ABSTRACT
Parental cannabis use has been associated with adverse neurodevelopmental outcomes in offspring, but how such phenotypes are transmitted is largely unknown. Using reduced representation bisulphite sequencing (RRBS), we recently demonstrated that cannabis use is associated with widespread DNA methylation changes in human and rat sperm. Discs-Large Associated Protein 2 (DLGAP2), involved in synapse organization, neuronal signaling, and strongly implicated in autism, exhibited significant hypomethylation (p < 0.05) at 17 CpG sites in human sperm. We successfully validated the differential methylation present in DLGAP2 for nine CpG sites located in intron seven (p < 0.05) using quantitative bisulphite pyrosequencing. Intron 7 DNA methylation and DLGAP2 expression in human conceptal brain tissue were inversely correlated (p < 0.01). Adult male rats exposed to delta-9-tetrahydrocannabinol (THC) showed differential DNA methylation at Dlgap2 in sperm (p < 0.03), as did the nucleus accumbens of rats whose fathers were exposed to THC prior to conception (p < 0.05). Altogether, these results warrant further investigation into the effects of preconception cannabis use in males and the potential effects on subsequent generations.
Introduction
Cannabis sativa is the most commonly used illicit psychoactive drug in the United States (U.S.) and Europe [Citation1]. In the U.S., 11 states and Washington D.C. have legalized the recreational use of cannabis and 33 states have legalized the use of medicinal cannabis [Citation2,Citation3]. Since 1995, cannabis potency (defined as the concentration of the psychoactive cannabis component delta-9-tetrahydrocannabinol, or THC, in the sample [Citation4]) has consistently risen from ~4% to as high as 32% in some states [Citation2,Citation5,Citation6]. Changes in cannabis potency have been accompanied by changes in attitudes about cannabis and patterns of cannabis use. Between 2002 and 2014, the percentage of adults in the U.S. who perceived cannabis use as risky declined from 50% to 33% [Citation6]. During this same period, the percentage of U.S. adults who believed cannabis to have no risk rose from 6% to 15% [Citation6]. According to a 2015 Survey on Drug Use and Health, 52.5% of men in the U.S. of reproductive age (≥18) have reported cannabis use at some point in their lives, making cannabis exposure especially relevant for potential future fathers [Citation7–Citation11].
Given the increased prevalence of cannabis use in the U.S., studies are beginning to focus on the effects of use on the health and development of offspring. Prenatal cannabis exposure via maternal use during pregnancy is associated with decreased infant birth weight, an increased likelihood to require the neonatal intensive care unit, and the potential for an impaired fetal immune system compared to those infants who are not exposed during gestation [Citation1,Citation12]. In rodent studies, rat pups born to parents who were both exposed to THC during adolescence had increased heroin-seeking behaviour later in life, a phenotype that was accompanied by epigenetic changes in the nucleus accumbens [Citation13–Citation15]. These studies and others have begun to highlight the potential for intergenerational consequences of cannabis exposure [Citation16]. Identifying the mechanism that underlies these changes is critical as cannabis use continues to increase across the U.S.
The environment impacts the integrity and maintenance of the epigenome such that it is now viewed as a molecular archive of past exposures [Citation17]. While the majority of environmental epigenetic studies are focused on the impact of the inutero environment on the epigenome and health of the child, it has become apparent that the exposure history of the father must also be considered – specifically the impact of his exposures on the sperm epigenome. Studies have shown that exposure to phthalates, pesticides, nutritional deficiencies, and obesity can all induce potentially heritable changes in the sperm epigenome [Citation18–Citation24]. It is likely that other common and emerging exposures, including cannabis, may also contribute to disruption of sperm DNA methylation in a similar fashion, and that such changes could be transmitted to the subsequent generation.
Using reduced representation bisulphite sequencing (RRBS) our group recently demonstrated that cannabis use in humans, and THC exposure in rats, is associated with decreased sperm concentrations and widespread changes in sperm DNA methylation [Citation25]. Of the regions identified in humans, Discs-Large Associated Protein 2 (DLGAP2) exhibited significant hypomethylation in the sperm of cannabis-exposed men compared to controls (p < 0.05). DLGAP2, a membrane-associated protein located in the post-synaptic density of neurons, plays a key role in synapse organization and neuronal signaling [Citation26]. Dysregulation of DLGAP2 is associated with various neurological and psychiatric disorders, such as autism spectrum disorder (ASD) and schizophrenia [Citation26–Citation29]. In our prior screen, we identified 17 differentially methylated CpG sites within DLGAP2 in the sperm of cannabis-exposed men compared to controls. DLGAP2 was just one of 46 genes with greater than 10 CpG sites showing significantly altered DNA methylation in the sperm of cannabis users compared to controls, out of the 2,077 genes we identified as having altered DNA methylation. The first objective of this study was to validate our preliminary RRBS findings for DLGAP2 using quantitative bisulphite pyrosequencing. Our second objective was to determine the functional association between DNA methylation and gene expression of DLGAP2 to better understand how cannabis use might affect this relationship. To determine the possible intergenerational effects of paternal cannabis use, our third objective was to determine if Dlgap2 was differentially methylated in the sperm of rats exposed to THC versus controls, and if so, whether or not these changes were intergenerationally heritable.
Results
DLGAP2 is hypomethylated in sperm from cannabis users versus controls by Reduced Representation Bisulphite Sequencing (RRBS)
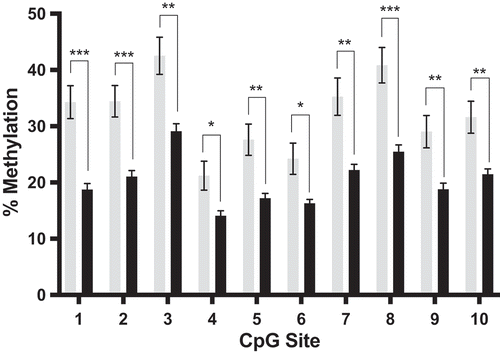
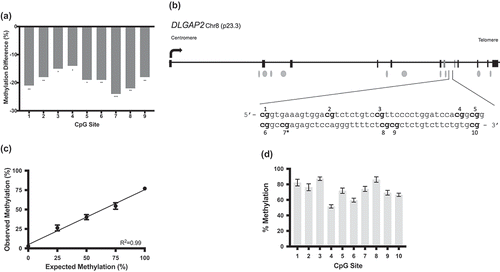
Our prior study [Citation25] revealed 17 differentially methylated sites by RRBS in the sperm of cannabis users compared to controls for the DLGAP2 gene. Table S1 lists all 17 of these sites and their genomic coordinates. graphically demonstrates the significant hypomethylation of nine of these sites that are clustered together in the seventh intron of this gene. DLGAP2 is schematically shown in , including the exon-intron structure, position of CpG islands, transcription start site and the region of interest in intron 7 within the context of the gene body, with an inset showing the nucleotide sequence analysed in this study.
Figure 1. Reduced representation bisulphite sequencing data and DLGAP2 schematic demonstrating the region used for bisulphite pyrosequencing. (a) RRBS data for DLGAP2 from our prior study. This genome-scale analysis identified CpG sites that were differentially methylated in the sperm of cannabis users compared to controls. Results for nine CpG sites, intronically located in DLGAP2, that were significantly hypomethylated in the sperm of cannabis users compared to controls. (b) A gene schematic of DLGAP2. Exons are represented by the rectangular boxes, while the ovals represent CpG islands. The sequence identified by RRBS that was subsequently analysed here is identified in the inset. CpG site 7 (7*) was not initially identified in the RRBS data but included in further analyses, for a total of 10 CpG sites analysed. (c) Pyrosequencing assay validation (performed in triplicate) for DLGAP2. Data points and error bars represent the mean ± SEM, respectively. Error bars not shown had deviations too small for inclusion on the graph. (d) Pyrosequencing results showing the average level of DNA methylation for each of the ten CpG sites of DLGAP2 in human conceptal diploid testes tissues demonstrate that the intron 7 region of DLGAP2 is not an imprint control region. Shown are the mean ± SEM.* = p < 0.05; ** = p < 0.01; *** = p < 0.001.
Validation of DLGAP2 RRBS methylation data
To confirm the methylation differences that were initially detected using RRBS, we designed a bisulphite pyrosequencing assay for the DLGAP2 intron 7 region (see ) which captures 10 CpG sites, nine of which were identified as significantly differentially methylated using RRBS. We first validated pyrosequencing assay performance using defined mixtures of fully methylated and unmethylated human genomic DNAs. The measured levels of methylation by pyrosequencing showed good agreement between the amount of input methylation levels and the amount of methylation detected (r2 = 0.99 and p = 0.0003) (). These results confirmed the linearity of the assay in the ability to detect increasing amounts of DNA methylation at this region across the full range of possible methylation values, and indicate that the assay is suitable for use with biological specimens.
The DLGAP2 intron 7 region is not an imprinting control region (ICR)
DLGAP2 is paternally expressed in the testis, biallelically expressed in the brain, and has low expression elsewhere in the body [Citation30]. Since DLGAP2 is known to be genomically imprinted in testis [Citation30], and since the imprint control region for this gene has not yet been defined, we sought to determine if the region of interest in intron 7 is part of the DLGAP2 imprint control region (ICR). The methylation at ICRs is established during epigenome reprogramming in the primordial germ cells in embryonic development. Male and female gametes exhibit divergent methylation at ICRs, and this methylation profile is maintained through subsequent post-fertilization epigenetic reprogramming and in somatic cells throughout the life course. Therefore, we expected that if the DLGAP2 intron 7 region is an ICR, the diploid testis tissues from human conceptuses would exhibit approximately 50% methylation due to the complete methylation of one allele at this region and the complete lack of methylation at the other allele. Human conceptal testes tissues (n = 3) showed an average of 72.5% methylation at the DLGAP2 intron 7 region (). This finding, of higher than anticipated and variable levels of methylation, is inconsistent with ICR status.
Bisulphite pyrosequencing validates the RRBS methylation data in human sperm
We next performed quantitative bisulphite pyrosequencing on the same sperm DNA samples from cannabis users and controls as those used to generate the RRBS data to confirm the loss of methylation present at the intron 7 region of DLGAP2. All nine CpG sites that were hypomethylated in the cannabis users by RRBS were also found to be hypomethylated by bisulphite pyrosequencing, as well as an additional CpG site that was captured in the assay design (p < 0.05 for all 10 sites) (). Following Bonferroni correction of the p value to adjust for multiple comparisons (p < 0.005), CpG sites 1,2,3,5,7,8,9, and 10 remained significant. From this pyrosequencing assay we observed methylation differences of 7–15% between the sperm of the cannabis users (n = 8) compared to controls (n = 7). Correlation of the RRBS and pyrosequencing data for each individual CpG site showed significant agreement at all sites analysed (p < 0.02 for all sites; Figure S1). All CpG sites showed a significant loss of methylation in accordance with the direction of change observed by RRBS for these same CpG sites.
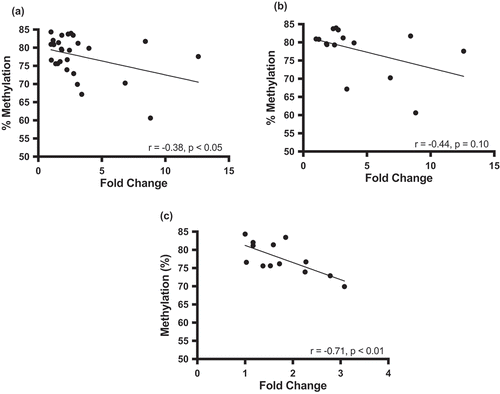
Methylation of DLGAP2 intron 7 is inversely correlated with DLGAP2 expression
Given that we observed significant loss of intron 7 DLGAP2 DNA methylation in sperm of cannabis users relative to non-users, we next examined the relationship between DNA methylation and gene expression in the brain, where this gene’s function is critical. We used 28 conceptal brain tissues to examine the relationship between DNA methylation and mRNA expression. Expression levels were normalized to the lowest expressing sample, and the relationship between DNA methylation and mRNA expression was calculated with a Pearson correlation. We found that as methylation increased in this region, mRNA expression decreased significantly (p < 0.05) (). Knowing that there are sex differences in autism spectrum disorder (ASD), and that dysregulation of DLGAP2 is associated with ASD [Citation26], we sought to determine if there were any sex differences in the methylation-expression relationship in these tissues. To investigate this, we ran the correlation for males (n = 15) and females (n = 13) independently. The inverse relationship between methylation and expression was evident for both males and females, but this relationship was significant only in females (p = 0.006) ().
Intergenerational inheritance of altered Dlgap2 DNA methylation
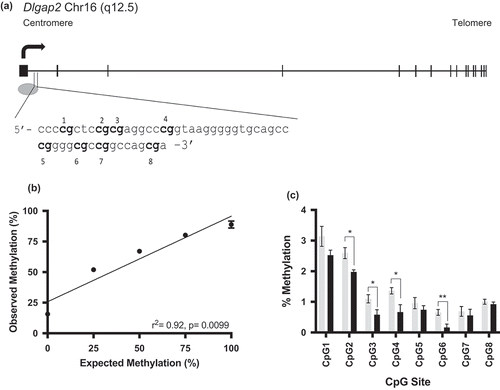
We next sought to investigate Dlgap2 using data obtained from our prior study [Citation25] to determine if there was any differential methylation of Dlgap2 in THC versus control rats that was not initially identified using the imposed thresholds of that study. We were particularly interested in the potential for intergenerational transmission and to determine if route of THC exposure affected DNA methylation at this gene. The pilot study rats [Citation25] were given THC via oral gavage (to mimic oral ingestion of drug) while subsequent studies dosed rats via intraperitoneal injection (to mimic inhalation of drug). From the rats administered THC via oral gavage versus controls, we identified a region of Dlgap2 that showed differential methylation by the RRBS analysis that contains eight CpG sites. This region is in the first intron of Dlgap2, in a CpG island that spans the first exon of this gene as well (schematic of the gene structure and sequence of this region shown in ). We validated the rat Dlgap2 pyrosequencing assay using commercially available rat DNA of defined methylation status. The results showed good agreement between the input methylation and the amount of methylation detected by pyrosequencing (r2 = 0.92, p = 0.01) ().
Figure 4. Rat Dlgap2 gene schematic and pyrosequencing assay validation. (a) A gene schematic of Dlgap2. Exons are represented by the rectangular boxes, while the oval represents a CpG island. The sequence of interest is identified in the inset. (b) Pyrosequencing assay validation performed in triplicate for Dlgap2. Error bars represent the SEM. Error bars not shown had deviations too small for inclusion on the graph. (c) Bisulphite pyrosequencing data from control (gray) or 4mg/kg THC exposed (black) rat sperm for Dlgap2. Shown is the mean +/- SEM for each group. * = p < 0.05; ** = p < 0.01.
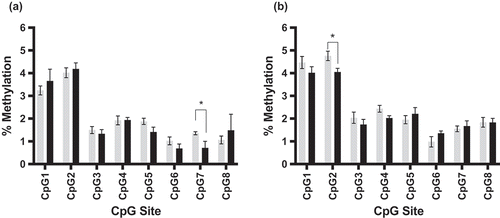
We were able to demonstrate intergenerational inheritance of an altered DNA methylation pattern in Dlgap2. Comparing the average methylation for exposed and unexposed sperm for each CpG site revealed that sites 2,3,4 and 6 of the eight CpG sites analysed were significantly hypomethylated in the sperm of rats exposed via injection to 4mg/kg THC compared to controls (p = 0.03 to p = 0.005) (). CpG site 6 remained significant after Bonferroni correction (p < 0.006). The same region of Dlgap2 was then analysed in the hippocampus and nucleus accumbens of rats whose fathers were exposed to control or 4mg/kg THC. While CpG site 7 was significantly hypomethylated (p < 0.05) in the hippocampus of the offspring (), this site was not identified as differentially methylated in the sperm of THC exposed rats, and therefore we could not conclude that this change was transmitted as the result of changes present in the exposed sperm. In the nucleus accumbens, however, significant hypomethylation (p = 0.02) at CpG site 2 was detected in the offspring (), one of the same sites identified in the sperm of THC exposed rats. We also found that there was an inverse relationship between DNA methylation and expression of Dlgap2 in the nucleus accumbens, though not statistically significant likely due to the small sample size available in this study (n = 6 exposed, n = 8 unexposed; Figure S2).
Discussion
In this study, we examined the effects of regular male cannabis use on human sperm DNA methylation, at DLGAP2. Our RRBS study initially identified 17 CpG sites in DLGAP2 that were differentially methylated in the sperm of cannabis users compared to controls. Of the sites that were initially identified, nine of them all reside together in the seventh intron of this gene, though not in a defined CpG island. To first confirm the RRBS data, we performed quantitative bisulphite pyrosequencing for the nine clustered CpG sites. We were able to capture an additional CpG site with careful assay design for a total of ten CpG sites analysed via bisulphite pyrosequencing. We successfully validated the RRBS findings, confirming that there was significant hypomethylation among these ten sites with cannabis use. We confirmed a significant inverse correlation between methylation and expression at this region in human conceptal brain tissues.
To begin to determine whether or not the effects of cannabis on sperm are heritable, we analysed sperm from THC exposed and control male rats, as well as the hippocampus and nucleus accumbens from offspring of THC exposed and control males for changes in DNA methylation at Dlgap2. Rats exposed to THC were given a dose (4mg/kg THC for 28 days) that is pharmacodynamically equivalent to daily cannabis use to resemble frequent use in humans. We identified significant hypomethylation at Dlgap2 in the sperm of exposed rats as compared to controls. This hypomethylated state was also detected in the nucleus accumbens of rats born to THC exposed fathers compared to controls, supporting the potential for intergenerational inheritance of an altered sperm DNA methylation pattern. While the changes in the degree of methylation are small in the rats (0.5–0.7%), we previously reported that fractional changes in methylation can significantly influence the degree to which the gene’s expression is altered [Citation31].
DLGAP2 is a member of the DLGAP family of scaffolding proteins located in the post-synaptic density (PSD) of neurons. The PSD is a protein-dense web that lies under the postsynaptic membrane of neurons and facilitates excitatory glutamatergic signaling in the central nervous system [Citation26,Citation32]. DLGAP2 functions to transmit neuronal signals across synaptic junctions and helps control downstream signaling events [Citation26,Citation32]. Due to its important role in PSD signaling, even small changes in the expression of DLGAP2 can have severe consequences [Citation26,Citation32]. Of particular relevance, DLGAP2 has been linked to schizophrenia and importantly, has been identified as an autism candidate gene [Citation27,Citation28,Citation33,Citation34]. Differential methylation of DLGAP2 is reported in the brain of individuals with autism, and has been linked to post-traumatic stress disorder in rats [Citation27,Citation35]. Knockout of Dlgap2 in mice results in abnormal social behaviour, increased aggressive behaviour, and learning deficits [Citation36].
Studies are increasingly showing associations between cannabis use and various neuropsychiatric and behavioural disorders including anxiety, depression, cognitive deficits, autism, psychosis, and addiction [Citation2,Citation6,Citation7,Citation9,Citation14,Citation37–Citation39]. Research looking into the effects of THC exposure found that rat pups born to parents who were exposed to THC during adolescence showed increased effort to self-administer heroin compared to those born to unexposed parents [Citation13]. This increase in addictive behaviour was driven by THC-induced changes in DNA methylation, occurring in the striatum, including the nucleus accumbens [Citation14,Citation15]. One of the genes whose methylation was altered by parental THC exposure was Dlgap2 [Citation15]. Recently, a group from Australia analysed datasets from two independent cohorts to examine the relationship between cannabis legalization in the U.S. and ASD incidence. They determined there was a strikingly significant positive association between cannabis legalization and increased ASD incidence. Further, the study authors predicted that there will be a 60% increase in excess ASD cases in states with legal cannabis by 2030, and deemed ASD the most common form of cannabis-associated clinical teratology [Citation40].
It is estimated that the ratio of boys with ASD to girls with ASD is 4:1 which led us to stratify our analysis looking at the relationship between DNA methylation and gene expression by sex [Citation41,Citation42]. The results of our methylation-expression analyses demonstrated a significant association in females but not males. While we don’t know the ASD status of these samples, there are several reasons why this may be the case. First, there are certain genes that confer a stronger ASD phenotype in girls compared to boys [Citation41,Citation42]. Thus, while we see the trend in both sexes, it is possible that dysregulation of this gene may manifest phenotypically more in girls. Alternatively, it may be that the regulatory relationship between methylation and expression is retained in females while altered methylation further exacerbates an already fragile relationship in males. Overall, this data confirms that the region of DNA methylation within DLGAP2 that was differentially methylated in the sperm of cannabis users compared to controls is functionally important in the brain.
DLGAP2 is an imprinted gene that exhibits paternal expression in the testis, biallelic expression in the brain, and low expression elsewhere in the body [Citation30]. Because the methylation established at imprinted genes resists post-fertilization epigenetic reprogramming [Citation43–Citation45], this supports the possibility that changes in methylation at DLGAP2 in sperm could be transmitted to the next generation. However, given that the region in intron 7 is not an ICR, it is unlikely that this would be a potential mechanism for intergenerational inheritance of an altered methylation pattern at this region. However, it has recently been discovered that a subset of genes termed ‘escapees’ are able to escape primordial germ cell (PGC) and post-fertilization reprogramming events [Citation46,Citation47], providing a mechanism for epigenetic changes incurred by sperm to be passed on to the subsequent generation.
Processes in the PSD are sensitive to endocannabinoids [Citation26,Citation48–Citation51], which suggests that these processes are potentially sensitive to exogenous cannabinoids, such as THC and cannabis. This is especially important as cannabis legalization and use are increasing dramatically across the U.S. It is estimated that 22% of American adults currently use cannabis, of which 63% are regular users (≥1–2 times per month) [Citation7–Citation10]. Among regular users 55% are males and over half of all men over 18 have reported cannabis use in their lifetime [Citation7–Citation10]. Importantly, this age range includes individuals of reproductive age. Since almost half of all pregnancies in the U.S. are unplanned, there is concern that many pregnancies may occur during a time when one, or both, parents are using or are exposed to cannabis [Citation52].
Our results provide novel findings about the effects of paternal cannabis use on the methylation status of an ASD candidate gene, a disorder whose rates continue to climb, but whose precise aetiologies remain unknown. Studies are beginning to show that there is a potential for paternal intergenerational inheritance. In particular, epigenetic changes in umbilical cord blood of babies born to obese fathers were also found in the sperm of obese men. This study is the first to demonstrate that there are changes present in the sperm epigenome of cannabis users at a gene involved in ASD.
The results of this study have several limitations. The sample size was small, which might limit generalization of the study findings. However, even though our sample size was small, we were able to identify common pathways that were differentially methylated in both human and rat sperm, highlighting the potential specificity of these effects [Citation25]. We did not account for a wide variety of potential confounders such as various lifestyle habits, sleep, diet/nutrition, exercise, etc, given that their influence on the sperm DNA methylome is largely unknown. Larger studies are required to confirm these findings. In the conceptal tissues we were only able to analyse whole brain, rather than the areas where DLGAP2 is most highly expressed such as the hippocampus and the striatum, which could have diluted the strength of the results.
Strengths of the study included that we used a highly quantitative method to confirm the methylation status that was measured by RRBS. This study was the first demonstration of the association between cannabis use and substantial hypomethylation of DLGAP2 in human sperm. Additionally, we are able to confirm a functional relationship between methylation and expression in a relevant target tissue, and have shown that the relationship between methylation and expression is weakened in males, which could bear relevance to the sexual dimorphism in the prevalence of autism. This is the first demonstration of potential heritability of altered methylation resulting from preconceptional paternal THC exposure. Given the increasing legalization and use of cannabis in the U.S., our results underscore a need for larger studies to determine the potential for heritability of DLGAP2 methylation changes in the human F1 generation and beyond. It will also be important to examine how cannabis-associated methylation changes relate to neurobehavioral phenotypes
Methods
Participants
Participants were recruited as described in Murphy et al, 2018 and all aspects of the study were carried out in accordance with the Duke Institutional Review Board [Citation25]. Twenty-four males of age 18–40 years who met all inclusion and no exclusion criteria participated in the study. In brief, participants were excluded if there was a positive result for drugs of abuse on a rapid urine test (except for cannabis in the user group), if they were currently prescribed any psychoactive medication, if there was self-reported nicotine or tobacco use within the past 12 weeks (verified by urine cotinine levels), or if there was an estimated IQ < 80 (see Murphy et al 2018 for more detailed participant characteristics). At screening, users (n = 12) reported ≥weekly cannabis use for the past six months, were positive for cannabis use on a rapid urine test and had unadjusted urinary 11-nor-9-carboxy-delta9-THC (THCCOOH) concentrations ≥50 ng/ml. Non-users (n = 12) self-reported no cannabis use in the past six months and fewer than ten lifetime uses at screening, with negative results for THCCOOH on a rapid urinary test and a verified urinary THCCOOH level of 0 ng/ml. Participants were scheduled for a visit to the Duke University fertility clinic to provide a semen specimen using procedures as previously described [Citation25]. Human conceptal tissues from elective pregnancy termination procedures including brain and testes were analysed from 28 individuals. Female samples (n = 13) had gestational ages ranging from 67d-122d, and male samples (n = 15 brain, n = 3 testes) had gestational ages ranging from 78d-108d. Samples were obtained between 1999 and 2010 from the NIH-funded Laboratory of Developmental Biology at the University of Washington and used with approval from the Duke Institutional Review Board.
Rat exposure to tetrahydrocannabinol (THC)
Nine-week-old, sexually mature male Sprague-Dawley rats were purchased from Charles River Laboratories and were housed 2–3 per cage. Rats were randomized to two groups that were dosed daily for 28 days via subcutaneous injection with vehicle only (4% TWEEN-80 in saline) (n = 8) or 4mg/kg THC (n = 7). Rats were sacrificed and the epididymis was placed in sterile PBS where the swim out method enriched the solution for mature (motile) sperm. Sperm were washed with PBS and were flash frozen in liquid nitrogen before being transferred to −80°C for further use. To study the intergenerational effects of paternal THC exposure, young adult Sprague-Dawley rats were dosed with 4mg/kg/day of THC by subcutaneous injection. Controls received vehicle (4% TWEEN-80 in saline). The rats were then bred with drug-naïve females. Subjects were maintained on a reversed 12/12 day-night cycle and had ad libitum access to food and water. Offspring (n = 8 control offspring and n = 6 4mg/kg/day offspring) were sacrificed at adulthood and brains were dissected to isolate the hippocampus and nucleus accumbens. After isolation tissues were flash frozen in liquid nitrogen before being stored in −80°C for further use. All study protocols were approved by the Institutional Animal Care and Use Committee and Duke University and conducted in accordance with federal guidelines.
DNA isolation from sperm
DNA was extracted from human and rat sperm samples according to Qiagen’s Puregene DNA Purification Protocol (Qiagen, Germantown, MD). Briefly, sperm samples underwent cell lysis and Proteinase K digestion, followed by treatment with RNase A solution. Following protein precipitation and removal, DNA was isolated and eluted in 30μL of nuclease-free water. Genomic DNA (gDNA) was quantified on the NanoDrop 2000 and quality was determined through measurement of the A260/280 and A260/230 ratios. gDNA was stored at −20°C for subsequent studies.
Reduced representation bisulphite sequencing (RRBS)
gDNA (200-500ng) from sperm of 12 cannabis users and 12 non-user controls was digested with MspI enzyme and was subsequently used to construct DNA libraries that were sequenced by Zymo Research on the Illumina HiSeq Platform. Full details of the methodology and analytical approach are available in Murphy et al 2018 [Citation25].
Nucleic acid isolation from tissues
DNA and RNA were extracted from conceptal brain (n = 28) and testis (n = 3) tissues, and rat hippocampus (n = 20) and nucleus accumbens (n = 20) using the column-based isolation and purification Qiagen All Prep DNA/RNA Mini Kit according to the manufacturer's recommendations. DNA was eluted in 50-100μL of TE buffer, while RNA was eluted in 35-50μL of nuclease-free water. DNA and RNA were quantified on the NanoDrop 2000 immediately following extraction. Following quantification, DNA was stored at −20°C and RNA was stored at −80°C for further use.
Bisulphite pyrosequencing
Pyrosequencing assay design was performed using the PyroMark CpG Assay Design Software (Qiagen) and sequencing was carried out using the Pyromark Q96 MD Pyrosequencing Instrument (Qiagen) using 800ng of gDNA. PCR amplification and pyrosequencing were performed as previously described [Citation53]. Briefly, bisulphite DNA (bsDNA) (20ng) was used as a template for PCR amplification that was optimized to produce a single robust band. Primers and PCR conditions can be found in Table S2. Assay validation was performed in triplicate using defined mixtures of fully methylated and fully unmethylated DNAs (0%, 25%, 50%, 75%, and 100% methylated) in order to demonstrate the ability for increasing amounts of methylated DNA to be accurately measured. Once assay performance was validated, samples were analysed using the optimized assay conditions.
Quantitative real-time reverse transcription-PCR
Quantitative real-time RT-PCR was performed for DLGAP2 expression in conceptal brain tissues, as well as Dlgap2 expression in rat nucleus accumbens using the QuantStudio 6 Flex Real-Time PCR System from Thermo Fisher Scientific (Waltham, MA). DLGAP2 (Taqman probe ID: Hs01109777_m1), and the 18S rRNA Endogenous Control (probe ID: Hs99999901_s1) probes were multiplexed in a 1:1 ratio for each sample. PCR mastermix contained a final volume of 100ng/μL RNA, 1μL of each probe, 10μl of Quantabio qScript one-step RT-qPCR ToughMix (QuantaBio, Beverly, MA), and was brought to a total reaction volume of 20μL with nuclease-free water. Samples were run in duplicate and PCR cycling conditions were as follows: 50°C for 10 minutes, followed by 95°C for one minute, and finally followed by 40 cycles of 95°C for 10 seconds and 60°C for one minute. For rat tissues, Dlgap2 (Taqman probe ID: Rn00588099_m1) and Gapdh (Taqman probe ID: Rn01775763_g1) were multiplexed in a 1:1 ratio for each sample and samples were prepared and run as described above.
Statistical analyses
GraphPad Prism Version 8 was used for statistical analyses (GraphPad Software, San Diego, CA). For bisulphite pyrosequencing of human sperm samples for DLGAP2, as well as bisulphite pyrosequencing of rat sperm and hippocampus and nucleus accumbens of rats born to THC exposed fathers, a two-tailed Student’s t-test was run for each CpG site, comparing the means of the users and controls. Unadjusted p-values are reported, and those that remain significant after Bonferroni corrections were identified. To quantitatively examine the relationship between methylation and expression in brain tissues, delta delta CT was calculated and fold change expression was determined with expression levels normalized to the lowest expressing tissue. A Pearson correlation was run to identify the relationship between the level of DNA methylation and the fold change expression for each sample, which are the reported R and P values. This analysis was run for all 28 brain samples together, and then was calculated for males (n = 15) and females (n = 13) independently due to existing prior knowledge of sex differences in autism. To quantitatively examine the relationship between DNA methylation and expression in rat F1 offspring nucleus accumbens tissues (n = 6 exposed, n = 8 unexposed) of F0 THC-exposed/unexposed fathers, delta Ct was calculated and relative expression was determined (using levels of Gapdh to normalize) and multiplied by a constant value of 10 for ease of presentation. These values were correlated to Dlgap2 CpG site 2 methylation using Pearson correlation.
Author contributions
RS contributed to study conception and design, acquisition and interpretation of data, and drafting and revising the manuscript. KA contributed to acquisition of data and revising the manuscript. NIR was involved with study design, acquisition of data, and revising the manuscript. ABH was involved in acquisition of data and revising the manuscript. EP was involved in acquisition of data and revising the manuscript. JTM contributed to study design, acquisition of data, and revising the manuscript. SHK contributed to study conception, study design, acquisition, analysis and interpretation of data, and revising the manuscript. EDL contributed to study conception and design, acquisition, analysis and interpretation of data, and revising the manuscript. SKM contributed to study conception, study design, acquisition, analysis and interpretation of data, and drafting and revising the manuscript.
Supplemental Material
Download Zip (251.5 KB)Acknowledgments
We thank the CIPHERS study participants for their generosity and willingness to be involved in this study. We also thank Zhiqing Huang and Carole Grenier for their expert assistance with this project.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Gunn JK, Rosales CB, Center KE, et al. Prenatal exposure to cannabis and maternal and child health outcomes: a systematic review and meta-analysis. BMJ Open. 2016 Apr 5;6(4):e009986.
- Smart R, Caulkins JP, Kilmer B, et al. Variation in cannabis potency and prices in a newly legal market: evidence from 30 million cannabis sales in Washington state. Addiction. 2017 Dec;112(12):2167–2177.
- Berke J, Gould S. Illinois just became the first state to legalize marijuana sales through the legislature – here are all the states where marijuana is legal. 2018 cited 2019 Jun 25. Available from: https://www.businessinsider.com/legal-marijuana-states-2018-1.
- Elsohly MA, Slade D. Chemical constituents of marijuana: the complex mixture of natural cannabinoids. Life Sci. 2005 Dec 22;78(5):539–548.
- ElSohly MA, Mehmedic Z, Foster S, et al. Changes in cannabis potency over the last 2 decades (1995-2014): Analysis of current data in the United States. Biol Psychiatry. 2016 Apr 1;79(7):613–619.
- Hasin DS. US epidemiology of cannabis use and associated problems. Neuropsychopharmacology. 2018 Jan;43(1):195–212.
- Budney AJ, Roffman R, Stephens RS, et al. Marijuana dependence and its treatment. Addict Sci Clin Pract. 2007 Dec;4(1):4–16.
- Lloyd D. JRAM, O’Malley PM, Bachman JG, et al. Monitoring the future. National survey results on drug use: The University of Michigan Institute for Social Research. 2017 cited 2018 Oct 29. Available from: http://www.monitoringthefuture.org/pubs/monographs/mtf-overview2017.pdf.
- Azofeifa A, Mattson ME, Schauer G, et al. National estimates of marijuana use and related indicators — National survey on drug use and health. 2016 [cited 2018 oct]. Available from: https://www.cdc.gov/mmwr/volumes/65/ss/ss6511a1.htm.
- Yahoo News/Marist Poll. Weed and the American family. 2017 cited 2018 Oct 29. Available from: http://maristpoll.marist.edu/wp-content/misc/Yahoo%20News/20170417_Banners%20of%20Americans_Yahoo%20News-Marist%20Poll_Weed%20and%20The%20American%20Family.pdf.
- Illum LRH, Bak ST, Lund S, et al. DNA methylation in epigenetic inheritance of metabolic diseases through the male germ line. J Mol Endocrinol. 2018 Feb;60(2):R39–R56.
- Dong C, Chen J, Harrington A, et al. Cannabinoid exposure during pregnancy and its impact on immune function. Cell Mol Life Sci. 2019;76(4):729–743.
- Szutorisz H, DiNieri JA, Sweet E, et al. Parental THC exposure leads to compulsive heroin-seeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology. 2014 May;39(6):1315–1323.
- Szutorisz H, Hurd YL. Epigenetic effects of cannabis exposure. Biol Psychiatry. 2016 Apr 1;79(7):586–594.
- Watson CT, Szutorisz H, Garg P, et al. Genome-Wide DNA methylation profiling reveals epigenetic changes in the rat nucleus accumbens associated with cross-generational effects of adolescent THC exposure. Neuropsychopharmacology. 2015 Dec;40(13):2993–3005.
- Ibn Lahmar Andaloussi Z, Taghzouti K, Abboussi O. Behavioural and epigenetic effects of paternal exposure to cannabinoids during adolescence on offspring vulnerability to stress. Int J Dev Neurosci. 2019 Feb;72:48–54.
- Heijmans BT, Tobi EW, Lumey LH, et al. The epigenome: archive of the prenatal environment. Epigenetics. 2009 Nov 16;4(8):526–531.
- Wu H, Hauser R, Krawetz SA, et al. Environmental susceptibility of the sperm epigenome during windows of male germ cell development. Curr Environ Health Rep. 2015 Dec;2(4):356–366.
- Day J, Savani S, Krempley BD, et al. Influence of paternal preconception exposures on their offspring: through epigenetics to phenotype. Am J Stem Cells. 2016;5(1):11–18.
- Soubry A, Murphy SK, Wang F, et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. Int J Obes (Lond). 2015 Apr;39(4):650–657.
- Abbasi J. The paternal epigenome makes its mark. JAMA. 2017 May 23;317(20):2049–2051.
- Donkin I, Barres R. Sperm epigenetics and influence of environmental factors. Mol Metab. 2018 Feb 27;14:1–11.
- Schuster A, Skinner MK, Yan W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet. 2016;2:1.
- Leter G, Consales C, Eleuteri P, et al. Exposure to perfluoroalkyl substances and sperm DNA global methylation in Arctic and European populations. Environ Mol Mutagen. 2014 Aug;55(7):591–600.
- Murphy SK, Itchon-Ramos N, Visco Z, et al. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics. 2018;13(12):1208–1221.
- Rasmussen AH, Rasmussen HB, Silahtaroglu A. The DLGAP family: neuronal expression, function and role in brain disorders. Mol Brain. 2017 Sep 4;10(1):43.
- Nardone S, Sams DS, Reuveni E, et al. DNA methylation analysis of the autistic brain reveals multiple dysregulated biological pathways. Transl Psychiatry. 2014 Sep;2(4):e433.
- Soler J, Fananas L, Parellada M, et al. Genetic variability in scaffolding proteins and risk for schizophrenia and autism-spectrum disorders: a systematic review. J Psychiatry Neurosci. 2018 Jul;43(4):223–244.
- Lee SE, Kim JA, Chang S. nArgBP2-SAPAP-SHANK, the core postsynaptic triad associated with psychiatric disorders. Exp Mol Med. 2018 Apr 9;50(4):2.
- Luedi PP, Dietrich FS, Weidman JR, et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007 Dec;17(12):1723–1730.
- Murphy SK, Adigun A, Huang Z, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012 Feb 15;494(1):36–43.
- Dosemeci A, Weinberg RJ, Reese TS, et al. The postsynaptic density: There is more than meets the eye. Front Synaptic Neurosci. 2016;8:23.
- Chien WH, Gau SS, Liao HM, et al. Deep exon resequencing of DLGAP2 as a candidate gene of autism spectrum disorders. Mol Autism. 2013 Aug 1;4(1):26.
- Xing J, Kimura H, Wang C, et al. Resequencing and association analysis of six PSD-95-related genes as possible susceptibility genes for schizophrenia and autism spectrum disorders. Sci Rep. 2016 Jun 7;6:27491.
- Chertkow-Deutsher Y, Cohen H, Klein E, et al. DNA methylation in vulnerability to post-traumatic stress in rats: evidence for the role of the post-synaptic density protein Dlgap2. Int J Neuropsychopharmacol. 2010 Apr;13(3):347–359.
- Jiang-Xie LF, Liao HM, Chen CH, et al. Autism-associated gene Dlgap2 mutant mice demonstrate exacerbated aggressive behaviors and orbitofrontal cortex deficits. Mol Autism. 2014;5:32.
- Rey JM, Tennant CC. Cannabis and mental health. BMJ. 2002 Nov 23;325(7374):1183–1184.
- David A, AJS G, Richard H. Cannabis use and disorder: epidemiology, comorbidity, health consequences, and medico-legal status. 2016 cited 2018 Nov 28. Available from: https://www.uptodate.com/contents/cannabis-use-and-disorder-epidemiology-comorbidity-health-consequences-and-medico-legal-status.
- Szutorisz H, Hurd YL. High times for cannabis: Epigenetic imprint and its legacy on brain and behavior. Neurosci Biobehav Rev. 2018 Feb;85:93–101.
- Reece AS, Hulse GK. Effect of cannabis legalization on US autism incidence and medium term projections. Clin Pediatr Open Access. 2019;4(2):17.
- Centers of Disease Control and Prevention. Autism Spectrum Disorder (ASD) Data & Statistics. 2014 cited 2018 Oct 30. Available from: https://www.cdc.gov/ncbddd/autism/data.html.
- Werling DM, Geschwind DH. Sex differences in autism spectrum disorders. Curr Opin Neurol. 2013 Apr;26(2):146–153.
- Ly L, Chan D, Trasler JM. Developmental windows of susceptibility for epigenetic inheritance through the male germline. Semin Cell Dev Biol. 2015 Jul;43:96–105.
- Murphy SK, Huang Z, Hoyo C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS One. 2012;7(7):e40924.
- Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525.
- Gkountela S, Zhang KX, Shafiq TA, et al. DNA demethylation dynamics in the human prenatal germline. Cell. 2015 Jun 4;161(6):1425–1436.
- Tang WW, Dietmann S, Irie N, et al. A unique gene regulatory network resets the human germline epigenome for development. Cell. 2015 Jun 4;161(6):1453–1467.
- Shin SM, Zhang N, Hansen J, et al. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci. 2012 Dec;15(12):1655–1666.
- Roloff AM, Anderson GR, Martemyanov KA, et al. Homer 1a gates the induction mechanism for endocannabinoid-mediated synaptic plasticity. J Neurosci. 2010 Feb 24;30(8):3072–3081.
- Howlett AC, Barth F, Bonner TI, et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002 Jun;54(2):161–202.
- Chen M, Wan Y, Ade K, et al. Sapap3 deletion anomalously activates short-term endocannabinoid-mediated synaptic plasticity. J Neurosci. 2011 Jun 29;31(26):9563–9573.
- Rey RA, Grinspon RP. Normal male sexual differentiation and aetiology of disorders of sex development. Best Pract Res Clin Endocrinol Metab. 2011 Apr;25(2):221–238.
- Bassil CF, Huang Z, Murphy SK. Bisulfite pyrosequencing. Methods Mol Biol. 2013;1049:95–107.