-specific DNA methylation changes noted for PFOS in the present study. In short, the persistent nature of these DNA methylation-associated chemicals raises concerns of their long-lasting health effects from birth to later-life.
Compared with other genome-wide techniques, Illumina 450K is one of the most cost-effective options for genome-wide methylation discovery for modern epidemiology studies. Still, the coverage of CpG sites is far from ideal, comprising less than 2% of the 28 million CpGs within the human methylome. In other words, the majority of methylation sites have not been included in this unbiased discovery. Another limitation of this study is the small sample size, especially in sex-specific analyses, which resulted in non-equivalent sample sizes in boys versus girls. Reduced power may explain in part the lack of significant associations, especially in the case of MeHg, for which, even though it is a known epigenetic toxicant, associations in this study did not reach the level of statistical significance. Also, the epidemiological findings in this populations show that MeHg is the main risk indicator for neurodevelopmental deficits [Citation19]. However, given that MeHg has a much shorter half-life than the POPs measured, associations with exposure indicators measured at the time of birth may require considerations of the timing of epigenetic and functional changes during pregnancy. Hence, special caution should be applied when interpreting absence of associations suggested from this study. Future investigations in the full cohort will help throw new light on these preliminary findings.
In conclusion, this pilot study, in spite of its small sample size, has unveiled potentially key epigenetic toxicants amidst a complex mixture of 15 environmental chemicals found in the cord blood of Faroese newborns. Our data show chemical-and sex-specific methylation changes in UCWBC DNA located in genes associated with networks of reproductive, cardiovascular, and neural/behavioral disruption. Of great interest, significantly more methylome changes were found in male infant DNA samples, with enrichment in the X-chromosome. Future larger-scale studies are warranted to further expand these preliminary observations that are likely of high relevance to the impact of environmental chemicals on the developmental origins of disease and health.
Disclosure of potential conflicts of interest
The authors report no conflict of interest
Supplemental Material
Download MS Power Point (149.9 KB)Supplemental Material
Download MS Excel (891.1 KB)Acknowledgements
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health (R01-ES009797, R01-ES021477, P30-ES006096, U01-ES020988) and a VA Merit Award (I01-BX000675).
Additional information
Funding
References
- Ladd-Acosta C. Epigenetic signatures as biomarkers of exposure. Curr Environ Health Rep. 2015;2:117–125. doi:10.1007/s40572-015-0051-2. PMID:26231361
- Hanna CW, Bloom MS, Robinson WP, et al. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum Reprod. 2012;27:1401–1410. doi:10.1093/humrep/des038. PMID:22381621
- Pilsner JR, Hu H, Ettinger A, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–1471. doi:10.1289/ehp.0800497. PMID:19750115
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi:10.1038/35057062. PMID:11237011
- Jordan IK, Rogozin IB, Glazko GV, et al. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi:10.1016/S0168-9525(02)00006-9. PMID:12547512
- Ho SM, Johnson A, Tarapore P, et al. Environmental epigenetics and its implication on disease risk and health outcomes. ILAR J. 2012;53:289–305. doi:10.1093/ilar.53.3-4.289. PMID:23744968
- Ho SM, Cheong A, Adgent MA, et al. Environmental factors, epigenetics, and developmental origin of reproductive disorders. Reprod Toxicol. 2017;68:85–104. doi:10.1016/j.reprotox.2016.07.011. PMID:27421580
- Perera F, Tang WY, Herbstman J, et al. Relation of DNA methylation of 5'-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One. 2009;4:e4488. doi:10.1371/journal.pone.0004488. PMID:19221603
- Tang WY, Levin L, Talaska G, et al. Maternal exposure to polycyclic aromatic hydrocarbons and 5'-CpG methylation of interferon-gamma in cord white blood cells. Environ Health Perspect. 2012;120:1195–1200. doi:10.1289/ehp.1103744. PMID:22562770
- Joubert BR, Haberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–1431. doi:10.1289/ehp.1205412. PMID:22851337
- Joubert BR, Haberg SE, Bell DA, et al. Maternal smoking and DNA methylation in newborns: in utero effect or epigenetic inheritance?. Cancer Epidemiol Biomarkers Prev. 2014;23:1007–1017. doi:10.1158/1055-9965.EPI-13-1256. PMID:24740201
- Richmond RC, Simpkin AJ, Woodward G, et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the avon longitudinal study of parents and children (ALSPAC). Hum Mol Genet. 2015;24:2201–2217. doi:10.1093/hmg/ddu739. PMID:25552657
- Koestler DC, Avissar-Whiting M, Houseman EA, et al. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect. 2013;121:971–977. doi:10.1289/ehp.1205925. PMID:23757598
- Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412–417. doi:10.1111/j.1365-2796.2007.01809.x. PMID:17444880
- Wu S, Hivert MF, Cardenas A, et al. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in project viva: an epigenome-wide association study. Environ Health Perspect. 2017;125:087019-1–087019-10. doi:10.1289/EHP1246. PMID:28858830
- Grandjean P, Budtz-Jorgensen E, White RF, et al. Methylmercury exposure biomarkers as indicators of neurotoxicity in children aged 7 years. Am J Epidemiol. 1999;150:301–305. doi:10.1093/oxfordjournals.aje.a010002. PMID:10430235
- Grandjean P, Weihe P, Burse VW, et al. Neurobehavioral deficits associated with PCB in 7-year-old children prenatally exposed to seafood neurotoxicants. Neurotoxicol Teratol. 2001;23:305–317. doi:10.1016/S0892-0362(01)00155-6. PMID:11485834
- Weihe P, Joensen HD. Dietary recommendations regarding pilot whale meat and blubber in the Faroe Islands. Int J Circumpolar Health. 2012;71:18594-1–18594-5. doi:10.3402/ijch.v71i0.18594.
- Grandjean P, Weihe P, Nielsen F, et al. Neurobehavioral deficits at age 7 years associated with prenatal exposure to toxicants from maternal seafood diet. Neurotoxicol Teratol. 2012;34:466–472. doi:10.1016/j.ntt.2012.06.001. PMID:22705177
- Grandjean P, Weihe P, Jorgensen PJ, et al. Impact of maternal seafood diet on fetal exposure to mercury, selenium, and lead. Arch Environ Health. 1992;47:185–195. doi:10.1080/00039896.1992.9938348. PMID:1596101
- Johansen T, Olaffson A. The faroe islands: a brief introduction. Copenhagen: Faroese Government Office; 1999.
- Weihe P, Grandjean P, Debes F, et al. Health implications for Faroe islanders of heavy metals and PCBs from pilot whales. Sci Total Environ. 1996;186:141–148. doi:10.1016/0048-9697(96)05094-2. PMID:8685706
- Bloch D, Desportes G, Hoydal K, et al. Pilot whaling in the Faroe Islands. North Atlantic Studies. 1990;2:36–44.
- Borrell A, Aguilar A. Pollution by DDT and PCB in blubber and muscle of long-finned pilot whales from the Faroe Islands. In: Donovan GP, Lockyer CH, Martin AR, editors. Biology of northern hemisphere pilot whales. Cambridge: International Whaling Commission; 1993. p. 351–367.
- Dam M. Mercury in the Faroe Islands: a review of available data. Frodskaparrit. 2004;52:85–133.
- Rotander A, Karrman A, van Bavel B, et al. Increasing levels of long-chain perfluorocarboxylic acids (PFCAs) in Arctic and North Atlantic marine mammals, 1984–2009. Chemosphere. 2012;86:278–285. doi:10.1016/j.chemosphere.2011.09.054. PMID:22051347
- Grandjean P, Henriksen JE, Choi AL, et al. Marine food pollutants as a risk factor for hypoinsulinemia and type 2 diabetes. Epidemiology (Cambridge, Mass). 2011;22:410–417. doi:10.1097/EDE.0b013e318212fab9. PMID:21364465
- Weihe P, Kato K, Calafat AM, et al. Serum concentrations of polyfluoroalkyl compounds in Faroese whale meat consumers. Environ Sci Technol. 2008;42:6291–6295. doi:10.1021/es800695m. PMID:18767701
- Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi:10.1093/bioinformatics/btu049. PMID:24478339
- Xu Z, Niu L, Li L, et al. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44:e20-1–e20-6. doi:10.1093/nar/gkv907. PMID:26384415
- Xu Z, Langie SA, De Boever P, et al. RELIC: a novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics. 2017;18:4, 1–7. doi:10.1186/s12864-016-3426-3. PMID:28049437
- Niu L, Xu Z, Taylor JA. RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics. 2016;32:2659–2663. doi:10.1093/bioinformatics/btw285. PMID:27153672
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi:10.1371/journal.pgen.0030161. PMID:17907809
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3: Article3, 1–25. doi:10.2202/1544-6115.1027. PMID:16646809
- McGregor K, Bernatsky S, Colmegna I, et al. An evaluation of methods correcting for cell-type heterogeneity in DNA methylation studies. Genome Biol. 2016;17:84, 1–17. doi:10.1186/s13059-016-0935-y. PMID:27142380
- Wang H, Ding N, Guo J, et al. Dysregulation of TTP and HuR plays an important role in cancers. Tumour Biol. 2016;37:14451–14461. doi:10.1007/s13277-016-5397-z. PMID:27644249
- Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi:10.4161/epi.6.6.16196. PMID:21593595
- Giorda R, Bonaglia MC, Beri S, et al. Complex segmental duplications mediate a recurrent dup(X)(p11.22-p11.23) associated with mental retardation, speech delay, and EEG anomalies in males and females. Am J Hum Genet. 2009;85:394–400. doi:10.1016/j.ajhg.2009.08.001. PMID:19716111
- Butler MG, Rafi SK, Manzardo AM. High-resolution chromosome ideogram representation of currently recognized genes for autism spectrum disorders. Int J Mol Sci. 2015;16:6464–6495. doi:10.3390/ijms16036464. PMID:25803107
- Gutierrez B, Arias B, Gasto C, et al. Association analysis between a functional polymorphism in the monoamine oxidase A gene promoter and severe mood disorders. Psychiatr Genet. 2004;14:203–208. doi:10.1097/00041444-200412000-00007. PMID:15564894
- Chatila TA, Blaeser F, Ho N, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi:10.1172/JCI11679. PMID:11120765
- Bennett CL, Ochs HD. IPEX is a unique X-linked syndrome characterized by immune dysfunction, polyendocrinopathy, enteropathy, and a variety of autoimmune phenomena. Curr Opin Pediatr. 2001;13:533–538. doi:10.1097/00008480-200112000-00007. PMID:11753102
- Rajender S, Thangaraj K, Gupta NJ, et al. A novel human sex-determining gene linked to Xp11.21-11.23. J Clin Endocrinol Metab. 2006;91:4028–4036. doi:10.1210/jc.2006-0950. PMID:16868052
- Cardenas A, Koestler DC, Houseman EA, et al. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics. 2015;10:508–515. doi:10.1080/15592294.2015.1046026. PMID:25923418
- Huen K, Yousefi P, Bradman A, et al. Effects of age, sex, and persistent organic pollutants on DNA methylation in children. Environ Mol Mutagen. 2014;55:209–222. doi:10.1002/em.21845. PMID:24375655
- Kobayashi S, Azumi K, Goudarzi H, et al. Effects of prenatal perfluoroalkyl acid exposure on cord blood IGF2/H19 methylation and ponderal index: the Hokkaido study. J Expo Sci Environ Epidemiol. 2017;27:251–259. doi:10.1038/jes.2016.50. PMID:27553991
- Reaves DK, Ginsburg E, Bang JJ, et al. Persistent organic pollutants and obesity: are they potential mechanisms for breast cancer promotion?. Endocr Relat Cancer. 2015;22:R69–R86. doi:10.1530/ERC-14-0411. PMID:25624167
- Deierlein AL, Rock S, Park S. Persistent endocrine-disrupting chemicals and fatty liver disease. Curr Environ Health Rep. 2017;4(4):439–449. doi:10.1007/s40572-017-0166-8. PMID: 28980219
- Kennedy SM, Koehoorn M. Exposure assessment in epidemiology: does gender matter?. Am J Ind Med. 2003;44:576–583. doi:10.1002/ajim.10297. PMID: 14635234
- Sharp AJ, Stathaki E, Migliavacca E, et al. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21:1592–1600. doi:10.1101/gr.112680.110. PMID: 21862626
- Deng X, Berletch JB, Nguyen DK, et al. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi:10.1038/nrg3687. PMID: 24733023
- Pasque V, Plath K. X chromosome reactivation in reprogramming and in development. Curr Opin Cell Biol. 2015;37:75–83. doi:10.1016/j.ceb.2015.10.006. PMID: 26540406
- Reue K. Sex differences in obesity: X chromosome dosage as a risk factor for increased food intake, adiposity and co-morbidities. Physiol Behav. 2017;176:174–182. doi:10.1016/j.physbeh.2017.02.040. PMID: 28284880
- Arnold AP, Cassis LA, Eghbali M, et al. Sex hormones and sex chromosomes cause sex differences in the development of cardiovascular diseases. Arterioscler Thromb Vasc Biol. 2017;37:746–756. doi:10.1161/ATVBAHA.116.307301. PMID: 28279969
- Reed L, Buchner V, Tchounwou PB. Environmental toxicology and health effects associated with hexachlorobenzene exposure. Rev Environ Health. 2007;22:213–243. doi:10.1515/REVEH.2007.22.3.213. PMID: 18078005
- Karlsen M, Grandjean P, Weihe P, et al. Early-life exposures to persistent organic pollutants in relation to overweight in preschool children. Reprod Toxicol. 2017;68:145–153. doi:10.1016/j.reprotox.2016.08.002. PMID: 27496715
- Valvi D, Mendez MA, Garcia-Esteban R, et al. Prenatal exposure to persistent organic pollutants and rapid weight gain and overweight in infancy. Obesity (Silver Spring). 2014;22:488–496. doi:10.1002/oby.20603. PMID: 23963708
- Jarrell JF, Gocmen A, Akyol D, et al. Hexachlorobenzene exposure and the proportion of male births in Turkey 1935–1990. Reprod Toxicol. 2002;16:65–70. doi:10.1016/S0890-6238(01)00196-4. PMID: 11934533
- Kelce WR, Wilson EM. Environmental antiandrogens: developmental effects, molecular mechanisms, and clinical implications. J Mol Med (Berl). 1997;75:198–207. doi:10.1007/s001090050104. PMID: 9106076
- De Jager C, Farias P, Barraza-Villarreal A, et al. Reduced seminal parameters associated with environmental DDT exposure and p,p'-DDE concentrations in men in Chiapas, Mexico: a cross-sectional study. J Androl. 2006;27:16–27. doi:10.2164/jandrol.05121. PMID: 16400073
- Charlier CJ, Foidart JM. Comparative study of dichlorodiphenyldichloroethylene in blood and semen of two young male populations: lack of relationship to infertility, but evidence of high exposure of the mothers. Reprod Toxicol. 2005;20:215–220. doi:10.1016/j.reprotox.2005.03.007. PMID: 15907656
- Quan C, Shi Y, Wang C, et al. p,p'-DDE damages spermatogenesis via phospholipid hydroperoxide glutathione peroxidase depletion and mitochondria apoptosis pathway. Environ Toxicol. 2016;31:593–600. PMID: 25410718
- Eskenazi B, Marks AR, Bradman A, et al. In utero exposure to dichlorodiphenyltrichloroethane (DDT) and dichlorodiphenyldichloroethylene (DDE) and neurodevelopment among young Mexican American children. Pediatrics. 2006;118:233–241. doi:10.1542/peds.2005-3117. PMID: 16818570
- Ribas-Fito N, Cardo E, Sala M, et al. Breastfeeding, exposure to organochlorine compounds, and neurodevelopment in infants. Pediatrics. 2003;111:e580–e585. doi:10.1542/peds.111.5.e580. PMID: 12728113
- Torres-Sanchez L, Rothenberg SJ, Schnaas L, et al. In utero p,p'-DDE exposure and infant neurodevelopment: a perinatal cohort in Mexico. Environ Health Perspect. 2007;115:435–439. doi:10.1289/ehp.9566. PMID: 17431495
- Torres-Sanchez L, Schnaas L, Rothenberg SJ, et al. Prenatal p,p -DDE exposure and neurodevelopment among children 3.5-5 years of age. Environ Health Perspect. 2013;121:263–268. PMID: 23151722
- Wnuk A, Rzemieniec J, Litwa E, et al. The crucial involvement of retinoid X receptors in DDE neurotoxicity. Neurotox Res. 2016;29:155–172. doi:10.1007/s12640-015-9572-6. PMID: 26563996
- Richardson JR, Roy A, Shalat SL, et al. Elevated serum pesticide levels and risk for Alzheimer disease. JAMA Neurol. 2014;71:284–290. doi:10.1001/jamaneurol.2013.6030. PMID: 24473795
- Valvi D, Oulhote Y, Weihe P, et al. Gestational diabetes and offspring birth size at elevated environmental pollutant exposures. Environ Int. 2017;107:205–215. doi:10.1016/j.envint.2017.07.016. PMID: 28753482
- Betts K. PFOS and PFOA in humans: new study links prenatal exposure to lower birth weight. Environ Health Perspect. 2007;115:A550. doi:10.1289/ehp.115-a550a. PMID: 18007977
- Chen MH, Ha EH, Wen TW, et al. Perfluorinated compounds in umbilical cord blood and adverse birth outcomes. PLoS One. 2012;7:e42474-1–e42474-8. doi:10.1371/journal.pone.0042474. PMID: 22879996
- Apelberg BJ, Witter FR, Herbstman JB, et al. Cord serum concentrations of perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA) in relation to weight and size at birth. Environ Health Perspect. 2007;115:1670–1676. doi:10.1289/ehp.10334. PMID: 18008002
