ABSTRACT
Post-transcriptional regulation by miRNAs plays an important role in the pathogenesis of rheumatoid arthritis (RA), however, the roles of specific miRNAs in RA pathogenesis remain largely unclear. This study performed dual-omics (miRNA and mRNA) integration analysis and in-depth cellular and molecular functional exploration to identify novel RA-associated miRNAs and to understand their underlying pathogenic mechanism. Based on the miRNA and mRNA expression profiles in peripheral blood mononuclear cells (PBMCs) from a discovery sample set (25 RA cases and 18 healthy controls), 18 differentially expressed miRNAs (DEMIRs) (|Fold-change|>2 and P < 0.05) were identified and corresponding interaction networks of DEMIRs and mRNA were constructed. After the expression validation of the DEMIRs in a validation sample set (35 RA cases and 35 healthy controls), miR-99b-5p was highlighted. The over-expression of newly discovered miR-99b-5p is able to suppress T cell apoptosis, promote cell proliferation and activation, increase expression of proinflammatory cytokines (IL-2, IL-6, TNF-α, and IFN-γ), and inhibit expression of its target genes mTOR and RASSF4. This study comprehensively identified PBMC-expressed miRNAs along with corresponding regulatory networks significant for RA and discovered miR-99b-5p as a novel post-transcriptional mediator involved in RA pathogenesis. The findings improved our understanding of RA pathogenesis and provided novel insights into the molecular mechanisms underlying RA pathogenesis.
KEYWORDS:
Introduction
Rheumatoid arthritis (RA) is a serious autoimmune disease characterized by inflammatory synovitis and progressive impairment of joints [Citation1]. Previous studies have shown multiple genetic and environmental factors to be associated with RA, however, the role of genetic and environmental factors in RA pathogenesis remains largely unclear [Citation2]. Epigenetic factors not only serve as a bridge between genotype and phenotype but also as a reflection of the specific mechanism of environmental factors in pathogenesis [Citation3]. Post-transcriptional regulation by miRNAs, as an important epigenetic mechanism, exerts essential effects on gene expression through binding to mRNA 3ʹ untranslated region (3ʹUTR), influences mRNA degradation or translation repression, and contributes to the variation in cell development process including proliferation, differentiation, apoptosis, oxidative stress, and so on. Accumulating evidence has suggested that miRNAs exert important effects on the RA pathogenesis [Citation4–Citation6].
Previous studies have identified several RA-associated miRNAs [Citation6–Citation10]. These miRNAs participate in the regulation of multiple RA-related cytokine signalling pathways [Citation11], which leads to the development of synovial tissue lesions, the dysregulation of immune cells [Citation12–Citation14], and the aggravation of joints by inducing osteoclastogenesis and the secretion of metalloproteinase [Citation15,Citation16]. However, due to relatively lower coverage of miRNA screening in previous studies [Citation9,Citation10], the miRNAs identified for RA are thus far limited. Furthermore, most previous studies on miRNA lacked in-depth investigation of target genes or regulatory networks. Therefore, comprehensive and in-depth studies are needed for better understanding the significance of miRNA in RA pathogenesis.
To identify novel RA-associated miRNAs and understand their underlying pathogenic mechanism, this study generated dual-omics datasets (miRNA and mRNA) from peripheral blood mononuclear cells (PBMCs) of a discovery sample. Subsequently, we identified and validated DEMIRs along with corresponding target genes in a validation sample and constructed RA-associated expression regulatory network in PBMCs. We evaluated the clinical significance of all DEMIRs and explored the functional effects of highlighted miRNA on RA-relevant immune cells (Jurkat T cells). Concisely, we found that PBMC-expressed miRNAs, together with their target genes, were interacted and significantly regulated with RA. The miR-99b-5p targets RASSF4 and mTOR, influences immune cell behaviour, and contributes to inflammation and rheumatoid arthritis. The workflow of the present study is schematically presented in Figure S1.
Results
Identification of differentially expressed miRNAs between RA cases and controls
Among the 2,578 miRNAs tested by Affymetrix miRNA 4.0, 18 differentially expressed miRNAs (DEMIRs) (17 down-regulated and 1 up-regulated) were identified between 25 RA cases and 18 controls included in the discovery sample set (|Fold-change|>2 and P < 0.05). (, Figure S2). Hierarchical clustering analysis showed that these DEMIRs have the ability to discriminate most of the RA cases and controls (Figure S3).
Table 1. Differentially expressed miRNAs between RA cases and controls.
Integrative analysis (miRNA &mRNA) and interaction network construction
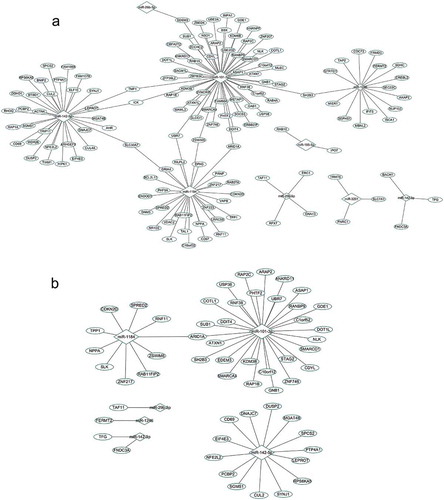
To explore the role of the identified miRNAs in the pathogenesis of RA, we integrated miRNA and mRNA data generated from the discovery sample and constructed interaction networks for significant miRNAs and mRNAs. We predicted target genes of DEMIR using three publicly available bioinformatics databases (TargetScan, miRDB and miRanda). A total of 1,243 target genes were simultaneously predicted by using the 3 databases. Among them, 921 genes were covered and quantified in the miRNA microarray. Among the 921 genes, 592 were differentially expressed between RA cases and controls (P < 0.05). Pearson correlation analysis further showed that 141 of the 592 DEGs were significantly correlated with 9 DEMIRs, which constituted 149 significant pairs (P < 0.05) for subsequent network analysis.
The 9 DEMIRs and 141 putative target genes contributed to multiple complex regulatory networks ()). Among the 149 pairs of DEMIRs and DEGs, miR-101-3p and its putative target ARID1A demonstrated the strongest correlation (r = −0.66, Table S1). The primary network was comprised of 4 miRNAs (miR-101-3p, miR-1184, miR-1246, and miR-142-5p) and their target genes (the total is 129) with each miRNA targeting at least 16 genes. Network analyses helped to find 8 common target genes (TMF1, ICK, SLC30A7, UBR7, ZSWIM6, ARID1A, SH2B3 and EDEM3) of different miRNAs. When focused on strong correlation pairs (r<-0.4), 2 miRNAs (miR-101-3p and miR-1184) and their target genes (the total is 36) formed a big network ()) with ARID1A as the common target gene. The target genes have known RA associated functions (e.g. SH2B3, COTL1 and SPRED2) [Citation17–Citation19]. Mining the literature, we found 11 genes were also involved in the immune inflammation (Table S2).
Clinical evaluation for the identified miRNAs
Receiver-operating characteristic (ROC) analyses were performed to assess the discriminative capacity of the 18 DEMIRs, respectively (Table S3). The area under the curve (AUC) ranged from 0.698 to 0.876 for the 18 DEMIRs, most of which showed great power in discriminating between RA patients and healthy subjects.
As shown in Table S3, significant correlations were observed between hsa-miR-26b-5p and C-reactive protein (CRP; r = −0.425, P = 0.034), between hsa-miR-3201, hsa-miR-8084, and erythrocyte sedimentation rate (ESR; r = −0.409 and −0.407, P = 0.043 and 0.043), between hsa-miR-3613-3p and 28-joint Disease Activity Score (DAS28; r = 0.423, P = 0.035), between hsa-miR-4448 and swollen joint count (SJC; r = 0.398, P = 0.049), and between hsa-miR-1184, hsa-miR-3613-3p, hsa-miR-4668-5p and tender joint count (TJC; r = 0.459, 0.498 and 0.496, P = 0.021, 0.011 and 0.012), respectively.
Validation of the DEMIRs in additional sample
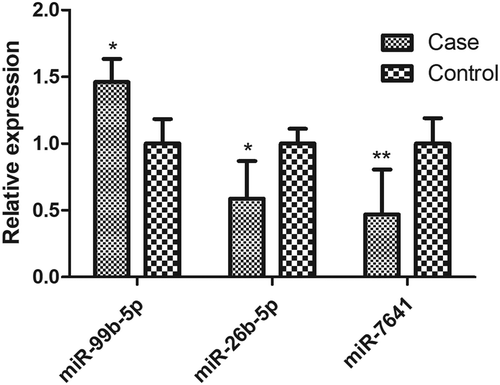
RT-qPCR assays in a validation sample (35 RA cases and 35 healthy controls) showed that 3 (miR-99b-5p, miR-26b-5p and miR-7641) of the 9 selected miRNAs presented significant differential expression between RA cases and controls (P < 0.05, ), as well as consistent expression trends with the discovery sample.
Figure 2. Differential expressions of miRNAs in the validation sample.
The sample consisted of 35 RA cases and 35 healthy subjects. The expression level of each miRNA had been normalized against RNU48. P values were calculated by the two-sided Student’s t-test. * P < 0.05, ** P < 0.01. The miRNAs with non-significant differential expression or inconsistent direction with the discovery sample were shown in Figure S6.
Functional role of miR-99b-5p in Jurkat T cells
Among the 18 DEMIRs, the miR-99b-5p was the only up-regulated miRNA in RA in the discovery sample. Additionally, the miR-99b-5p was verified to have significantly higher expression in T lymphocytes among RA patients in contrast to healthy controls (P = 0.039, Figure S4). Therefore, the Jurkat cells (T cells) were selected as target cells in investigating the in-deep functions of miR-99b-5p. We first constructed the miR-99b-5p over-expression (OE) Jurkat cell line (using lentiviral particles) and the empty vector (negative control, OE-NC) cell line. The percentage of GFP-positive cells determined by flow cytometry was 84.6% in the stable transfected Jurkat cell culture with miR-99b-5p lentivirus. The relative amount of miR-99b-5p was over 5.23-fold higher in OE cells than that in NC cells ()). We further investigated the functional effect on behaviours of Jurkat T cells after treatment with miR-99b-5p inhibitor. Compared with the treatment with inhibitor negative control, the expression of miR-99b-5p reduced to 0.3-fold after treating with miR-99b-5p inhibitor in Jurkat cells ()).
Figure 3. Functional effect of miR-99b-5p overexpression on behaviours of Jurkat T cells.
(a). Relative expression of miR-99b-5p in OE cells and OE-NC cells using RT-qPCR. The expression was normalized against U6. (b). Cell proliferation using cell counting kit-8 assay. (c). Cell apoptosis using Annexin V/PI double staining. (d). Cell cycle using PI staining. (e). Cell activation assay with (+) or without (-) phytohemagglutinin (PHA) stimulation. (f). Expression changes of inflammatory cytokines. The expression level of each mRNA was normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Two-sided Student’s t-test was used for the comparisons between groups. * P < 0.05, ** P < 0.01, *** P < 0.001. OE: over-expression cells, OE-NC: negative control cells.
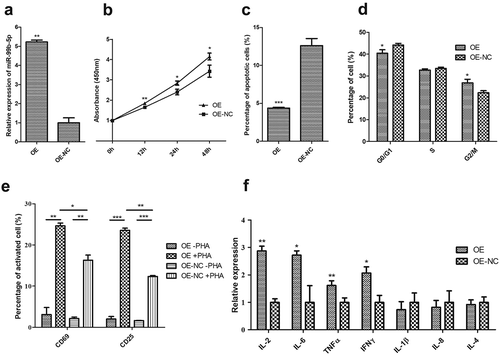
Figure 4. Functional effect on behaviours of Jurkat T cells by using miR-99b-5p inhibitor.
(a). Relative expression of miR-99b-5p in miR-99b-5p inhibitor cells and inhibitor-NC cells using RT-qPCR. The expression was normalized against U6. (b). Cell proliferation using cell counting kit-8 assay. Two-sided Student’s t-test was used for the comparisons between groups. ** P < 0.01, *** P < 0.001.
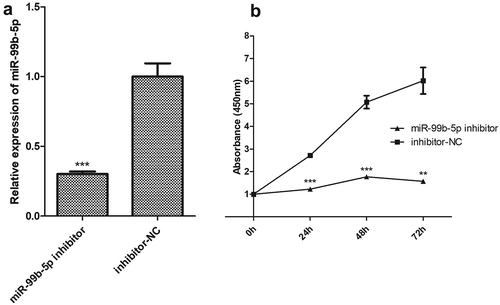
CCK-8 assays revealed that the proliferative ability of Jurkat cells was significantly increased in OE cells compared with NC cells (P < 0.05, )). Conversely, miR-99b-5p-inhibitor cells exhibited lower proliferation rate compared with the negative controls (P < 0.01) ()). Flow cytometry analysis showed the effects of miR-99b-5p on cell apoptosis and cell cycle (raw data shown in Figure S5). As shown in ), the overexpression of miR-99b-5p significantly decreased the percentage of apoptotic cells compared with the NC cells (P < 0.001). Furthermore, cell cycle analysis revealed that the overexpression of miR-99b-5p significantly increased cells at G2/M phase ()) suggesting that the miR-99b-5p stimulated cell division and proliferation. Together, these results suggested that miR-99b-5p promotes Jurkat T cell growth by stimulating proliferation and inhibiting apoptosis.
Cell surface CD69 and CD25 are early and middle T lymphocyte activation markers. Without phytohemagglutinin (PHA) stimulation, the positive rates of CD69 and CD25 Jurkat cells were low in both miR-99b-5p OE cells and NC cells (), Figure S5). With PHA stimulation, the positive rates of both antigens significantly increased in both OE and NC cells (P < 0.01). Moreover, the overexpression of miR-99b-5p caused a significant increase in the CD69 and CD25 positive rate compared with the negative controls (P < 0.05). The results indicated that the overexpression of miR-99b-5p could effectively promote Jurkat T cell activation.
The overexpression of miR-99b-5p significantly elevated the expression of proinflammatory cytokines, including IL-2, IL-6, TNF-α and IFN-γ (P < 0.05, )). However, no significant difference was suggested for IL-1β, IL-4 and IL-8.
Prediction and validation of target genes for miR-99b-5p
Based on the 3 publicly available bioinformatics databases (TargetScan, miRDB and miRanda), a total of 10 target genes of miR-99b-5p were predicted from either one of the 3 databases for subsequent analysis.
The 10 genes were all significantly differentially expressed between RA cases and controls in the discovery sample (P < 0.05) and were negatively correlated with miR-99b-5p (P < 0.05, r = −0.32~−0.62) (Table S4). Furthermore, RT-qPCR of the 10 target genes in Jurkat cells showed that, 6 genes were significantly down-regulated in the OE cells out of 7 successfully quantified genes compared to the NC cells ()).
Figure 5. Effect of miR-99b-5p on target gene expression.
(a). Relative mRNA levels of 7 target genes in miR-99b-5p OE cells and OE-NC cells using RT-qPCR. The expression was normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (b). Relative mRNA levels of 2 target genes in human PBMC in 35 RA cases and 35 controls using RT-qPCR. The expression was normalized against beta-2-microglobulin (B2M). (c). Dual luciferase assay of Jurkat T cells co-transfected with the luciferase reporter plasmid containing the wild-type or mutant-type RASSF4-3ʹUTR and miR-99b-5p overexpression lentivirus or negative vector. Two-sided Student’s t-test was used for the comparisons between groups. * P < 0.05, ** P < 0.01, *** P < 0.001. OE: over-expression cells, OE-NC: negative control cells.
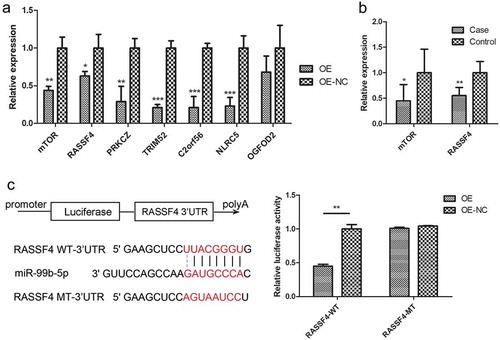
According to the correlation coefficient of target gene mRNA level with miR-99b-5p level and the fold change with RA in the microarray analyses of the discovery sample, 2 of the 7 validated target genes (mTOR with highest correlation and RASSF4 with highest fold change) were further tested the differential expression in an additional human sample (35 RA cases vs. 35 controls). The differential expression between RA cases and controls for both mTOR and RASSF4 genes were replicated in PBMC in vivo ()).
Previous study has validated the miR-99b-5p could decrease the expression of mTOR by directly targeting to its 3ʹUTR [Citation20]. The dual luciferase assay of 293T cells was performed with co-transfecting the wild-type or mutant-type 3ʹUTR of mTOR and miR-99b-5p mimics. The relative luciferase activity of the wild-type mTOR 3ʹ UTR construct was found significantly lower than controls, whereas that of the mutated site was not significantly changed. In the present study, the dual-luciferase assays using pmirGLO vector showed that compared with NC cells, the overexpression of miR-99b-5p significantly inhibited the luciferase activity of wild-type 3ʹUTR of RASSF4, which was rescued when the 3ʹUTRs of RASSF4 was mutated. The data suggested that miR-99b-5p indeed inhibits RASSF4 mRNA level through binding to the 3ʹUTR of RASSF4 ()).
To further explore the function of miR-99b-5p target gene, we conducted the Pearson correlations between the target genes and inflammatory cytokines by using the gene expression levels detected in Jurkat T cells and PBMCs (Table S5). Highly negative correlations were found between IL-2 and the 2 targets including mTOR and RASSF4 in Jurkat T cells (P < 0.05). Significant negative correlations were also found between RASSF4 and IL-6 in both Jurkat and PBMCs (P < 0.05). The above results taken together gave us clues that miR-99b-5p may induce the expression of inflammatory cytokines through regulating the targets negatively. More functional studies are called for the further underlying mechanisms.
Discussion
By performing high-throughput miRNA and mRNA expression microarray analyses for RA, this study comprehensively identified PBMC-expressed miRNAs and their regulatory networks significant for RA, and discovered miR-99b-5p as a novel post-transcriptional mediator involved in RA pathogenesis. The miRNA-mRNA integrative analysis improved our understanding of the targeted regulation mechanism of miRNA and provided novel candidate miRNA biomarkers for RA diagnosis, as supported by both in vivo and in vitro validations, high discriminative capacity, and significant correlation with the RA-associated clinical variables. Pursuant molecular biological assays dissected the miR-99b-5p-mediated regulatory effects on target gene expression, inflammatory cytokine expression, T cell growth (proliferation, apoptosis, cycle), and activation. The above dual-omics integration analysis and in-depth cellular and molecular functional exploration shed new lights on the underlying association mechanisms between key miRNA and RA, highlighted specific biological pathways, and provided novel biomarkers for developing molecular diagnosis and treatment strategies for RA.
Previous studies have reported that RA is associated with altered miRNA levels in various tissues or cells [Citation6–Citation8,Citation21]. Our study has found 18 aberrantly expressed miRNAs in PBMCs of RA cases, most of which had not been reported previously. Interestingly, three significant miRNAs (miR-29b-3p, miR-29c-3p and miR-26b-5p), identified herein in PBMCs, were non-significant in whole blood [Citation9]. The miR-1246 was down-regulated in PBMC in RA cases in the present study, but up-regulated in serum of RA cases as reported previously [Citation22]. These conflicting results imply cell- and tissue- specific expression pattern of disease-related miRNAs.
This study demonstrated the significance and functional relevance of newly discovered miR-99b-5p to RA. Located in the non-coding regions of Sperm Acrosome Associated 6 (SPACA6) on chromosomes 19, miR-99b-5p is a member of the miR-99b family that is highly conserved among 58 different species [Citation23]. Previous studies have reported that the miR-99b-5p regulates the oncogenesis process in various cancers and has the potential to serve as an important tumour biomarker for cancer diagnosis, prognosis and therapy [Citation20,Citation23–Citation25]. Yet, this study is the first to reveal that miR-99b-5p is involved in RA pathogenesis. Specifically, the miR-99b-5p is significantly up-regulated in RA cases, which could significantly promote T cell growth, activation, and expression of multiple proinflammatory cytokines (IL-2, IL-6, TNF-α and IFN-γ), and therefore play an important role in inflammatory response and RA development. Previous research about the effect of miR-99b-5p on osteoclast development and differentiation [Citation26] serves as additional evidence supporting the role of miR-99b-5p in RA pathogenesis and progression, which manifests bone and joint damage.
Supported by in vivo and in vitro evidences, we identified mTOR and RASSF4 as potential target genes of miR-99b-5p. Specifically, both gene expression levels were negatively correlated with miR-99b-5p level in Jurkat T cell culture as well as circulating PBMC and significantly down-regulated in PBMC in RA cases. The mTOR is a serine/threonine kinase that regulates cell growth, proliferation and survival via two distinctive multi-protein complexes, mTOR complex-1 (mTORC1) and mTOR complex-2 (mTORC2) [Citation27]. The frequent hyperactivity and promotion on aberrant cell survival, via the specific PI3K/AKT/mTOR signalling pathway, in various cancers makes mTOR a desirable target for anticancer drug discovery [Citation27–Citation29]. However, the role of mTOR in RA pathogenesis was unclear. Due to the complexity of immune and structural cells, including T cells, B cells, macrophages and fibroblasts [Citation30–Citation33], mTOR inhibition was not necessarily applicable for RA treatment. The newly disclosed mechanism, i.e. mTOR is regulated by miR-99b-5p, provides supportive evidences for research and development on RA treatment. RASSF4 belongs to the RAS-associated domain family, which has the degraded expression and acts as tumour suppressor in different tumour types [Citation34–Citation36]. With the CpG islands spanning across the promoter and exon of RASSF4, the gene was frequently inactivated by promoter methylation and contributed to inhibition of cell growth, interference of tumorigenesis-related signal transduction, and consequently suppression of tumour development [Citation36]. The present study examined RASSF4 expression in RA and demonstrated that miR-99b-5p inhibited the expression of RASSF4 by directly targeting its 3ʹUTR. The mRNA level of RASSF4 was found significantly decreased by miR-99b-5p over-expression in T cells in vitro and reduced in PBMC of RA populations. Taken together, the above evidences suggested that RASSF4 mRNA expression level could be regulated by DNA methylation and miRNA expression and play important roles in pathogenesis of various diseases.
Target genes that shared with the different DEMIRs in the miRNA regulatory network also attracted our attention. Among the whole correlation pairs, the target gene ARID1A was detected to have the strongest negative correlation with miR-101-3p and remained to be the only one common gene in the higher correlation pairs. The gene is implicated in transcriptional activation and repression of select genes by acting as a subunit within the SWI/SNF chromatin remodelling complex [Citation37]. Knockdown of the gene resulted in the reduction of the anti-inflammatory cytokine IL-4 [Citation38]. The ARID1A mutations have been found to cooperate with PI3K/AKT/mTOR pathway mutation on pro-tumorigenic cytokine signalling [Citation39] through which contributing to several cancer aetiology [Citation40–Citation42]. The common gene SH2B3 encoded for the lymphocyte adaptor protein LNK and participated in T cell growth, cytokine signalling, and immune responses [Citation43,Citation44]. The gene has been found to have the SNP site (rs10774624) associated with RA [Citation17]. Enrichment analysis performed in the present study also found SH2B3 was involved in the neurotrophin signalling pathway, which exerted vital roles in the aetiology of immune disorders [Citation45–Citation48].
In summary, this study identified 18 newly discovered miRNAs and their regulated gene expression networks significantly regulated in PBMC during RA pathogenesis. Furthermore, the present study highlighted that miR-99b-5p, via targeting and inhibiting mTOR and RASSF4 mRNA expression, inhibited T cell apoptosis, stimulated T cell proliferation, and promoted T cell activation and proinflammatory cytokine expression thus playing a significant role in inflammation and RA pathogenesis. The findings improved our understanding of RA pathogenesis and provided novel insights into the molecular mechanisms underlying RA pathogenesis.
Materials and methods
Study population and sample preparation
A discovery sample of 43 subjects (25 RA cases and 18 controls) was recruited for miRNA and mRNA microarray profiling in PBMCs. Another sample of 35 RA cases and 35 controls was recruited for validation purpose (Table S6). All the RA patients met the 2010 criteria of the American College of Rheumatology and the European Union League Against Rheumatism. All patients had an examination of tender and swollen joints and disease activity score of 28 joints (DAS28) recorded. Laboratory investigations included C-reactive protein (CRP) and erythrocyte sedimentation rate (ESR). Active RA was defined as having a 28-joint disease activity score (DAS28) of 2.6 or higher. For health controls, subjects with severe cardiovascular diseases, liver and kidney dysfunction, malignant tumour and other immune diseases including systemic lupus erythematosus, ankylosing spondylitis were excluded. No significant differences in age and BMI were detected between the cases and controls in either discovery or validation sample (Table S6). The study was approved by the ethical committee of Soochow University. All the study subjects signed informed consents before enrollment.
Transcriptome-wide miRNA and mRNA expression profiling
The transcriptome-wide miRNA and mRNA expression profiles were determined using Affymetrix miRNA 4.0 and lncRNA&mRNA Human Gene Expression Microarray V4.0 (CapitalBio, Beijing, China), respectively. Peripheral blood samples were collected from each subject and total RNA was then extracted from PBMCs which separated by density gradient centrifugation from blood samples. About 1.0 ug of total RNA per sample was converted in microarray to profile miRNA and mRNA expression. Fold-change and/or P-value of two-sided Student’s t-test were calculated to demonstrate the expression difference of miRNAs and mRNAs.
Network construction based on the expression levels of miRNA and mRNA
This study used 3 online prediction tools (TargetScan, miRDB, and miRanda) to predict target genes for the identified miRNAs. Pearson’s correlation analysis was conducted, and the significant and negatively correlated pairs of miRNA-mRNA were retained. The target genes were annotated through PUBMED literature searching or through DAVID (https://david.ncifcrf.gov/). Based on the correlation, the regulatory network of miRNA-mRNA was constructed and visualized using Cytoscape3.2.1 software.
Validation for the expressions of the identified miRNA and corresponding targets
The expression of the miRNAs and mRNA was validated using real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR). Total RNA was extracted from PBMCs or CD3 + T lymphocytes using TRIzol reagent. Each sample was analysed in triplicate. All the specific primers were designed and purchased from CT Bioscience (CT Bioscience, Changzhou, China). The gene expression levels were relatively quantified by using the comparative cycle threshold (Ct) method. The miRNA and mRNA expression levels were normalized against RNU48/U6 and beta-2-microglobulin (B2M), respectively.
Functional effects of miR-99b-5p on RA-relevant immune cells
To study the role of miR-99b-5p in RA, we constructed the miR-99b-5p overexpression and low-expression Jurkat cell lines. Jurkat cell is an immortalized human lymphocyte cell line, which has been commonly used as a cell model in the research field of autoimmune diseases [Citation49,Citation50]. The full-length and 1000bp up- and downstream sequences of miR-99b-5p was synthesized and cloned in to the pCDH-CMV-MCS-EF1-copGFP lentiviral expression vector. The miR-99b-5p inhibitor was synthesized by GenePharma (Shanghai, China). The resulting construct of miR-99b-5p overexpression plasma was verified by sequencing. The miRNA expression of miR-99b-5p was detected by RT-PCR according to standard protocols as previously described.
For the cell proliferation assay, the cell counting kit-8 (CCK-8) was used to detect cell proliferation. The cell apoptosis was assessed using PE Annexin V Apoptosis Detection Kit. The cell cycle was assessed using propidium iodide (PI) staining. For the cell activation assay, we determined the positive rate of T cell activation biomarkers (CD69 and CD25) by flow cytometry and expression of cytokines (IL-2, IL-6, TNF-α, IFN-γ, IL-1β, IL-8, proinflammatory cytokines; IL-4 anti-inflammatory cytokine) by RT-qPCR in Jurkat cells. The luciferase reporter plasmids (wild-type or mutant-type RASSF4 3ʹUTR) were constructed using the pmirGLO vector. 2.0µg of pmirGLO vector with wild-type or mutant-type RASSF4 3ʹUTR was co-transfected with miR-99b-5p overexpression lentivirus into Jurkat T cells using DharmaFECT Duo Transfection Reagent (Active Motif, USA). The relative luciferase activity was assessed using Dual-Luciferase Reporter Assay system. The primer information was shown in Table S7.
Statistical analyses
Two-sided Student’s t-test was used to compare the differential expression between RA cases and controls, as well as between miRNA over-expression cell lines and controls. Receiver-operating characteristic (ROC) analysis was conducted to determine the cut-off points that yielded the highest sensitivity, specificity, and accuracy in molecular diagnosis of RA. Pearson’s correlation analysis was performed to test the association between miRNA expression levels and the clinical variables, including C-reactive protein, erythrocyte sedimentation rate, 28-joint Disease Activity Score, swollen joint count, and tender joint count. P < 0.05 was considered as statistically significant.
Data statement
The microarray data for miRNA expression has been submitted to the GEO database with accession number (GSE124373).
Authors’ contributions
XZ, LW and SL designed the study. XZ, LW, WX, YG, MW, KZ, JW, YQ, XLin and XLu collected the data. XZ, LW, XM and WX were involved in data cleaning and data analysis. XZ drafted the manuscript. LW, FD and SL contributed to data interpretation, critical revision of the manuscript for important intellectual content, and approved the final version of the manuscript. All authors have read and approved the final manuscript.
Supplemental Material
Download MS Word (2 MB)Acknowledgments
We thank the help from Mr. Amanda Hall Pendegraft (The University of Alabama at Birmingham, USA) for fixing the typos and grammar errors.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- O’shea JJ, Laurence A, Mcinnes IB. Back to the future: oral targeted therapy for RA and other autoimmune diseases. Nat Rev Rheumatol. 2013;9:173–182.
- Arend WP, Firestein GS. Pre-rheumatoid arthritis: predisposition and transition to clinical synovitis. Nat Rev Rheumatol. 2012;8:573–586.
- Viatte S, Plant D, Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol. 2013;9:141–153.
- Chen JQ, Papp G, Szodoray P, et al. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15:1171–1180.
- Chen XM, Huang QC, Yang SL, et al. Role of micro RNAs in the pathogenesis of rheumatoid arthritis: novel perspectives based on review of the literature. Medicine (Baltimore). 2015;94:e1326.
- Churov AV, Oleinik EK, Knip M. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun Rev. 2015;14:1029–1037.
- Miao CG, Yang YY, He X, et al. New advances of microRNAs in the pathogenesis of rheumatoid arthritis, with a focus on the crosstalk between DNA methylation and the microRNA machinery. Cell Signal. 2013;25:1118–1125.
- Long L, Yu P, Liu Y, et al. Upregulated microRNA-155 expression in peripheral blood mononuclear cells and fibroblast-like synoviocytes in rheumatoid arthritis. Clin Dev Immunol. 2013;2013:296139.
- Anaparti V, Smolik I, Meng X, et al. Whole blood microRNA expression pattern differentiates patients with rheumatoid arthritis, their seropositive first-degree relatives, and healthy unrelated control subjects. Arthritis Res Ther. 2017;19:249.
- Wang W, Zhang Y, Zhu B, et al. Plasma microRNA expression profiles in Chinese patients with rheumatoid arthritis. Oncotarget. 2015;6:42557–42568.
- Sharma AR, Sharma G, Lee SS, et al. miRNA-regulated key components of cytokine signaling pathways and inflammation in rheumatoid arthritis. Med Res Rev. 2016;36:425–439.
- Guo Y, Tan LJ, Lei SF, et al. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806.
- Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009.
- Stanczyk J, Ospelt C, Karouzakis E, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63:373–381.
- Nakasa T, Shibuya H, Nagata Y, et al. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63:1582–1590.
- Philippe L, Alsaleh G, Suffert G, et al. TLR2 expression is regulated by microRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J Immunol. 2012;188:454–461.
- Jansen H, Willenborg C, Lieb W, et al. Rheumatoid arthritis and coronary artery disease: genetic analyses do not support a causal relation. J Rheumatol. 2017;44:4–10.
- Jin EH, Shim SC, Kim HG, et al. Polymorphisms of COTL1 gene identified by proteomic approach and their association with autoimmune disorders. Exp Mol Med. 2009;41:354–361.
- Lopez Herraez D, Martinez-Bueno M, Riba L, et al. Rheumatoid arthritis in Latin Americans enriched for Amerindian ancestry is associated with loci in chromosomes 1, 12, and 13, and the HLA class II region. Arthritis Rheum. 2013;65:1457–1467.
- Li W, Chang J, Wang S, et al. miRNA-99b-5p suppresses liver metastasis of colorectal cancer by down-regulating mTOR. Oncotarget. 2015;6:24448–24462.
- Khalifa O, Pers YM, Ferreira R, et al. X-Linked miRNAs associated with gender differences in rheumatoid arthritis. Int J Mol Sci. 2016;17:1852.
- Krissansen GW, Yang Y, Mcqueen FM, et al. Overexpression of miR-595 and miR-1246 in the sera of patients with active forms of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:520–530.
- Lukamowicz-Rajska M, Mittmann C, Prummer M, et al. MiR-99b-5p expression and response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients. Oncotarget. 2016;7:78433–78447.
- Sun D, Lee YS, Malhotra A, et al. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71:1313–1324.
- Slattery ML, Pellatt AJ, Lee FY, et al. Infrequently expressed miRNAs influence survival after diagnosis with colorectal cancer. Oncotarget. 2017;8:83845–83859.
- De La Rica L, Garcia-Gomez A, Comet NR, et al. NF-kappaB-direct activation of microRNAs with repressive effects on monocyte-specific genes is critical for osteoclast differentiation. Genome Biol. 2015;16:2.
- Shor B, Gibbons JJ, Abraham RT, et al. Targeting mTOR globally in cancer: thinking beyond rapamycin. Cell Cycle. 2009;8:3831–3837.
- Baselga J. Targeting the phosphoinositide-3 (PI3) kinase pathway in breast cancer. Oncologist. 2011;16(Suppl 1):12–19.
- Sudarsanam S, Johnson DE. Functional consequences of mTOR inhibition. Curr Opin Drug Discovery Dev. 2010;13:31–40.
- Malemud CJ. The PI3K/Akt/PTEN/mTOR pathway: a fruitful target for inducing cell death in rheumatoid arthritis? Future Med Chem. 2015;7:1137–1147.
- Yang Z, Fujii H, Mohan SV, et al. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210:2119–2134.
- Laragione T, Gulko PS. mTOR regulates the invasive properties of synovial fibroblasts in rheumatoid arthritis. Mol Med. 2010;16:352–358.
- Jones RG, Pearce EJ. MenTORing immunity: mTOR signaling in the development and function of tissue-resident immune cells. Immunity. 2017;46:730–742.
- De Smedt E, Maes K, Verhulst S, et al. Loss of RASSF4 expression in multiple myeloma promotes RAS-driven malignant progression. Cancer Res. 2018;78(5):1155–1168.
- Han Y, Dong Q, Hao J, et al. RASSF4 is downregulated in nonsmall cell lung cancer and inhibits cancer cell proliferation and invasion. Tumour Biol. 2016;37:4865–4871.
- Guo W, Dong Z, Guo Y, et al. Decreased expression and frequent promoter hypermethylation of RASSF2 and RASSF6 correlate with malignant progression and poor prognosis of gastric cardia adenocarcinoma. Mol Carcinog. 2016;55:1655–1666.
- Chandler RL, Brennan J, Schisler JC, et al. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 2013;33:265–280.
- Wurster AL, Pazin MJ. BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol Cell Biol. 2008;28:7274–7285.
- Chandler RL, Damrauer JS, Raab JR, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat Commun. 2015;6:6118.
- Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698.
- Jones S, Wang TL, Ie M S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231.
- Buglioni S, Melucci E, Sperati F, et al. The clinical significance of PD-L1 in advanced gastric cancer is dependent on ARID1A mutations and ATM expression. Oncoimmunology. 2018;7:e1457602.
- Saleh MA, Mcmaster WG, Wu J, et al. Lymphocyte adaptor protein LNK deficiency exacerbates hypertension and end-organ inflammation. J Clin Invest. 2015;125:1189–1202.
- Devalliere J, Charreau B. The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem Pharmacol. 2011;82:1391–1402.
- Bandola J, Richter C, Ryser M, et al. Neurotrophin receptor p75NTR regulates immune function of plasmacytoid dendritic cells. Front Immunol. 2017;8:981.
- Virchow JC, Julius P, Lommatzsch M, et al. Neurotrophins are increased in bronchoalveolar lavage fluid after segmental allergen provocation. Am J Respir Crit Care Med. 1998;158:2002–2005.
- Copray S, Kust B, Emmer B, et al. Deficient p75 low-affinity neurotrophin receptor expression exacerbates experimental allergic encephalomyelitis in C57/BL6 mice. J Neuroimmunol. 2004;148:41–53.
- Anand P, Terenghi G, Warner G, et al. The role of endogenous nerve growth factor in human diabetic neuropathy. Nat Med. 1996;2:703–707.
- Lai NS, Yu HC, Tung CH, et al. The role of aberrant expression of T cell miRNAs affected by TNF-alpha in the immunopathogenesis of rheumatoid arthritis. Arthritis Res Ther. 2017;19:261.
- Houtman M, Shchetynsky K, Chemin K, et al. T cells are influenced by a long non-coding RNA in the autoimmune associated PTPN2 locus. J Autoimmun. 2018;90:28–38.