 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Bullying among children is ubiquitous and associated with pervasive mental health problems. However, little is known about the biological pathways that change after exposure to bullying. Epigenome-wide changes in DNA methylation in peripheral blood were studied from pre- to post measurement of bullying exposure, in a longitudinal study of the population-based Generation R Study and Avon Longitudinal Study of Parents and Children (combined n = 1,352). Linear mixed-model results were meta-analysed to estimate how DNA methylation changed as a function of exposure to bullying. Sensitivity analyses including co-occurring child characteristics and risks were performed, as well as a Gene Ontology analysis. A candidate follow-up was employed for CpG (cytosine-phosphate-guanine) sites annotated to 5-HTT and NR3C1. One site, cg17312179, showed small changes in DNA methylation associated to bullying exposure (b = −2.67e-03, SE = 4.97e-04, p = 7.17e-08). This site is annotated to RAB14, an oncogene related to Golgi apparatus functioning, and its methylation levels decreased for exposed but increased for non-exposed. This result was consistent across sensitivity analyses. Enriched Gene Ontology pathways for differentially methylated sites included cardiac function and neurodevelopmental processes. Top CpG sites tended to have overall low levels of DNA methylation, decreasing in exposed, increasing in non-exposed individuals. There were no gene-wide corrected findings for 5-HTT and NR3C1. This is the first study to identify changes in DNA methylation associated with bullying exposure at the epigenome-wide significance level. Consistent with other population-based studies, we do not find evidence for strong associations between bullying exposure and DNA methylation.
Introduction
The social environment is a major contributor to mental health. Bullying is a ubiquitous social stressor, with worldwide estimates ranging from one in ten to almost half of all children that are exposed[Citation1]. Following Olweus’ definition, a person is being bullied ‘when he or she is exposed, repeatedly and over time, to negative actions on the part of one or more other persons’. Such negative actions should be intentional and performed by someone perceived be more powerful than the subject. Actions can include physical behaviours, such as hitting and kicking, verbal behaviours, such as calling names, as well as indirect or relational behaviours, such as social exclusion [Citation2]. Bullying exposure (i.e. bullying victimization) has been associated with numerous mental health issues including behavioural problems, depressive symptoms [Citation3–Citation6] and suicidal ideation [Citation7]. However, whereas a myriad of harmful and persistent psychiatric consequences of being bullied have been identified, the biological pathways that change after exposure to bullying remain largely uncharted. Identifying these pathways is a pivotal step in understanding how peer-inflicted stress affects the human body.
Research on other environmental stressors, such as parental abuse [Citation8], prenatal maternal stress [Citation9,Citation10] or childhood trauma in general [Citation11–Citation15] has incorporated epigenetic data to investigate the hypothesis that stressors affect the molecular configuration on and around the DNA, thereby influencing its functionality, with potential downstream effects on stress reactivity and mental health [Citation16–Citation19]. One often studied epigenetic mechanism is DNA methylation, in which a methyl-group binds to a cytosine nucleotide of the DNA (cytosine-phosphate-guanine site or CpG site). Whereas early epigenetic studies focused on DNA methylation of a single candidate gene, there has been an increase in hypothesis-free epigenome-wide methylation studies (EWASs) investigating DNA methylation levels of hundreds of thousands of CpG sites (CpGs) across the genome. One study [Citation11], for example, found multiple epigenome-wide significant differentially methylated CpGs related to different types of childhood maltreatment.
In contrast to other forms of adversity, research on bullying exposure and epigenetics is markedly scarce. To the best of our knowledge, only three such studies have been performed. In 28 monozygotic twin pairs discordant for bullying exposure [Citation20], increased levels of methylation were observed in the serotonin transporter gene (5-HTT) promoter region for the exposed twin siblings from 5 to 10 years, but not for the non-exposed twin siblings. Another study in 1,149 13 to 14 year old children found bullying exposure to be associated with increased methylation levels of exon 1F of the glucocorticoid receptor gene (NR3C1) [Citation21]. Further, an EWAS was performed on bullying exposure in 1,658 twins [Citation13], thereby expanding the search for differentially methylated sites beyond the ‘usual suspects’, i.e. candidate genes that have been firmly implicated in neurotransmitter and hormonal functions, to enable the identification of potentially novel biological pathways. Bullying exposure during childhood as reported by mother and child at age 7–12 years, and bullying exposure during adolescence, retrospectively reported by the child at 18 years, were however not related to differential methylation. Given that DNA methylation is expected to change over time [Citation22] due to both extrinsic as well as intrinsic factors, a model in which DNA methylation both before and after bullying exposure is taken into account should be more sensitive to the effects of exposure.
In the current study, we made use of two population-based cohorts featuring repeated measures of DNA methylation to characterize longitudinal epigenome-wide associations with bullying exposure. Longitudinal mixed models were performed separately in the two cohorts to identify associations between exposure to bullying and changes in DNA methylation from pre- to post bullying report. Results were then meta-analysed to maximise statistical power and to evaluate coherence among the estimates derived from the two populations. Epigenome-wide associations with bullying were studied in a hypothesis-free analysis. In a secondary candidate gene follow-up analysis we examined DNA methylation at 5-HTT and NR3C1 for gene-wide associations with bullying.
Results
Sample characteristics
Sample characteristics are described in . In Generation R, bullying exposure was reported by the mother at the mean (SD) age of 8.1 (0.1) years, and DNA methylation was measured at 6.0 (0.3) years and 9.8 (0.3) years of age. In the Avon Longitudinal Study of Parents and Children (ALSPAC), bullying exposure was reported by the child at the mean (SD) age of 8.6 (0.2) years, and DNA methylation was measured at 7.5 (0.1) and 17.1 (1.0) years of age (Supplemental Figure 1). In the main analysis 45.5% of children in the Generation R sample and 39.4% of children in the ALSPAC sample were categorized as exposed to bullying victimization. In the sensitivity analysis with a more stringent definition of bullying, these numbers are 9.9% and 12.1%, respectively. The current selected samples for each cohort were compared with (i) a set of participants with complete data on covariates, and (ii) a set of participants with complete data on both covariates and bullying exposure (full details in Supplemental Table 1). This showed that children in the current sets had a higher gestational age and higher SES, had mothers who were older, had a higher non-verbal intelligence quotient (IQ) score and were older at bullying exposure report. In Generation R, but not ALSPAC, children in the selected sample also had a lower Body Mass Index (BMI), less behavioural problems prior to exposure, and had less reported stressful experiences other than bullying exposure. No differences were found for child sex, or bullying exposure.
Table 1. Sample characteristics.
Comparison of longitudinal epigenome-wide association studies
The separate longitudinal EWASs for the two cohorts (Q-Q plots in Supplemental Figure 3), identified no epigenome-wide Bonferroni-significant associations, with a lowest obtained p-value of p = 5.93x10−06 (CpG site cg034529555, annotated to NAV2) for Generation R and of p = 1.08x10−06 (CpG site cg24506221, annotated to GSTM1) in ALSPAC. Estimates for bullying exposure among the top 1000 CpG sites in each cohort more often had a negative direction (79.8% in Generation R, 66.3% in ALSAPC) than would be expected by chance (Χ2(1) = 355.22, p < 2.20x10−16 for Generation R, Χ2(1) = 106.28, p < 2.20x10−16 for ALSPAC). Bullying exposure estimates among the top 1000 in Generation R were not significantly correlated with those in ALSPAC (r(998) = −0.05, p = 1.09x10−01). The correlation between the top 1000 in ALSPAC was slight but significantly negative (r(998) = −0.07, p = 1.09x10−02) with those in Generation R.
Meta-analysis
In the meta-analysis, one CpG site was significantly associated with bullying exposure: cg17312179 (b = −2.67x10−03, SE = 4.97x10−04, p = 7.17x10−08; Supplemental Figure 4), a site in the leader sequence (5'UTR) of the RAB14 gene, located on chromosome 9. In Generation R (b = −2.47x10−03, SE = 6.17x10−04, p = 6.25x10−05), DNA methylation of this CpG site increased on average 0.13% from the mean age of 6.0 to 9.8 years in non-exposed, but decreased −0.12% in exposed. In ALSPAC (b = −3.05x10−03, SE = 8.36x10−04, p = 2.64x10−04) methylation of this CpG site increased 0.09% from the mean age of 7.5 to 17.1 years in non-exposed, whereas it decreased −0.21% in exposed (). DNA methylation differences at cg17312179 between the non-exposed and the exposed group were not present before exposure measurement, but were so after (Supplemental Analysis). The ten CpGs with the lowest p-values are shown in (see Supplemental Table 2 for associated functions).
Table 2. Ten CpG sites with lowest p-values in meta-analysis of epigenome-wide associations with bullying exposure.
Figure 1. Change in DNA methylation pre- and post- bullying exposure measurement for exposed and non-exposed in Generation R and ALSPAC. Data are residualized for covariates present in linear mixed model.
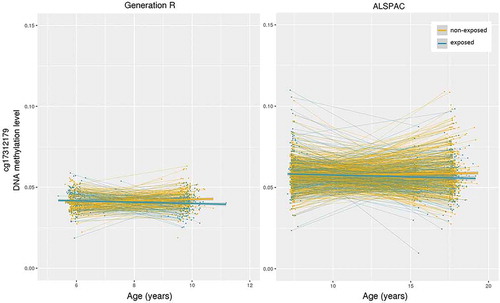
The top 1000 CpGs from the meta-analysis had a higher representation of CpGs with a negative estimate for bullying exposure (83.5%) than all other CpGs (58.2%, Χ2(1) = 261.07, p < 2.20x10−16). Moreover, the top 1000 CpGs had a higher representation of CpGs with a positive age estimate (representing change in non-exposed, 79.2%) than the other CpGs (65.1%, Χ2(1) = 87.29, p < 2.20x10−16), and a higher representation of CpGs with low levels of DNA methylation (mean β value<0.2 in both Generation R and ALSPAC, threshold as elsewhere [Citation23]) at both time points (55.7% versus 37.0%, Χ2(1) = 149.63, p < 2.20x10−16), and more often located in promoters (28.8% versus 20.1%, Χ2(1) = 46.66, p = 8.45x10−12) and CpG islands (41.8% versus 30.8%, Χ2(1) = 56.63, p = 5.25x10−14). Together, this indicates that top sites tended to be located in promoters and CpG islands, and to have overall low levels of DNA methylation, decreasing in exposed, while increasing in non-exposed. See for these characteristics at multiple p-thresholds.
Table 3. Characteristics of CpG sites selected for various levels of significance in meta-analysis.
Follow-up analyses
Sensitivity analyses
A series of sensitivity analyses were performed on cg17312179 in each cohort and then meta-analysed. First, an analysis was performed with a more stringent definition of bulling exposure. Second, we reran separate analyses or additionally adjusting for (i) BMI; (ii) pre-existing behaviour problems; (iii) non-verbal intelligence quotient (IQ); (iv) stressful experiences other than bullying exposure; and (v) alcohol use. The bullying exposure coefficients from sensitivity analyses were not different from the bullying exposure coefficient from the main analysis (lowest p = 7.42x10−02). ‘Other stressful experiences’ was the only added variable that independently associated with cg17312179 (b = 2.20x10−04, SE = 9.96x10−05, p = 2.73x10−02).
Genetic associations
A triad of look-ups did not show evidence of genetic associations with cg17312179 methylation. First, the probe was not present in a list of polymorphic probes [Citation24]. Second, no cis or trans meQTLs were found to associate with this probe [Citation25], and third, low additive genetic influences (1.79x10−10%) and high shared (34.9%) and non-shared (65.1%) environmental influences have been reported for this probe based on twin heritability analyses [Citation26].
Look-up of previous findings in the literature
Results from eleven EWASs on childhood adversity [Citation11,Citation13,Citation15,Citation16,Citation27–Citation32] were searched for cg17312179 or other CpGs annotated to RAB14. No RAB14-associated probes were reported. Since cg17312179 was not reported in these studies, we could not establish if the direction of association was congruent with the one currently reported.
Candidate gene-wide analyses
Results from the meta-analysis were separately studied for probes annotated to 5-HTT and NR3C1 (Supplemental Figure 5). None of the probes reached gene-wide Bonferroni-significance (thresholds p = 3.13x10−03 for 5-HTT and p = 1.22x10−03 for NR3C1).
Functional associations
Gene Ontology (GO) analysis on CpGs with p < 0.001 (n = 644 CpGs, n = 396 genes) yielded 126 pathways, 25 of which were confirmed by a GO analysis on CpGs with p < 0.01 (n = 5997 CpGs, n = 3722 genes) and 43 of which were confirmed by a GO analysis on CpGs with p < 0.0001 (n = 66 CpGs, n = 53 genes). Ryanodine-sensitive calcium-release channel activity as the most enriched (p = 9.99x10−08) biological process (Supplemental Tables 4–6, Supplemental Figure 6). Three isoforms of the ryanodine receptors exist [Citation33], RYR1, RYR2, and RYR3, each present in a different tissue. Here, RYR2 was part of the GO pathway, a gene specifically active in the heart tissue. Other enriched terms for biological processes involve various neurodevelopmental processes, such as astrocyte differentiation and action potential regulation, as well as processes such as muscle fibre development.
Discussion
The current study is the first to characterize epigenome-wide intra-individual changes in DNA methylation related to bullying exposure. Our meta-analysis identified a CpG site with increasing levels of DNA methylation in non-exposed but decreasing levels in the exposed group. Other research [Citation24–Citation26] on this probe suggests that variance in DNA methylation at this CpG is primarily explained by environmental influences, with weak evidence of genetic effects. Sensitivity analyses showed that this association was not explained by co-occurring child characteristics, co-occurring risks, or consequences of bullying, including pre-existing behavioural problems, IQ, BMI, alcohol use or exposure to stressful experiences other than bullying. The site is located in the 5' untranslated region of RAB14, a member of the Ras oncogene family of GTPases. Ras GTPases are important in cellular signalling and RAB14 is involved in vesicle transport and Golgi apparatus functioning [Citation34], and is expressed in multiple tissues (Supplemental Figure 7). No RAB14-associated probes have been reported in previous EWAS on childhood adversity.
RAB14 expression has however been associated to stress in different tissues. In rat hippocampus it was shown to be downregulated after prenatal stress [Citation35] and upregulated after mild chronic stress in stress-resilient rats [Citation36], possibly marking an adaptive response. In humans, its expression was found to be reduced in the prostate of men with prostate cancer after nutrition and lifestyle intervention focused on stress reduction [Citation37]. Also, RAB14 expression in human brain tissue has been linked to depression and suicide [Citation38], as was Syntaphilin, a protein that regulates synaptic vesicle processing [Citation39] and encoded by SYNPH, a gene associated to one of the top 10 CpG sites from the meta-analysis. How the observed changes in RAB14 methylation might relate to expression levels in this gene and what the downstream effects of these changes might be, however, remains to be elucidated in future functional studies.
GO analysis showed enrichment of the biological process of ryanodine-sensitive calcium release channel activity; these channels are a pathway important in cardiac functioning and the fight-or-flight response [Citation40]. This finding is congruent with several other enriched pathways associated with cardiac functioning, and fits with GO findings from other research on epigenetics and physical abuse [Citation11]. Further, GO analysis showed many neurodevelopmental processes, such as neuron differentiation, a biological pathway in which two of the associated genes, SYNPH and ST8SIA4, were among the top meta-analysis hits. ST8SIA4 (CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase) is present in the Golgi apparatus, involved in neural plasticity [Citation41], and ST8SIA4 knockout mice have been shown to display a decreased motivation for social interaction [Citation42]. Functioning of both genes has been associated with brain disorders, such as schizophrenia [Citation43,Citation44] and Alzheimer’s disease [Citation45,Citation46]. Interestingly, in contrast to previous studies [Citation20,Citation21], no associations with bullying exposure were found at candidate genes 5-HTT and NR3C1. Failure to replicate candidate epigenetic studies with epigenome-wide analyses is not uncommon [Citation13]. This discrepancy may be explained by the stricter multiple testing correction applied in (candidate gene analyses as part of) epigenome-wide studies, or in the different specific regions tested by targeted gene approaches and microarray studies, rendering direct comparison unfeasible.
A pattern emerged of enrichment for CpGs with low overall methylation levels, increased over time, but decreased for exposed individuals. Furthermore, the top CpGs from the meta-analysis were more often located in CpG islands and promoter regions than would be expected by chance. Together this might indicate that bullying exposure is associated with an overall delayed downregulation of gene expression. However, promoter regions typically have low levels of methylation, and the enrichment of CpGs with low overall methylation levels in general seems to be more pronounced than the enrichment of promoter CpGs. In an EWAS on childhood abuse and promoter DNA methylation in adulthood [Citation8], the stressor was also more often negatively than positively associated with DNA methylation. In another EWAS on childhood maltreatment and DNA methylation around 10 years of age (range 5–14 years) [Citation23], researchers found an enrichment of CpGs with low methylation levels, often located in promoter regions as we did in the current study, but the association with maltreatment was more often positive. Unfortunately, direct comparison among studies is not straightforward because of timing differences in the measurement of DNA methylation, as well as the inherently unclear timing of often retrospectively reported stressors [Citation32]. In the current study, effect estimates for the top ranking CpGs were incongruent and even seemed slightly oppositional between the two cohorts. One explanation might be the longer time period between bullying exposure and the DNA methylation measurement in ALSPAC. One study on timing differences in ALSPAC for example, found that recency of adversity exposure was more important in explaining DNA methylation levels than accumulation of adversity, regardless of timing [Citation32]. On the other hand, even among top ranking CpGs associations were weak. Such effect sizes are in line however, with other epigenome-wide studies in population-based samples [Citation13,Citation15,Citation47–Citation49], where exposures are generally less extreme and abundant than in risk samples that typically encounter larger effect sizes [Citation11,Citation23]. In any case, thorough knowledge of normative development of DNA methylation levels is currently lacking and needed to interpret dissimilar estimates in the face of different measurement periods. Regarding the interpretation of the top hit, we further highlight that while stringent significance thresholds were used to reduce the risk of false positives, our current results may still reflect a chance finding and will need to be replicated in future studies.
To facilitate harmonization of the bullying exposure measurements in both cohorts, bullying exposure was defined with a lenient threshold. This implies that the difference in DNA methylation found for the CpG in RAB14 is associated with exposure to bullying that is prevalent for children in the normal population. A more stringent definition of bullying might have brought forward different results, but a larger sample would be preferential for such an analysis. Additionally, with the current design we were unable to control for bullying exposure that participants might have been subjected to outside of the moment of measurement. More measurements of bullying exposure would likely lead to more precise estimates. Further, more questions on the different types of bullying in the Generation R Study would have permitted us to differentiate between specific bullying exposures. Multiple reporters of bullying would have been preferable as well, especially the current use of mother report in one cohort and child report in the other is suboptimal. For the RAB14 CpG site, there was converging agreement however. Another constraint of the study was that the current selected samples were more affluent than the fuller populations of their respective cohorts, where ideally the full spectrum of characteristics for the children in our cohorts would be represented. Last, we do not know if changes in DNA methylation are the consequence of bullying exposure, or that such changes are associated with children who are more at risk of being bullied [Citation50]. An experimental set-up, for example with an anti-bullying intervention [Citation51], would shed more light on this.
In conclusion, the current study is the first to report an epigenome-wide hit related to bullying exposure. This CpG site is located in the RAB14 gene and suggests that exposure bullying might be associated with Golgi apparatus functioning. The effect size was small, but in line with other population-based studies. Further, we found an enrichment for CpGs related to cardiac functioning and neurodevelopment, as well as for CpGs with low levels of methylation and sites for which DNA methylation decreased in exposed but increased in non-exposed. We believe that experimental and longitudinal research into DNA methylation is the path to a broader understanding of social stress and its effect on biological pathways.
Methods
Setting
Data were drawn from two population-based prospective birth cohorts, the Dutch Generation R Study (Generation R) and the British Avon Longitudinal Study of Parents and Children (ALSPAC). Pregnant women residing in the municipality of Rotterdam, the Netherlands, with an expected delivery date between April 2002 and January 2006 were invited to enrol in the Generation R Study. A more extensive description of the study can be found elsewhere [Citation52]. The Generation R Study is conducted in accordance with the World Medical Association Declaration of Helsinki and has been approved by the Medical Ethics Committee of the Erasmus Medical Centre, Rotterdam. Written informed consent was obtained for all participants.
Pregnant women residing in the study area of former county Avon, United Kingdom, with an expected delivery date between April 1991 and December 1992 were invited to enrol in the ALSPAC study. Detailed information on the study design has been published previously [Citation53,Citation54]. The ALSPAC website contains details of all available data through a fully searchable data dictionary and variable search tool (http://www.bristol.ac.uk/alspac/researchers/our-data/). Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. Consent for biological samples has been collected in accordance with the Human Tissue Act (2004). Informed consent for the use of data collected via questionnaires and clinics was obtained from participants following the recommendations of the ALSPAC Ethics and Law Committee at the time.
In both cohorts DNA methylation was studied before and after reported bullying exposure. A timeline can be found in Supplemental Figure 1.
Study population
In the Generation R Study, 9,778 pregnant mothers gave birth to 9,749 live-born children. For a subsample of 608 singletons DNA methylation data was collected at 6 and/or 10 years old (343 at both time points). Of these, 506 children had information available on bullying exposure and relevant covariates, including 289 children with DNA methylation available for both time points (Supplemental Figure 2(a)). This subsample consisted of participants with parents born in the Netherlands, with European ancestry confirmed based on genetic principle component analysis for all children with genetic data available (99.6% of the current sample).
In ALSPAC, the inclusion of 14,541 pregnant mothers resulted in 14,062 live births. DNA methylation was available at 7 and/or 17 years old for a subsample of 936 European singletons (877 at both time points) as part of the Accessible Resource for Integrated Epigenomic Studies (ARIES) study [Citation55]. For 846 of these children data on bullying exposure and relevant covariates was available, including 793 children with DNA methylation data at both time points (Supplemental Figure 2(b)), leading to a combined sample size of 1,352 children in the meta-analysis. In each cohort, bullying exposure and covariates were compared between the selected sample and (i) a set of participants with complete data on covariates, irrespective of availability of data on bullying exposure or DNA methylation (n = 8,528 in Generation R and n = 12,393 in ALSPAC), and (ii) a set of participants with complete data on both covariates and bullying exposure, irrespective of availability of DNA methylation data (n = 4,336 in Generation R and n = 6,347 in ALSPAC).
Bullying exposure
In Generation R, mothers filled out a questionnaire (adapted [Citation56]) containing three questions on bullying exposure in the past few months, covering physical (‘In the past few months, how often has your child been bullied by way of spitting, hitting, kicking, or pinching?’), verbal (‘In the past few months, how often has your child been bullied by insulting, calling names or laughed at?’), and relational bullying (‘In the past few months, how often has your child been bullied by being excluded from activities, ignored by other children, or gossiped about?’). Items were rated on a 5-point scale (ranging from never to several times a week). In ALSPAC, bullying exposure was measured through self-report with an adapted version of the Bullying and Friendship Interview Schedule (BFIS) [Citation57]. Nine questions covered physical (being hit or beaten up/belongings taken), verbal (threatened or blackmailed/tricked/called nasty names), and relational forms of bullying (others would not play with them/being made to do things they did not want to do/others told lies or nasty things about them/had games spoilt) in the preceding six months on a 4-point scale (ranging from never to at least once a week). Internal reliability of both measures was acceptable (Generation R: α = 0.74, ALSPAC: α = 0.73). Scores were dichotomized to harmonize the two bullying scales and avoid issues arising from extreme skewness of the data. Children were classified as being exposed to bullying if they were bullied ‘at least once or twice in the past few months’ on at least one of the items in Generation R, and at least ‘1–3 times in the past six months’ in ALSPAC [Citation58–Citation61].
Variables sensitivity analyses
A more stringent bullying exposure variable was defined, in which children were classified as exposed when at least indicated to be bullied ‘2 or 3 times a month’ in Generation R, and they at least indicated to be bullied ‘about once a week’ in ALSPAC.
Body Mass Index (BMI) (kg/m2) was measured at 6 and 10 years in the Generation R and 7 and 17 years in the ALSPAC. Values were standardized to SD scores, adjusted for age and sex.
Child behavioural problems were measured at age 3 years with the mother-reported Child Behaviour Checklist for toddlers (CBCL1½-5) [Citation62] in Generation R. Ninety-nine items were scored one a 3-point scale (range 0–2), regarding symptoms of anxiety, sadness, withdrawn behaviour, attention problems, and aggressive behaviours (α = 0.92). Items were summed into a weighed total problem behaviour scale, with 25% missing allowed. In ALSPAC, the mother-reported Strengths and Difficulties Questionnaire (SDQ) [Citation63] at 4 years was used. The scales for emotional, conduct, and hyperactivity problems were used as a total problem behaviour score, consisting of 15 items, each rated on a 3-point scale (range 0–2) (α = 0.74). The remaining problem scale of the SDQ, the ‘peer problems’ scale, was excluded from the total score due to content overlap with bullying exposure.
Child non-verbal intelligence quotient (IQ) was measured by testing visuospatial abilities (Mosaics) and abstract reasoning (Categories) with the Snijders-Oomen Niet-verbale Intelligentie Test-Revisie (SON-R 2½-7) [Citation64] at age 6 years in Generation R. In ALSPAC, a shortened version of the Wechsler Intelligence Scale for Children (3rd UK edition (WISC-III)) [Citation65] was measured at age 9 years.
Other stressful experiences were measured in Generation R with a major life events inventory [Citation66], reported by the mother, when the child was 10 years. This inventory covers stressful life events spanning the lifetime of the child, such as physical abuse, sexual abuse, conflict in the household, illness or death in the family, and parental separation. Three items related to bullying exposure were excluded, leaving 21 items (range 0–1). In ALSPAC, we used an Adverse Child Experiences (ACE) lifetime composite score [Citation15,Citation67]. This score is based on 541 questions mapping on to 10 ACEs up to age 16 years. Participants were included if there was at least 50% of the data available for each ACE. We excluded the ACE for bullying, leaving physical, sexual, and emotional abuse, emotional neglect, substance use in the household, violence between parents, parental mental health, parent conflict, parent offence, and parental separation (each range 0–1).
Last, alcohol use was measured in ALSPAC with Alcohol Use Disorders Identification Test (AUDIT) at age 17. This tests consists of 10 items (range 0–4); a total score of 8 or more is considered hazardous [Citation68].
DNA methylation
Both cohorts used the EZ-96 DNA Methylation kit (Shallow) (Zymo Research Corporation, Irvine, USA) for bisulphite conversion on the extracted DNA. DNA methylation profiles were generated using the Illumina Infinium HumanMethylation450 BeadChip (Illumina Inc., San Diego, USA). Quality control and normalization steps can be found in Supplemental Methods. Analyses were restricted to 473,864 autosomal CpGs. DNA methylation levels are characterized by beta values (β values), representing the ratio of methylated signal relative to the sum of methylated and unmethylated signal measured per CpG. Outlying data points outside the 3*interquartile range were winsorized to the nearest point for each CpG. White blood cell (WBC) composition was estimated using the reference-based Houseman method [Citation69]. Batch effects and additional unknown confounding were estimated using surrogate variable analysis (SVA) in meffil [Citation70,Citation71] in R version 3.4.3 [Citation72].
Statistical analyses
Associations between bullying exposure and changes in DNA methylation were analysed with a linear mixed model:
Here, M denotes DNA methylation level, β0 fixed intercept, u0i random intercept, β1 fixed age coefficient, β2 fixed bullying exposure coefficient, and ϵ random error. The Bullied variable was set to 0 for the first DNA methylation measurement and to 1 or 0 for the second measurement depending on whether the participant had been exposed to bullying, or not, respectively, and random intercept u0i allowed for inter-individual variation in DNA methylation at the first measurement. Participants are denoted by i and time points by j. Covariates included sex, gestational age, socio-economic status as indicated by highest attained educational level of the mother (low versus medium or high), surrogate variables (n = 20), WBCs (CD4 + T-lymphocytes, CD8 + T-lymphocytes, natural killer cells, B-lymphocytes, monocytes, and granulocytes). Current direct and second-hand smoking was adjusted for with the methylation level of AHRR cg05575921, which has proven to be a valid marker of tobacco exposure [Citation73–Citation76]. Methylation level for this CpG at both time points was entered into the equation, with levels divided into quintiles (as described elsewhere [Citation77]) and lower levels indicating more smoking. Linear mixed models were applied using the lme4 package [Citation78].
To compare congruency between results from the two cohorts, estimates of the top 1000 CpGs in each cohort were correlated with estimates of those CpGs in the other cohort (as elsewhere [Citation15]). Meta-analysis of estimates and standard errors of the two cohorts was performed using fixed models within the metafor [Citation79] R package. To account for multiple testing (n = 473,864 CpGs), the significance threshold was set at a Bonferroni-corrected p-value of 1.06 × 10−07.
Follow-up analyses
Sensitivity analyses
A series of sensitivity analyses were performed on CpGs with p < 1.06x10−07 in the meta-analysis. First, because the classification of bullying exposure is rather broad in the main analysis, we performed a sensitivity analysis with a more stringent dichotomization. Second, we reran analyses additionally adjusting for the following potential confounders or mediators that have been previously shown to associate with bullying exposure and DNA methylation. In each sensitivity analysis, one of the following variables was added to the main analysis: (i) Body Mass Index (BMI) (kg/m2) SD scores, adjusted for age and sex, at 6 and 10 years in Generation R and 7 and 17 years in ALSPAC (full sample size available) [Citation60,Citation80]; (ii) pre-existing behavioural problems (Generation R n = 451, ALSPAC n = 794) [Citation5,Citation50]; (iii) child non-verbal IQ (Generation R n = 465, ALSPAC n = 811) [Citation81]; (iv) stressful experiences other than bullying exposure (Generation R n = 482, ALSPAC n = 597) [Citation82,Citation83]; and (v) alcohol use in ALSPAC (n = 624), where children are older [Citation84,Citation85]. For each sensitivity analysis, the coefficient for bullying exposure was compared with that for the main analysis with a z-test [Citation86,Citation87].
Genetic associations
DNA methylation for CpGs with p < 1.06x10−07 in the meta-analysis were tested for genetic associations in three ways. First, a look-up was performed in a list of CpGs located on a single nucleotide polymorphism (SNP), e.g. polymorphic CpGs [Citation24]. Second, we tested for known associations with genetic variants, e.g. methylation quantitative trait loci, in cis (cis meQTLs) and in trans (trans meQTLs; http://www.mqtldb.org/; GCTA set) [Citation25]. Third, we tested for additive genetic influences versus shared and unique environmental influences on the DNA methylation, as based on twin heritability analyses [Citation26].
Look-up of previous findings in the literature
Previous studies on childhood adversity and DNA methylation, measured with the Illumina Infinium HumanMethylation450 BeadChip, were searched for current CpGs with p < 1.06x10−07 and their associated genes in the meta-analysis. Eleven studies were selected [Citation11,Citation13,Citation15,Citation16,Citation27–Citation32]. All studies examined childhood abuse or trauma, on study additionally examined bullying [Citation13].
Candidate gene-wide analyses
A candidate gene follow-up analysis was conducted on the results stemming from the meta-analysis, for sites annotated to 5-HTT and NR3C1. The significance threshold was set at a Bonferroni gene-level corrected p-value of 3.13 × 10−03 (n = 16 CpGs) and 1.22 × 10−03 (n = 41 CpGs), respectively.
Functional associations
Enrichment of Gene Ontology (GO) pathways was tested for genes associated with CpGs with p < 0.001 in the meta-analysis (cut-off described elsewhere [Citation11]), while adjusting for gene size and pruning for redundant terms (a full method description can be found elsewhere [Citation11]). Terms with p < 0.05 and more than one associated genes are reported, and highlighted if confirmed by near-identical terms from GO analyses with CpGs p < 0.01 and p < 0.001 in the meta-analysis.
Supplemental Material
Download MS Word (1.5 MB)Supplemental Material
Download MS Excel (25.7 KB)Acknowledgments
RHM is supported by the Van der Gaag Grant of the Royal Netherlands Academy of Arts and Sciences and The Netherlands Organization for Scientific Research (NWA Startimpuls 400.17.602). AN is supported by a Canadian Institutes of Health Research team grant and AN and HT are supported by a grant of the Dutch Ministry of Education, Culture, and Science and the Netherlands Organization for Scientific Research (NWO grant No. 024.001.003, Consortium on Individual Development). LCH is supported by ESRC/BBSRC Grant ES/N000382/1 for the Interpreting epigenetic signatures in studies of early life adversity project (Interstela Project). JFF has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 633595 (DynaHEALTH). This project received funding from the European Union’s Horizon 2020 research and innovation programme (733206, LIFECYCLE). MJBM is supported by the European Research Council (ERC AdG) and by the Dutch Ministry of Education, Culture, and Science and the Netherlands Organization for Scientific Research (NWO grant No. 024.001.003, Consortium on Individual Development). The work of CAMC has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 707404, and grant agreement No 848158 (EarlyCause).
The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University Rotterdam, the Municipal Health Service Rotterdam area, Rotterdam, the Rotterdam Homecare Foundation, Rotterdam and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives and pharmacies in Rotterdam. The study protocol was approved by the Medical Ethical Committee of the Erasmus Medical Centre, Rotterdam. Written informed consent was obtained for all participants. The generation and management of the Illumina 450K methylation array data (EWAS data) for the Generation R Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. We thank Mr Michael Verbiest, Ms Mila Jhamai, Ms Sarah Higgins, Mr Marijn Verkerk and Dr. Lisette Stolk for their help in creating the EWAS database. We thank Dr A.Teumer for his work on the quality control and normalization scripts. The general design of the Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam, the Netherlands Organization for Health Research and Development and the Ministry of Health, Welfare and Sport. The EWAS data was funded by a grant from the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA; project nr. 050-060-810), by funds from the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by a grant from the National Institute of Child and Human Development (R01HD068437).
We are extremely grateful to all the families who took part the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and Wellcome (grant number 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). This publication is the work of the authors and Dr Suderman will serve as guarantor for the ALSPAC-related contents of this paper. Analysis of the ALSPAC data was funded by UK Economic & Social Research Council grant (grant number ES/N000498/1). ARIES was funded by the BBSRC (BBI025751/1 and BB/I025263/1). Supplementary funding to generate DNA methylation data which are (or will be) included in ARIES has been obtained from the MRC, ESRC, NIH and other sources. ARIES is maintained under the auspices of the MRC Integrative Epidemiology Unit at the University of Bristol (grant numbers MC_UU_12013/2, MC_UU_12013/8 and MC_UU_12013/9).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Craig W, Harel-Fisch Y, Fogel-Grinvald H, et al. A cross-national profile of bullying and victimization among adolescents in 40 countries. Int J Public Health. 2009;54:216–224.
- Olweus D. Bullying at school. Aggressive behavior. Boston, MA: Springer; 1994. p. 97–130.
- Ringoot AP, Jansen PW, Rijlaarsdam J, et al. Self-reported problem behavior in young children with and without a DSM-disorder in the general population. Eur Psychiatry. 2017;40:110–115.
- Schoeler T, Duncan L, Cecil CM, et al. Quasi-experimental evidence on short-and long-term consequences of bullying victimization: A meta-analysis. Psychol Bull. 2018;144:1229.
- Zwierzynska K, Wolke D, Lereya TS. Peer victimization in childhood and internalizing problems in adolescence: a prospective longitudinal study. J Abnorm Child Psychol. 2013;41:309–323.
- Copeland WE, Wolke D, Angold A, et al. Adult psychiatric outcomes of bullying and being bullied by peers in childhood and adolescence. JAMA Psychiatry. 2013;70:419–426.
- Winsper C, Lereya T, Zanarini M, et al. Involvement in bullying and suicide-related behavior at 11 years: a prospective birth cohort study. J Am Acad Child Adolesc Psychiatry. 2012;51:271–82. e3.
- Suderman M, Borghol N, Pappas JJ, et al. Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Med Genomics. 2014;7:13.
- Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Nat Acad Sci. 2008;105:17046–17049.
- Rijlaarsdam J, Pappa I, Walton E, et al. An epigenome-wide association meta-analysis of prenatal maternal stress in neonates: A model approach for replication. Epigenetics. 2016;11:140–149.
- Cecil CAM, Smith RG, Walton E, et al. Epigenetic signatures of childhood abuse and neglect: implications for psychiatric vulnerability. J Psychiatr Res. 2016;83:184–194.
- Rusiecki JA, Byrne C, Galdzicki Z, et al. PTSD and DNA methylation in select immune function gene promoter regions: A repeated measures case-control study of U.S. military service members. Front Psychiatry. 2013;4:1–12.
- Marzi SJ, Sugden K, Arseneault L, et al. Analysis of DNA methylation in young people: limited evidence for an association between victimization stress and epigenetic variation in blood. Am J Psychiatry. 2018;175:517–529.
- Labonté B, Suderman M, Maussion G, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–731.
- Houtepen LC, Hardy R, Maddock J, et al. Childhood adversity and DNA methylation in two population-based cohorts. Transl Psychiatry. 2018;8:266.
- Houtepen LC, Vinkers CH, Carrillo-Roa T, et al. Genome-wide DNA methylation levels and altered cortisol stress reactivity following childhood trauma in humans. Nat Commun. 2016;7:10.
- Jovanova OS, Nedeljkovic I, Spieler D, et al. DNA methylation signatures of depressive symptoms in middle-aged and elderly persons: meta-analysis of multiethnic epigenome-wide studies. JAMA Psychiatry. 2018;75:949–959.
- Mulder RH, Rijlaarsdam J, Luijk MPCM, et al. Methylation matters: FK506 binding protein 51 (FKBP5) methylation moderates the associations of FKBP5 genotype and resistant attachment with stress regulation. Dev Psychopathol. 2017;29:491–503.
- van Dongen J, Zilhão NR, Sugden K, et al. Epigenome-wide association study of attention-deficit/hyperactivity disorder symptoms in adults. Biol Psychiatry. 2019;86:599–607.
- Ouellet-Morin I, Wong CC, Danese A, et al. Increased serotonin transporter gene (SERT) DNA methylation is associated with bullying victimization and blunted cortisol response to stress in childhood: a longitudinal study of discordant monozygotic twins. Psychol Med. 2013;43:1813–1823.
- Efstathopoulos P, Andersson F, Melas PA, et al. NR3C1 hypermethylation in depressed and bullied adolescents. Transl Psychiatry. 2018;8:121.
- Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. Jama. 2008;299:2877–2883.
- Yang BZ, Zhang H, Ge W, et al. Child abuse and epigenetic mechanisms of disease risk. Am J Prev Med. 2013;44:101–107.
- Chen Y-A, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the illumina infinium humanmethylation450 microarray. Epigenetics. 2013;8:203–209.
- Gaunt TR, Shihab HA, Hemani G, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61.
- Hannon E, Knox O, Sugden K, et al. Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet. 2018;14:e1007544.
- Mehta D, Klengel T, Conneely KN, et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013;110:8302–8307.
- Prados J, Stenz L, Courtet P, et al. Borderline personality disorder and childhood maltreatment: A genome-wide methylation analysis. Genes Brain Behav. 2015;14:177–188.
- Suderman M, Pappas JJ, Borghol N, et al. Lymphoblastoid cell lines reveal associations of adult DNA methylation with childhood and current adversity that are distinct from whole blood associations. Int J Epidemiol. 2015;44:1331–1340.
- Kumsta R, Kreppner J, Kennedy M, et al. Psychological consequences of early global deprivation an overview of findings from the English & Romanian adoptees study. Eur Psychol. 2015;20:138–151.
- Marinova Z, Maercker A, Küffer A, et al. DNA methylation profiles of elderly individuals subjected to indentured childhood labor and trauma. BMC Med Genet. 2017;18:21.
- Dunn EC, Soare TW, Zhu Y, et al. Sensitive periods for the effect of childhood adversity on DNA methylation: results from a prospective, longitudinal study. Biol Psychiatry. 2019;85:838–849.
- Lanner JT, Georgiou DK, Joshi AD, et al. Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb Perspect Biol. 2010;2:a003996.
- Junutula JR, De Maziére AM, Peden AA, et al. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol Biol Cell. 2004;15:2218–2229.
- Biala Y, Bogoch Y, Bejar C, et al. Prenatal stress diminishes gender differences in behavior and in expression of hippocampal synaptic genes and proteins in rats. Hippocampus. 2011;21:1114–1125.
- Zhou J, Liu Z, Yu J, et al. Quantitative proteomic analysis reveals molecular adaptations in the hippocampal synaptic active zone of chronic mild stress-unsusceptible rats. Int J Neuropsychopharmacol. 2015;19:pyv100.
- Ornish D, Magbanua MJM, Weidner G, et al. Changes in prostate gene expression in men undergoing an intensive nutrition and lifestyle intervention. Proc Nat Acad Sci. 2008;105:8369–8374.
- Sequeira A, Mamdani F, Ernst C, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PloS One. 2009;4:e6585.
- Das S, Boczan J, Gerwin C, et al. Regional and developmental regulation of syntaphilin expression in the brain: a candidate molecular element of synaptic functional differentiation. Mol Brain Res. 2003;116:38–49.
- Shan J, Kushnir A, Betzenhauser MJ, et al. Phosphorylation of the ryanodine receptor mediates the cardiac fight or flight response in mice. J Clin Invest. 2010;120:4388–4398.
- Oltmann-Norden I, Galuska SP, Hildebrandt H, et al. Impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid synthesis during postnatal mouse brain development. J Biol Chem. 2008;283:1463–1471.
- Calandreau L, Márquez C, Bisaz R, et al. Differential impact of polysialyltransferase ST8SiaII and ST8SiaIV knockout on social interaction and aggression. Genes Brain Behav. 2010;9:958–967.
- Park C, Lee S-A, Hong J-H, et al. Disrupted-in-schizophrenia 1 (DISC1) and Syntaphilin collaborate to modulate axonal mitochondrial anchoring. Mol Brain. 2016;9:69.
- Sato C, Kitajima K. Impact of structural aberrancy of polysialic acid and its synthetic enzyme ST8SIA2 in schizophrenia. Front Cell Neurosci. 2013;7:61.
- Lin M-Y, Cheng X-T, Tammineni P, et al. Releasing syntaphilin removes stressed mitochondria from axons independent of mitophagy under pathophysiological conditions. Neuron. 2017;94:595–610. e6.
- Mikkonen M, Soininen H, Tapiola T, et al. Hippocampal plasticity in Alzheimer’s disease: changes in highly polysialylated NCAM immunoreactivity in the hippocampal formation. Eur J Neurosci. 1999;11:1754–1764.
- Panni T, Mehta AJ, Schwartz JD, et al. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ Health Perspect. 2016;124:983–990.
- Green BB, Karagas MR, Punshon T, et al. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire Birth Cohort Study (USA). Environ Health Perspect. 2016;124:1253–1260.
- Sharp GC, Arathimos R, Reese SE, et al. Maternal alcohol consumption and offspring DNA methylation: findings from six general population-based birth cohorts. Cohorts for H, Aging Research in Genomic Epidemiology plus methylation alcohol working g, et al. Epigenomics. 2018;10: 27–42.
- Singham T, Viding E, Schoeler T, et al. Concurrent and longitudinal contribution of exposure to bullying in childhood to mental health: the role of vulnerability and resilience. JAMA Psychiatry. 2017;74:1112–1119.
- Waasdorp TE, Bradshaw CP, Leaf PJ. The impact of schoolwide positive behavioral interventions and supports on bullying and peer rejection: A randomized controlled effectiveness trial. Arch Pediatr Adolesc Med. 2012;166:149–156.
- Kooijman MN, Kruithof CJ, van Duijn CM, et al. The generation R Study: design and cohort update 2017. Eur J Epidemiol. 2016;31:1243–1264.
- Boyd A, Golding J, Macleod J, et al. Cohort profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of parents and children. Int J Epidemiol. 2013;42:111–127.
- Fraser A, Macdonald-Wallis C, Tilling K, et al. Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2012;42:97–110.
- Relton CL, Gaunt T, McArdle W, et al. Data resource profile: accessible resource for integrated epigenomic studies (ARIES). Int J Epidemiol. 2015;44:1181–1190.
- Perren S, Alsaker FD. Social behavior and peer relationships of victims, bully‐victims, and bullies in kindergarten. J Child Psychol Psychiatry. 2006;47:45–57.
- Wolke D, Woods S, Stanford K, et al. Bullying and victimization of primary school children in England and Germany: prevalence and school factors. Br J Psychol. 2001;92:673–696.
- Horwood J, Waylen A, Herrick D, et al. Common visual defects and peer victimization in children. Invest Ophthalmol Vis Sci. 2005;46:1177–1181.
- Paget A, Parker C, Heron J, et al. Which children and young people are excluded from school? Findings from a large B ritish birth cohort study, the A von L ongitudinal S tudy of P arents and C hildren (ALSPAC). Child Care Health Dev. 2018;44:285–296.
- Jansen PW, Verlinden M, Dommisse-van Berkel A, et al. Teacher and peer reports of overweight and bullying among young primary school children. Pediatrics. 2014;134:473–480.
- Verlinden M, Jansen PW, Veenstra R, et al. Preschool attention-deficit/hyperactivity and oppositional defiant problems as antecedents of school bullying. J Am Acad Child Adolesc Psychiatry. 2015;54:571–579.
- Achenbach TM, Rescorla LA. Manual for the ASEBA preschool forms and profiles. Burlington, VT: University of Vermont, Research center for children, youth; 2000.
- Goodman R. The Strengths and Difficulties Questionnaire: a research note. J Child Psychol Psychiatry. 1997;38:581–586.
- Tellegen PJ, Winkel M, Wijnbergen-Williams BJ. Snijders-Oomen Niet-verbale Intelligentietest-Revisie SON-R 2½–7. Lisse: Swets & Zeitlinger; 1996.
- Wechsler D. Wechsler Intelligence Scale for Children–3rd UK ed (WISC–III UK). Sidcup, UK: The Psychological Corporation; 1992.
- Dunn EC, Nishimi K, Neumann A, et al. Time-dependent effects of exposure to physical and sexual violence on psychopathology symptoms in late childhood: in search of sensitive periods in development. J Am Acad Child Adolesc Psychiatry. 2019;59:283–295.
- Houtepen LC, Heron J, Suderman MJ, et al. Adverse childhood experiences in the children of the Avon Longitudinal Study of Parents and Children (ALSPAC). Wellcome Open Res. 2018;3:106.
- Saunders JB, Aasland OG, Babor TF, et al. Development of the alcohol use disorders identification test (AUDIT): WHO collaborative project on early detection of persons with harmful alcohol consumption‐II. Addiction. 1993;88:791–804.
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:1.
- Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883.
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:e161.
- R Core Team. R: A language and environment for statistical computing, Vienna, Austria. 2013. https://www.R-project.org/.
- Zeilinger S, Kühnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PloS One. 2013;8:e63812.
- Philibert R, Hollenbeck N, Andersen E, et al. Reversion of AHRR demethylation is a quantitative biomarker of smoking cessation. Front Psychiatry. 2016;7:55.
- Reynolds LM, Magid HS, Chi GC, et al. Secondhand tobacco smoke exposure associations with DNA methylation of the aryl hydrocarbon receptor repressor. Nicotine Tob Res. 2016;19:442–451.
- Dogan MV, Lei M-K, Beach SRH, et al. Alcohol and tobacco consumption alter hypothalamic pituitary adrenal axis DNA methylation. Psychoneuroendocrinology. 2016;66:176–184.
- Bojesen SE, Timpson N, Relton C, et al. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax. 2017;72:646–653.
- Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. arXiv Preprint arXiv:14065823. 2014.
- Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1-48.
- Fradin D, P-Y B, Belot M-P, et al. Genome-wide methylation analysis identifies specific epigenetic marks in severely obese children. Sci Rep. 2017;7:46311.
- Verlinden M, Veenstra R, Ghassabian A, et al. Executive functioning and non-verbal intelligence as predictors of bullying in early elementary school. J Abnorm Child Psychol. 2014;42:953–966.
- Jansen PW, Verlinden M, Dommisse-van Berkel A, et al. Prevalence of bullying and victimization among children in early elementary school: do family and school neighbourhood socioeconomic status matter? BMC Public Health. 2012;12:494.
- Holt MK, Kaufman Kantor G, Finkelhor D. Parent/child concordance about bullying involvement and family characteristics related to bullying and peer victimization. J Sch Violence. 2008;8:42–63.
- Rospenda KM, Richman JA, Wolff JM, et al. Bullying victimization among college students: negative consequences for alcohol use. J Addict Dis. 2013;32:325–342.
- Zhang R, Miao Q, Wang C, et al. Genome‐wide DNA methylation analysis in alcohol dependence. Addict Biol. 2013;18:392–403.
- Paternoster R, Brame R, Mazerolle P, et al. Using the correct statistical test for the equality of regression coefficients. Criminology. 1998;36:859–866.
- Clogg CC, Petkova E, Haritou A. Statistical methods for comparing regression coefficients between models. Am J Sociol. 1995;100:1261–1293.
