ABSTRACT
The ten-eleven translocation (TET) family (TET1/2/3) initiates conversion of 5-methylcytosine to 5-hydroxymethylcytosine, thereby orchestrating the DNA demethylation process and changes in epigenetic marks during early embryogenesis. In this study, CRISPR/Cas9 technology and a TET-specific inhibitor were applied to elucidate the role of TET family in regulating pluripotency in preimplantation embryos using porcine embryos as a model. Disruption of TET1 unexpectedly resulted in the upregulation of NANOG and ESRRB transcripts, although there was no change to the level of DNA methylation in the promoter of NANOG. Surprisingly, a threefold increase in the transcript level of TET3 was observed in blastocysts carrying modified TET1, which may explain the upregulation of NANOG and ESRRB. When the activity of TET enzymes was inhibited by dimethyloxalylglycine (DMOG) treatment, a dioxygenase inhibitor, to investigate the role of TET1 while eliminating the potential compensatory activation of TET3, reduced level of pluripotency genes including NANOG and ESRRB, and increased level of DNA methylation in the NANOG promoter was detected. Blastocysts treated with DMOG also presented a lower inner cell mass/TE ratio, implying the involvement of TET family in lineage specification in blastocysts. Our results indicate that the TET family modulates proper expression of NANOG, a key pluripotency marker, by controlling its DNA methylation profile in the promoter during embryogenesis. This study suggests that TET family is a critical component in pluripotency network of porcine embryos by regulating gene expression involved in pluripotency and early lineage specification.
Introduction
DNA methylation at CpG dinucleotides is an important epigenetic mark that regulates gene expression and its proper maintenance is essential for normal embryo development [Citation1–3]. Although DNA methylation patterns are stably preserved without dynamic changes in somatic cells, genome-wide reprogramming of DNA methylation occurs after fertilization and during germ-cell development [Citation4,Citation5]. Asymmetric global DNA demethylation in paternal and maternal genomes of fertilized zygotes has been reported in multiple species [Citation6–8]. Specifically, the paternal genome undergoes an active demethylation process in zygotes, while DNA methylation level is gradually decreased following embryo cleavage in the maternal genome through DNA replication-dependent manner in the absence of maintenance methyltransferase DNMT1 [Citation9,Citation10]. More recent studies challenge the theory and suggest the presence of active and passive demethylation processes in both maternal and paternal genomes of zygotes [Citation11,Citation12], indicating that the mechanism of the global reprogramming of DNA methylation marks upon fertilization is a complicated process. De novo methylation near or at the blastocyst stage establishes hypermethylation of inner cell mass (ICM) compared to trophectoderm (TE) [Citation13,Citation14], which is essential for establishing lineage-specific pluripotency of embryos.
Ten-eleven translocation (TET) proteins are 2-oxoglutarate (2OG)- and Fe(II)-dependent enzymes that catalyse successive conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine, and 5-carboxylcytosine [Citation15–17]. These 5mC oxidation products are implicated as intermediates of the DNA demethylation process since the oxidized 5mC derivatives are ultimately transformed to unmodified cytosine through thymine DNA glycosylase-mediated base excision repair [Citation17,Citation18]. The TET gene family includes three members, TET1, TET2, and TET3 [Citation15,Citation16], and their expression patterns are individually distinct during preimplantation development. TET3 is highly expressed in oocytes and fertilized zygotes and known to be responsible for active DNA demethylation in zygote genomes [Citation11,Citation12,Citation19,Citation20]. In contrast, the expression levels of TET1 and TET2 are very low in oocytes or zygotes but increase following preimplantation development and both are highly expressed in blastocysts [Citation16,Citation21].
To understand the physiological relevance of TET enzymes and 5hmC in development, mouse models carrying knockout (KO) alleles for TET genes have been generated. Mouse with an individual KO of TET1 [Citation22] or TET2 [Citation23,Citation24] gene develops to adults without critical developmental defects, whereas TET3 depletion leads to neonatal lethality for unknown reasons [Citation19]. In contrast, embryos deficient in both TET1 and TET2 present substantial perinatal lethality, while a fraction of the double KO embryos survives and develops normally in mice [Citation25]. Triple KO (TET1, TET2, and TET3) mouse ES cells fail to support embryonic development at E9.5 in tetraploid complementation assay [Citation26]. These suggest that the effects of single or dual KO of TET genes on development can be partially offset by the involvement of redundancy in the function among the TET enzymes.
Both TET1 and 5hmC are highly detected in mouse embryonic stem (ES) cells and the ICM of blastocyst. The role of TET1 in maintaining pluripotency and development has been studied using a TET1 knockdown model [Citation15,Citation16] and KO mouse ES cells [Citation22]. Using shRNA to knockdown TET1 gene in mouse ES cells, one study reported that TET1 is involved in maintaining NANOG gene expression and important for self-renewal of ES cells [Citation16]. However, in another study, TET1 KO leads to reduction of the 5hmC level but did not affect the pluripotency of mouse ES cells [Citation22]. Moreover, pluripotency is maintained in TET1 and TET2 double KO mouse ES cells, albeit with some defects in differentiation [Citation25]. These reports raise the possibility that activities of more than one TET family are involved in regulation of pluripotency, despite the higher abundance of TET1 than other TET proteins in ES cells.
Although the subtle effect of TET1 depletion on pluripotency and full-term development has been demonstrated using mouse ES cells, the role of TET1 during preimplantation embryo development, especially in blastocysts where embryonic blastomeres differentiate into ICM and TE, has not been well characterized. Here, we investigated the role of TET1 in regulating gene expression involved in pluripotency and preimplantation development using porcine embryos to expand our understanding of TET1-mediated regulation of pluripotency. To define the necessity of combined activities of TET family in pluripotency of developing embryos, gene expression profiles, as well as global and locus-specific DNA methylation (5hmC and 5mC) status, were examined in TET1 KO blastocysts and blastocysts impaired in overall activities of TET enzymes. Our results identify that a proper level of TET1 is critical for establishing proper expression of pluripotency genes in embryos by delineating the level of DNA methylation.
Results
Disruption of TET1 leads to an abnormal level of global 5hmC and 5mC in blastocysts
To disrupt TET1 function in blastocysts, TET1-specific CRISPR/Cas9 system containing three sgRNAs targeting different regions of TET1 was injected into one-cell stage embryos (). Efficiency of the targeted disruption was verified by genotyping 10 blastocysts that were injected by the CRISPR/Cas9 system. All of blastocysts tested (n = 10) possessed mutations on both alleles, demonstrating efficacy of the approach (Table S1). The total activity of TET enzymes was significantly decreased in TET1 KO blastocysts compared to that in controls, indicating successful disruption of TET1 by the CRISPR/Cas9 system (Fig. S1A). The frequency of blastocyst formation was reduced by the TET1 disruption; however, the average total cell number of blastocysts at d 7 was not affected (Table S2 ad Fig. S1B), suggesting that the loss of TET1 does not lead to developmental stall.
Figure 1. Global levels of 5hmC and 5mC in TET1 KO blastocysts. (a) Strategy to disrupt TET1 gene in porcine embryos. Three sgRNAs target different regions of TET1 gene; one on the immediate downstream of the presumable translation start site and two on the 5′ side of the 2-oxoglutarate-Fe(II)-oxygenase domain. (b) Global 5hmC level of blastocysts was dramatically decreased by TET1 KO. (c) Difference of global 5mC level between TET1 KO and control blastocysts was not detected by immune-staining. Numbers in parentheses indicate the number of embryos displaying a positive signal for 5hmC or 5mC out of the total number of embryos examined. (d) Bisulphite sequencing analysis of DNA methylation pattern in repetitive elements. Methylation level of PRE-1 was increased by TET1 KO whereas centromeric satellites were hypomethylated in both TET1 KO (n = 17) and control blastocysts (n = 17). Scale bar indicates 100 µm. Methylated and unmethylated CpG dinucleotides are indicated by filled circle and open circle, respectively
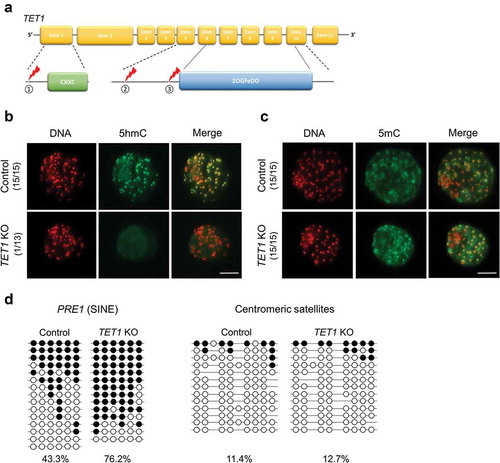
The global level of 5mC and 5hmC was monitored in TET1 KO blastocysts as TET family is known to oxidize 5mC to initiate DNA demethylation process. The disruption of TET1 dramatically reduced the level of global 5hmC in blastocysts (); however, distinct differences in the 5mC level between TET1 KO and control blastocysts were not observed in immunocytochemistry analysis (). To investigate the global level of 5mC at the nucleotide level, DNA methylation status of repetitive elements was quantified by using bisulphite sequencing analysis. Specifically, the DNA methylation level of PRE-1 (porcine-specific SINE retrotransposon) and centromeric satellite regions can capture the overview of methylation status on euchromatin and heterochromatin, respectively [Citation27,Citation28]. In the bisulphite sequencing analysis, DNA methylation on the PRE-1 region was increased in TET1 KO blastocysts compared to that in control blastocysts, while centromeric satellites were hypomethylated in both TET1 KO and control blastocysts (). Our findings from the immunocytochemistry and bisulphite sequencing suggest that TET1 is responsible for the formation of 5hmC in blastocysts and important for maintaining global DNA methylation levels, especially in the euchromatic region of the genome.
Disruption of TET1 leads to abnormal expression of pluripotency genes without converting lineage specification
Altered global 5hmC level and DNA methylation pattern by the TET1 KO implied that the KO embryos potentially carried abnormal gene expression patterns. To verify, transcript abundance of pluripotency-related genes and extra-embryonic lineage markers were quantified in TET1 KO blastocysts by RT-qPCR. The lack of functional TET1 did not alter the transcript level of pluripotency-related genes except for NANOG and ESRRB (). Unexpectedly, an elevated level of NANOG and ESRRB transcript was observed in TET1 KO blastocysts. There was no change in the transcript level of selected extra-embryonic lineage genes after TET1 KO except for GATA4 ()); the transcript level of GATA4 was higher in TET1 KO blastocysts compared to that in controls. RT-qPCR was also performed to monitor the level of TET family () as previous KO experiments in the mouse suggest functional redundancy of the TET enzymes. The expression level of TET1 transcript was significantly downregulated in TET1 KO blastocyst, indicating that the mutations introduced by CRISPR/Cas9 system interfered with transcription, as well as protein function via generation of premature stop codons. Interestingly, the disruption of TET1 induced activation of TET3 as indicated by the threefold increase in its transcript level compared to the control blastocysts. Expression level of TET2 was also numerically increased (1.5-fold), although the change was not statistically significant (p = 0.16). The overexpression of TET3 suggests that disruption of TET1 activated TET3 as compensatory action and could potentially contribute to the limited changes in gene expression profile of TET1 KO blastocysts.
Figure 2. Impact of TET1 KO on gene expression profile and lineage specification in blastocysts. (a) Expression levels of NANOG and ESRRB were increased by TET1 KO; however, levels of other pluripotency genes were not significantly changed. (b) Transcript levels of extra-embryonic lineage genes were not changed by TET1 KO, except GATA4. (c) The level of TET1 transcript was decreased; however, TET3 levels were increased approximately threefold. Relative expression levels were normalized to GAPDH level. Error bars represent SEM. A p-value < 0.05 was considered statistically significant. (d) CDX2 staining revealed that TET1 KO did not alter lineage commitment in blastocysts. The dotted white circle indicates ICM lineage. Scale bar indicates 100 µm. (e) ICM/TE cell number ratio was calculated by counting CDX2 negative and CDX2 positive cells. The ICM/trophectoderm ratio was not different between control and TET1 KO blastocysts. Error bars indicate SD. A p-value < 0.05 was considered statistically significant. TE: Trophectoderm
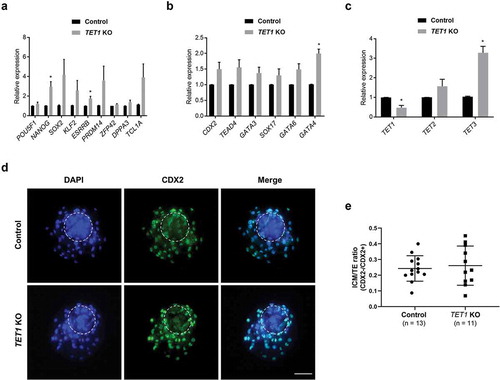
To examine the impact of TET1 on early lineage commitment, d 7 blastocysts derived from CRISPR/Cas9-injected and control group were stained with CDX2, a TE marker (). The ICM and TE lineages were distinguished by counting CDX2 negative and positive cells, respectively. In both control and TET1 KO blastocysts, a cluster of CDX2 negative cells was identified, indicating that ICM lineage formation was not affected by the disruption of TET1. Analysis of the ICM/TE ratio between control and TET1 KO embryos suggested that early lineage commitment in blastocysts was not affected by the lack of TET1 ().
Inhibition of catalytic activity of TET family and its impact on DNA methylation
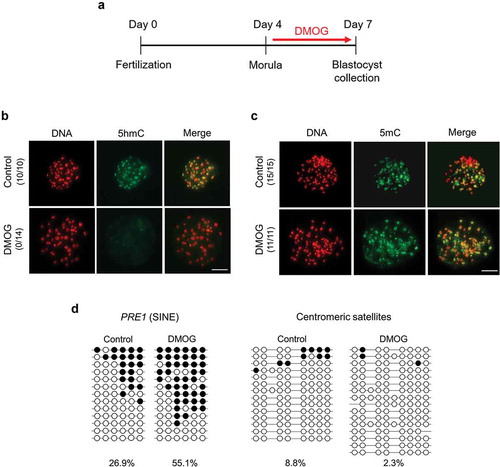
To investigate whether the findings from the TET1 KO blastocysts was influenced by the compensatory increase in other TET genes, overall inhibition of TET family was performed. Specifically, catalytic activity of TET enzymes was blocked by treating embryos with dimethyloxallyl glycine (DMOG), a small-molecule inhibitor of 2OG-dependent oxygenases. To confirm the effectiveness of DMOG in blocking oxygenase activity of TET, especially TET3, fertilized zygotes were incubated with 1 mM DMOG, then patterns of DNA demethylation after fertilization were monitored by uncovering the appearance of 5hmC after fertilization (Fig. S2A). Because the DNA demethylation process is orchestrated by TET3 [Citation11,Citation12,Citation19], the inhibitor would alter the level of 5hmC, if effective. The DMOG treatment diminished the appearance of 5hmC in both paternal and maternal pronuclei compared to the level of 5hmC in control zygotes, demonstrating the capability of DMOG to inhibit the conversion from 5mC to 5hmC by TET (Fig. S2B). To inhibit the activity of TET enzymes effectively at the blastocyst stage, embryos were incubated with 1 mM DMOG from morula (d 4) to blastocyst stage (d 7) (). The treatment would keep the activity of TET3 on initiating DNA demethylation process after fertilization but prevent activity of all TET family in blastocysts. Consistent with the TET1 KO experiment, frequency of blastocyst formation on d 7 was decreased in DMOG-treated embryos compared to that in control embryos (Table S3) while total cell number in blastocyst was not affected by the inhibition of TET activity (Fig. S1C). Similar to the findings in TET1 KO blastocysts, the global level of 5hmC was dramatically reduced in blastocysts treated with DMOG compared to the control blastocysts, indicating that active TET family is necessary for the formation of 5hmC at the blastocyst stage (). Dramatic changes of global level of 5mC were not observed by immunocytochemistry analysis in blastocysts treated with DMOG (). The DNA methylation pattern of repetitive elements in DMOG-treated blastocysts was also similar to that in TET1 KO blastocysts; methylation level of PRE-1 region increased when embryos were incubated with DMOG, while hypomethylation status of centromeric satellites was observed in both control and DMOG-treated blastocysts (). These data align with conclusions from the TET1 KO experiment that TET1 is responsible for the formation of 5hmC and maintenance of global DNA methylation level in the euchromatic region at the blastocyst stage.
Figure 3. Global levels of 5hmC and 5mC in DMOG-treated blastocysts. (a) Timeline of DMOG treatment to inhibit TET activities in blastocysts. To block TET activities at blastocyst stage, embryos were incubated with 1 mM DMOG from morula stage (d 4) to blastocyst stage (d 7); then, blastocysts were collected for further analysis. (b) Global 5hmC level of blastocysts was dramatically decreased by DMOG treatment. (c) Difference of global 5mC level between DMOG-treated and control blastocysts was not detected by immunocytochemistry. Numbers in parentheses indicate the number of embryos displaying a positive signal for 5hmC or 5mC out of the total number of embryos examined. (d) Bisulphite sequencing analysis of DNA methylation pattern in repetitive elements. Methylation level of PRE-1 was increased by DMOG treatment whereas centromeric satellites were hypomethylated in both DMOG-treated (n = 15) and control blastocysts (n = 16). Scale bar indicates 100 µm. Methylated and unmethylated CpG dinucleotides are indicated by filled circle and open circle, respectively
Inhibition of overall TET family activity impairs expression of pluripotency and extra-embryonic lineage genes and shifts lineage specification
To further investigate whether the abnormal level of NANOG and ESRRB in TET1 KO blastocysts was influenced by the compensatory increase of other TET genes, gene expression patterns in DMOG-treated blastocysts were compared to that in control blastocysts. A significant number of pluripotency-related genes were downregulated in DMOG-treated blastocysts; the transcript abundance of POU5F1, NANOG, ESRRB, PRDM14, ZFP42, and DPPA3 was lower in DMOG-treated blastocysts (). However, there was no significant change in the level of SOX2, KLF2, and TCL1A after the DMOG treatment. The transcript abundance of trophoblast markers (CDX2, TEAD4, and GATA3) and primitive endoderm marker (SOX17, GATA6, and GATA4) was downregulated in DMOG-treated blastocysts (). The DMOG treatment also affected the level of TET family genes; the level of TET1, 2, and 3 transcripts was downregulated by the DMOG treatment in blastocysts (). The gene expression analysis indicates that TET enzymes are critical for proper expression of pluripotency and extraembryonic lineage markers. In addition, the dynamic changes in transcripts after the DMOG treatment but not after TET1 KO suggest that compensatory action of TET family (TET2 or TET3) in the absence of TET1 assisted in maintaining the normal level of selected markers.
Figure 4. Relative mRNA levels of pluripotency genes, extra-embryonic lineage genes, and TET family genes in DMOG-treated blastocysts. (a) Expression levels of pluripotency genes were downregulated in DMOG-treated blastocysts, except for SOX2, KLF2, and TCL1A. (b) Transcript levels of extra-embryonic lineage genes were reduced in DMOG-treated blastocysts. (c) Expression level of TET family genes was reduced in DMOG-treated blastocysts. Relative expression levels were normalized to GAPDH level. Error bars represent SEM. A p-value < 0.05 was considered statistically significant. (d) The number of CDX2 negative cells (ICM) was reduced in DMOG-treated blastocysts compared to that in control blastocysts. The dotted white circle indicates ICM lineage. Scale bar indicates 100 µm. (e) DMOG treatment lowered ICM/TE ratio. Error bars indicate SD. A p-value < 0.05 was considered statistically significant. TE: Trophectoderm
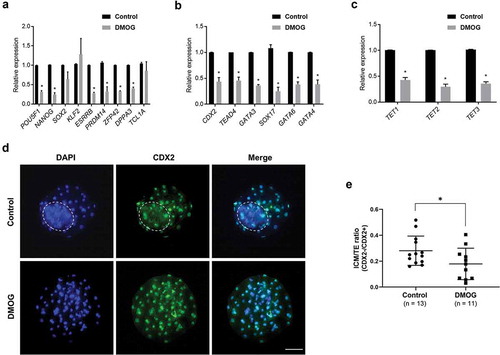
Intriguingly, capturing the number of CDX2 positive and negative cells through immunohistochemistry revealed that the DMOG treatment reduced the number of CDX2 negative cells, i.e., ICM, in blastocysts compared to that in control embryos (). The ICM/TE ratio was also decreased in DMOG-treated blastocysts ()), demonstrating that TET family is essential for normal lineage commitment in blastocysts.
TET enzymes regulate DNA methylation level in the promoter and gene-body regions of NANOG
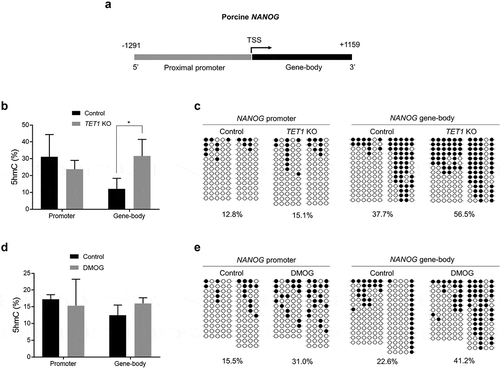
Contradicting effect between DMOG-treated and TET1 KO blastocysts on the level of NANOG implied a differential level of DNA methylation on the promoter of NANOG from the two treatments. To investigate that possibility, the levels of DNA methylation (5mC and/or 5hmC) levels in promoter and gene-body regions of NANOG gene were analysed (). The 5hmC level in NANOG promoter was not different between control and TET1 KO blastocysts (). However, the 5hmC level in gene-body regions was increased in TET1 KO blastocysts compared to that in controls. Bisulphite sequencing analysis revealed that there was no difference in the promoter methylation level between TET1 KO and control blastocysts, while higher methylation level in gene-body region was observed in TET1 KO blastocysts compared to that in controls (). When embryos were incubated with DMOG, 5hmC level of blastocysts was not changed in the promoter or gene-body regions (). However, the DMOG treatment increased DNA methylation level in both promoter and gene-body regions (). Bisulphite sequencing analysis cannot distinguish between 5mC and 5hmC; therefore, it is difficult to conclude whether the increase is due to the conversion of 5mC into 5hmC. Considering no alteration in 5hmC level by the DMOG treatment, it is arguable that the increase in DNA methylation level seen from bisulphite sequencing is due to 5mC and the elevated 5mC level in promoter and gene-body regions led to the downregulation of NANOG expression in DMOG-treated blastocysts.
Figure 5. DNA methylation (5hmC and 5mC) status of NANOG gene in TET1 KO and DMOG-treated blastocysts. (a) Analysis of methylation status of porcine NANOG gene. The proximal promoter region (1291 bp upstream the TSS) and gene-body region (1159 bp downstream the TSS) of NANOG gene were separately examined. (b) 5hmC level in promoter and gene-body regions of NANOG gene in TET1 KO blastocysts. (c) Bisulphite sequencing analysis of promoter and gene-body regions of NANOG gene in TET1 KO (n = 15) and control (n = 15) blastocysts. (d) 5hmC level in promoter and gene-body regions of NANOG gene in DMOG-treated blastocysts. (e) Bisulphite sequencing analysis of promoter and gene-body regions of NANOG gene in DMOG-treated (n = 15) and control (n = 15) blastocysts. 5hmC data were analysed with Student’s t-test for three replicates and a p-value < 0.05 was considered statistically significant
Discussion
Recent studies have expanded biological significance of the TET family on early development. Here, we demonstrated that the loss of TET activities alters the expression pattern of genes related to pluripotency and early lineage specification in porcine blastocysts; specific changes differed when activity of TET1 or overall TET family was interfered. Global reduction in the 5hmC level after TET1 KO suggests that TET1 is the main 5-methyl-dioxygenase responsible for establishing the 5hmC marks in porcine blastocysts. Studies using mouse ES cells indicate that while Tet1 is important for establishing 5hmC marks, Tet1 deletion alone has a relatively limited influence on the level of 5hmC whereas the lack of Tet1 and Tet2 leads to a complete loss of 5hmC [Citation15,Citation21,Citation22,Citation25]. Furthermore, Tet2 depletion resulted in a greater decrease in 5hmC level than Tet1 depletion in the mouse ES cells [Citation21,Citation29]. Concomitant with the decrease in global 5hmC level, DNA methylation level including 5mC and 5hmC was increased in euchromatic SINE, PRE-1, in both TET1 KO and DMOG-treated blastocysts, consistent with a previous report indicating that TET proteins play a protective role against de novo DNA methylation [Citation30]. A similar level of increase in DNA methylation level (about 30%) in PRE-1 regions between TET1 KO and DMOG-treated blastocysts suggests that TET1 is likely to be the main TET enzyme responsible for maintaining global methylation level in blastocysts among the TET family members (TET1-3). Hypomethylation of the centromeric region is common in germ cells and preimplantation embryos [Citation31] and disruption of TET1 or overall TET family did not change the level, indicating that TET proteins are not involved in methylation maintenance in heterochromatic regions. The result is consistent with previous studies in the mouse where 5hmC modifications are rarely detected in heterochromatic regions, whereas euchromatic regions are enriched with Tet1 binding sites and a wide distribution of 5hmC in the mouse ES cells [Citation32–34].
The role of TET1 in cellular pluripotency and differentiation has been extensively studied in ES cells and blastocysts because its level is highly enriched in the cells and embryos compared to other TET family genes. Impaired or skewed differentiation of ES cells caused by TET1 depletion has been demonstrated [Citation21,Citation22,Citation26,Citation33]; however, the impact of TET1 loss on the pluripotency of ES cells has not been consistent in the previous reports. Using siRNA or shRNA approach, multiple studies reported downregulation of pluripotency-associated genes and impaired self-renewal in ES cells after targeted disruption of TET1 [Citation16,Citation33–35]. However, other studies argue that TET1 depletion did not alter the expression of pluripotency markers [Citation21,Citation22,Citation36]. This variance could be caused by the compensatory role of other TET family members under the absence of functional TET1. Similarly, we observed a compensatory and overlapping function of TET family. The disruption of TET1 did not lead to total repression of pluripotency genes, potentially due to the compensatory activation of TET3. Transcript levels of pluripotency genes (SOX2, KLF2, PRDM14, TCL1A) were numerically increased in TET1 KO blastocysts. However, the differences were not statistically significant due to high variations among biological replications, which may be explained by a mouse study in which Tet1/Tet3 double KO mouse embryos presented a significant transcriptome variability among embryos [Citation37]. Blocking overall TET activity using DMOG repressed the expression of pluripotency-related genes, confirming an overlapping function between TET1 and TET3. The finding is consistent with the result from Tet1/Tet3 double KO mouse blastocysts which displayed severe loss in Nanog expression and dysregulation of extra-embryonic lineage markers [Citation37]. Another possibility that cannot be fully excluded is that the DMOG is an inhibitor of 2OG-dependent oxygenases and decreased activity of other 2OG-dependent oxygenases such as Jumonji-domain-containing enzymes could also contribute to the repressed expression of pluripotency-related genes.
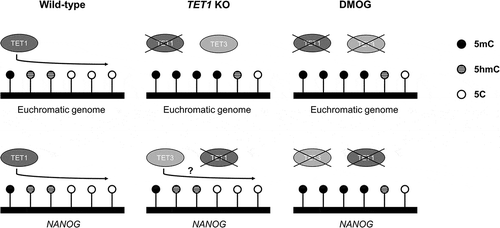
Although the majority of TET1 binding sites are located around transcription start sites of CpG-rich promoters and gene bodies, TET1 also binds to CpG-poor gene promoters, such as NANOG, TCL1, and ESRRB, important genes for maintaining pluripotency of ES cells [Citation34,Citation36]. In contrast to a mouse study where the lack of Tet1 reduced Nanog expression in ES cells [Citation34], the expression of NANOG was upregulated and proximal promoter regions were hypomethylated after the removal of TET1 in porcine blastocysts. Previous reports in the mouse suggest that Tet1 can act as a repressor to genes related to development, but not to pluripotency related genes [Citation34,Citation38]. Therefore, it is unlikely that the upregulation of NANOG in TET1 KO porcine blastocysts was the result of lifting TET1-mediated repression. Interestingly, the inhibition of overall TET by DMOG repressed NANOG expression and increased the methylation level of its promoter region, suggesting that the expression of NANOG is directly controlled by the level of DNA methylation in the promoter region, similar to transcriptionally active genes [Citation39]. The contrasting results between the TET1 KO and DMOG treatment suggest that other TET enzymes, such as TET2 and TET3, are involved in the regulation of NANOG expression. For instance, despite its low expression level, Tet3 contributes to a small portion of 5hmC in mouse ES cells, suggesting its action in pluripotent cells [Citation26,Citation40]. Maternal Tet3 is required for demethylation of the promoter region of paternal Nanog in mouse zygotes [Citation19], and knockdown of maternal TET3 leads to downregulation of NANOG expression in porcine blastocysts [Citation41]. In addition, TET3 activated by a hypoxia microenvironment regulates NANOG transcription by direct binding to promoter region in brain tumour cells [Citation42]. The previous publications and findings from current study imply that TET3 possesses a role in the transcriptional regulation of NANOG gene in blastocysts by controlling the level of DNA methylation on its promoter. Findings in this study lead us to a model of how TET family regulates global DNA methylation and NANOG expression in porcine blastocysts (). Under the presence of functional TET1, the DNA methylation level of the genome, including the promoter regions of NANOG, is regulated by TET1, thus maintaining the level of NANOG in the blastocysts. In the absence of functional TET1, a compensatory increase in TET3 potentially reprograms the promoter region of NANOG to ensure its expression; however, TET3 may have little effect on the genome-wide DNA methylation level, i.e., gene-specific regulation of DNA methylation rather than global level. Suppression of overall TET family led to the downregulation of NANOG and increased the level of DNA methylation on NANOG promoter and the euchromatic genome, indicating the involvement of TET family in establishing pluripotency through regulating the level of DNA methylation.
Figure 6. Graphical summary and proposed mechanism of TET actions on DNA methylation in porcine blastocysts under the TET1 KO or TET inhibition
The expression pattern of ESRRB was also aligned with NANOG: upregulation after TET1 KO and downregulation with DMOG treatment. The changes in the abundance of ESRRB are likely to be a secondary effect by the altered NANOG expression rather than the direct effect from the shift in TET family because ESRRB is known as a direct downstream target gene of NANOG in pluripotent cells [Citation43]. For example, depletion of Tet1 in mouse ES cells led to the downregulation of Esrrb and Nanog; however, the expression of Esrrb was restored by the overexpression of exogenous Nanog [Citation34], indicating that the expression of Esrrb is regulated by Nanog.
The transcription factors OCT4 (POU5F1) and NANOG are essential for the regulation of early embryo development and pluripotency in mammalian embryos. In porcine, similar to human blastocysts, OCT4 expression is maintained in both ICM and TE cells in blastocysts [Citation44,Citation45]. However, unlike mice and human, unique expression patterns of NANOG can be found in porcine blastocysts. For instance, although NANOG transcripts can be detected in early blastocyst stage, NANOG proteins are not detectable in early porcine blastocyst but begin to appear after epiblast formation when embryos arrive in uterus [Citation46]. The unique NANOG expression implies distinct pluripotency network in pig blastocysts and may render difficult derivation of ES cells from porcine embryos under the conventional ES cell culture condition. Therefore, an understanding of expressional regulation of NANOG gene is likely to unveil pluripotency regulation in porcine embryos and contribute to the establishment of porcine ES cells.
The CRISPR/Cas9 technology was used to disrupt TET1 in porcine embryos. Efficacy of the approach was high enough that no wild-type allele was identified in two target loci. The TET1 transcript level was also reduced in blastocysts carrying mutated TET1. One potential explanation could be that TET1 has a positive feedback to its transcription activity; thus, the lack of functional TET1 reduced the amount of TET1 transcripts. Another explanation could be that mutations on TET1, introduced by CRISPR/Cas9 system, could interfere with proper secondary structure formation of TET1 mRNA, which would lead to the degradation of TET1 transcript. The direct injection of CRISPR/Cas9 system has been successfully utilized in pigs to introduce targeted modifications at the high level [Citation47–49] and efficiency of the current study aligns with the previous reports. Pig embryos are suitable to model embryogenesis in humans because pig embryos mirror developmental events with human embryos. For example, dynamic changes in the level of DNA methylation on paternal DNA are observed in humans [Citation50] as well as pigs [Citation51,Citation52]. In addition, the timing of zygotic genome activation is similar between human [Citation53] and pig [Citation54]. Despite the similarities, pig models are rarely used for comparative studies due to the lack of tools available to induce genetic modifications. Unlike rodent models, the production of KO embryos in pigs through breeding founder animals is an unrealistic option considering cost associated with producing KO pigs and gestation period (114 d). As demonstrated in this study, utilization of CRISPR/Cas9 system allowed us to generate KO embryos without having to produce founder KO pigs. These technical advancements may expand the use of pig models to understand developmental events in embryos as a comparative study.
In summary, we demonstrate that the TET enzymes are closely involved in maintaining proper level of pluripotency genes in preimplantation embryos. Although TET1 has a major role in genome-wide 5hmC formation at the blastocyst stage, TET1 disruption has a minor impact on the pluripotency genes of preimplantation embryos. On the other hand, overall inhibition of TET family results in a defective expression of pluripotency genes, abnormal lineage specification, and methylation increase in NANOG promoter. Our data indicate that NANOG, a key gene in pluripotency, is delicately regulated by TET family through promoter methylation. Our findings propose that the TET1/3-mediated demethylation at promoter region is a key element for transcriptional activation of genes related to pluripotency and early lineage specification in mammalian blastocysts.
Materials and methods
Chemicals
All chemicals used in the experiments were purchased from Sigma Aldrich Chemical Company (St. Louis, MO, USA) unless indicated otherwise.
In vitro embryo production
Porcine oocytes were collected from ovaries obtained from an abattoir or purchased from Desoto Biosciences LLC (Seymour, TN, USA). Collected cumulus oocyte complexes were placed in 4-well dishes containing maturation medium, which was TCM-199 (Invitrogen) supplemented with 3.05-mM glucose, 0.91-mM sodium pyruvate, 0.57-mM cysteine, 10-ng/ml EGF, 0.5-mg/ml LH, 0.5-mg/ml FSH, 10-ng/ml gentamicin, and 0.1% polyvinyl alcohol at 38.5°C, 5% CO2 in humidified air. After 42–44 h of maturation, cumulus cells were removed by vortexing in the presence of 0.03% hyaluronidase. Oocytes with a polar body were collected in manipulation medium (TCM-199 supplemented with 0.6 mM NaHCO3, 2.9 mM Hepes, 30 mM NaCl, 10 ng/ml gentamicin, and 3 mg/ml bovine serum albumin [BSA]) and placed in 50 µl droplets of fertilization medium (modified Tris-buffered medium with 113. 1 mM NaCl, 3 mM KCl, 7.5 mM CaCl2, 11 mM glucose, 20 mM Tris, 2 mM caffeine, 5 mM sodium pyruvate, and 2 mg/ml BSA) in a group of 25–30 oocytes. Fresh semen obtained from boars was diluted with semen extender (MOFA, Verona, WI, USA) and stored at 17°C for 1 week. To wash semen, 1 ml of diluted semen was added to 9 ml of DPBS supplemented with 0.1% BSA and washed at 720 × g for 3 min by centrifugation. After three washing steps, the semen pellet was resuspended with fertilization medium to 0.5 × 106/ml. Fifty microlitres of diluted semen was added to the droplets with oocytes. Oocytes and sperms were co-incubated in fertilization medium at 38.5°C, 5% CO2 in humidified air for 5 h. After fertilization, embryos were cultured in PZM3 medium [Citation55] at 38.5°C, 5% CO2, 5% O2 in humidified air.
DMOG treatment
To test the ability of DMOG blocking catalytic activity of TET3 enzyme, oocytes were incubated in fertilization medium in the presence of 1 mM DMOG for 30 min before fertilization. Then, oocytes and sperms were co-incubated in fertilization medium with 1 mM DMOG for 5 h. Subsequently, oocytes were cultured in PZM3 medium with 1 mM DMOG for 16 h and collected for measurement of 5hmC level by immune-staining. For inhibition of TET family in blastocysts, oocytes and sperms were fertilized and cultured until d 4 without DMOG. At d 4, embryos were moved to PZM3 medium with 1 mM DMOG and cultured until d 7. Blastocysts were collected at d 7 for further examinations.
Microinjection
Three sgRNAs targeting different regions of TET1 gene were designed using the Zhang laboratory CRISPR design tool (http://crispr.mit.edu) () and Table S4). In vitro-synthesized Cas9 mRNA (20 ng/µl) and three sgRNAs (10 ng/µl each) were injected into the cytoplasm of presumable zygotes after in vitro fertilization (IVF) using the FemtoJet microinjector (Eppendorf) as previously described [Citation47,Citation48]. Embryos were microinjected in manipulation medium on a heated stage of a Nikon inverted microscope. After the microinjection, the zygotes were washed and then cultured in PZM3 media for 7 d.
Quantification of TET enzyme activity
Nuclear proteins were extracted from control and TET1 KO blastocysts (30 each) at d 7 using the Nuclear Extraction Kit (Abcam). The TET enzyme activity was quantified in triplicate using the TET Hydroxylase Activity Quantification Kit (Abcam) from the extracted nuclear proteins according to the manufacturer’s instruction. The activity was measured on a Tecan Infinite M200 Pro plate reader (Tecan) by measuring the fluorescence intensity at the excitation wavelength of 530 nm and the emission wavelength of 590 nm. The relative fluorescence units of control and TET1 KO samples were subtracted by that of blank and then compared each other.
Immunocytochemistry and blastomere counting
Zona-free embryos were fixed in 4% paraformaldehyde for 15 min at room temperature. The fixed embryos were washed and permeabilized in PBS containing 0.25% TritonX-100 for 1 h. Then, they were treated with 2 N HCl for 30 min and neutralized in Tris–HCl pH 8.5 for 10 min. The samples were incubated in PBS containing 0.1% Tween-20 and 2% BSA for 1 h at room temperature. After blocking, embryos were incubated in blocking solution together with 5hmC (dilution 1:100; Active Motif) or 5mC (dilution 1:100; Active Motif) or CDX2 (dilution 1:20; Biogenex) antibody overnight at 4°C. The next day, the samples were washed in blocking solution and stained with Fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (dilution 1:200; Santa Cruz Biotechnology or dilution 1:500; Thermo Fisher Scientific) for 1 h at room temperature. DNA was stained with 10 µg/ml propidiumiodide (PI) or 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI). The total cell number was calculated by counting PI/DAPI positive cells in d 7 blastocysts stained with 5hmC antibody; then, the average numbers were compared between control and TET-modified blastocysts. The number of ICM and TE cells was calculated by counting the numbers of CDX2 negative and positive cells in DAPI-stained blastocysts.
Quantitative RT-PCR (RT-qPCR)
Nine to 10 blastocysts per group were collected at d 7 to analyse gene expression patterns of pluripotency-related and extra-embryonic lineage genes using RT-qPCR. mRNA was immediately isolated from the pooled embryos using Dynabeads mRNA Direct Kit (Thermo Fisher Scientific), followed by cDNA synthesis using Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Amplification and detection were conducted with the ABI 7500 Real-Time PCR System (Applied Biosystems) using PerfeCTa SYBR Green SuperMix (Quantabio) under the following conditions: 95°C for 3 min, 40 cycles of denaturation at 95°C for 10 s, and annealing at 60°C for 60 s. Primers used in RT-qPCR analysis are listed in Table S5. All of the threshold cycle (CT) values of the tested genes were normalized to GAPDH level, and relative ratios were calculated using the 2−ΔΔCt method. Four biological and three experimental replications were used. Differences in the gene expression were evaluated by Student’s t-test. p < 0.05 was considered as statistically significant.
Bisulphite DNA sequencing
For analysis of DNA methylation status of repetitive elements and NANOG gene, 15–18 blastocysts at d 7 were collected and their DNA was treated with bisulphite using EZ DNA methylation kit (Zymo Research) following the manufacturer’s instruction. Subsequently, the bisulphite-treated DNA was PCR-amplified using specific primer sets. Details of primer information and PCR conditions are described in Tables S6 and S7. The PCR products were purified using GeneJet gel extraction kit (Thermo Fisher Scientific) and were then ligated into pCR 2.1 TA cloning vector (Invitrogen). Twelve to 16 colonies of each cloned sample were sequenced and evaluated.
Detection of locus-specific 5hmC
Detection of locus-specific 5hmC was done using Quest 5-hmC detection kit (Zymo Research). This kit enables detection of 5hmC in DNA sequence through glycosylation of 5hmC and treatment of restriction endonucleases; 5hmC in DNA is specifically tagged with a glucose moiety by glucosyltransferase yielding a modified base, glucosyl-5hmC, which is not digested with a glucosyl-5hmC sensitive restriction endonuclease (MspI) whereas 5C and 5mC are digested with the enzyme. To analyse 5hmC level in promoter and gene-body regions of NANOG gene, genomic DNA was isolated from 15 to 18 day 7 blastocysts using Purelink genomic DNA mini kit (Invitrogen) and then equal amounts of DNA from samples were divided into two groups; one group was treated with a glucosyltransferase and the other group did not receive the treatment. An internal control sample was included for reference. Glycosylation reaction was performed at 37°C for 2 h, and then both groups were digested with 15 units of MspI for 4 h following manufacturer’s instruction. The enzyme-digested DNA was purified with DNA clean and concentrator kit (Zymo Research) and used for qPCR. Test DNA that had not been processed was used as an internal control. NANOG promoter and gene-body regions were amplified from DNA using the following primers: 5′-ACAGACCAATGGAACAGAATAG-3′(forward) and 5′-CACTCATGTTGAGTTGAAGAG-3′(reverse) for promoter region and 5′-AGGACAGCCCTGATTCTTCCACAA-3′ (forward) and 5′-GTTGCTCCATGATGGGTTAT-3′ (reverse) for gene-body region. PCR amplification was performed with the ABI 7500 Real-Time PCR System (Applied Biosystems) using PerfeCTa SYBR Green SuperMix (Quantabio) under the following conditions: 95°C for 1 min, 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 15 s, and extension at 72°C for 25 s. Percentage of hydroxymethylation was obtained by the following formula: % = ((−control) − (+5hmC)/(−control) − (no treatment)) × 100, where ‘−control’ is unglucosylated but MspI digested, ‘+5hmC’ is glucosylated and MspI digested, and no treatment is unglycosylated and not digested with MspI.
Statistical analysis
Differences in the frequency of blastocyst formation were determined by the chi-square test. Average total cell numbers in IVF control, injection control, and TET1 KO blastocysts were compared using one-way ANOVA, and Student’s t-test was used to determine the difference in the average total cell numbers in control and DMOG-treated blastocysts. To determine the difference in the ICM/TE ratio between control and TET1 KO or DMOG treatment embryos, Student’s t-test was used. Statistical calculations of TET enzyme activity, RT-qPCR, and locus-specific 5hmC levels were performed using Student’s t-test. For calculations, the statistical software GraphPad Prism was used. Differences with p < 0.05 were considered significant.
List of abbreviations
| TET | = | ten-eleven translocation |
| 5mC | = | 5-metylcytosine |
| 5hmC | = | 5-hydroxymethylcytosine |
| 5fC | = | 5-formylcytosine |
| 5caC | = | 5-carboxylcytosine |
| ES cell | = | embryonic stem cell |
| DMOG | = | dimethyloxalylglycine |
| ICM | = | inner cell mass |
| TE | = | trophectoderm |
| TDG | = | thymine DNA glycosylase |
| KO | = | knockout |
Author’s contributions
KU and KL designed experiments and wrote the manuscript. KU, JR, KF, and NW performed experiment and data analysis. KL supervised and obtained funding for the project. All authors read and approved the final manuscript.
Supplemental Material
Download PDF (207.3 KB)Disclosure statement
The authors declare that they have no conflict of interests.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21.
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254.
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–19.
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432.
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762.
- Dean W, Santos F, Stojkovic M, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98(24):13734–13738.
- Zaitseva I, Zaitsev S, Alenina N, et al. Dynamics of DNA-demethylation in early mouse and rat embryos developed in vivo and in vitro. Mol Reprod Dev. 2007;74(10):1255–1261.
- Park JS, Jeong YS, Shin ST, et al. Dynamic DNA methylation reprogramming: active demethylation and immediate remethylation in the male pronucleus of bovine zygotes. Dev Dyn. 2007;236(9):2523–2533.
- Mayer W, Niveleau A, Walter J, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502.
- Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10(8):475–478.
- Shen L, Inoue A, He J, et al. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15(4):459–471.
- Guo F, Li X, Liang D, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–459.
- Santos F, Hendrich B, Reik W, et al. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182.
- Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127(6):643–651.
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, NY). 2009;324(5929):930–935.
- Ito S, D’Alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133.
- He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307.
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286(41):35334–35338.
- Gu TP, Guo F, Yang H, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610.
- Iqbal K, Jin SG, Pfeifer GP, et al. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108(9):3642–3647.
- Koh KP, Yabuuchi A, Rao S, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213.
- Dawlaty MM, Ganz K, Powell BE, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9(2):166–175.
- Quivoron C, Couronne L, Della Valle V, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38.
- Ko M, Bandukwala HS, An J, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA. 2011;108(35):14566–14571.
- Dawlaty MM, Breiling A, Le T, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24(3):310–323.
- Dawlaty MM, Breiling A, Le T, et al. Loss of tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29(1):102–111.
- Kang YK, Koo DB, Park JS, et al. Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome. J Biol Chem. 2001;276(43):39980–39984.
- Archer GS, Dindot S, Friend TH, et al. Hierarchical phenotypic and epigenetic variation in cloned swine. Biol Reprod. 2003;69(2):430–436.
- Huang Y, Chavez L, Chang X, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111(4):1361–1366.
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13(1):28–35.
- Yamagata K, Yamazaki T, Miki H, et al. Centromeric DNA hypomethylation as an epigenetic signature discriminates between germ and somatic cell lineages. Dev Biol. 2007;312(1):419–426.
- Pastor WA, Pape UJ, Huang Y, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397.
- Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402.
- Wu H, D’Alessio AC, Ito S, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473(7347):389–393.
- Freudenberg JM, Ghosh S, Lackford BL, et al. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012;40(8):3364–3377.
- Williams K, Christensen J, Pedersen MT, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473(7347):343–348.
- Kang J, Lienhard M, Pastor WA, et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc Natl Acad Sci USA. 2015;112(31):E4236–45.
- Shi FT, Kim H, Lu W, et al. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288(29):20776–20784.
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97.
- Lu F, Liu Y, Jiang L, et al. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014;28(19):2103–2119.
- Lee K, Hamm J, Whitworth K, et al. Dynamics of TET family expression in porcine preimplantation embryos is related to zygotic genome activation and required for the maintenance of NANOG. Dev Biol. 2014;386(1):86–95.
- Prasad P, Mittal SA, Chongtham J, et al. Hypoxia-mediated epigenetic regulation of stemness in brain tumor cells. Stem Cells. 2017;35(6):1468–1478.
- Festuccia N, Osorno R, Halbritter F, et al. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11(4):477–490.
- Kuijk EW, Du Puy L, Van Tol HT, et al. Differences in early lineage segregation between mammals. Dev Dyn. 2008;237(4):918–927.
- Hall VJ, Christensen J, Gao Y, et al. Porcine pluripotency cell signaling develops from the inner cell mass to the epiblast during early development. Dev Dyn. 2009;238(8):2014–2024.
- Du Puy L, Lopes SM, Haagsman HP, et al. Analysis of co-expression of OCT4, NANOG and SOX2 in pluripotent cells of the porcine embryo, in vivo and in vitro. Theriogenology. 2011;75(3):513–526.
- Lei S, Ryu J, Wen K, et al. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci Rep. 2016;6:25222.
- Yugo DM, Heffron CL, Ryu J, et al. Infection dynamics of hepatitis E virus in wild-type and immunoglobulin heavy chain knockout JH (-/-) gnotobiotic piglets. J Virol. 2018;92(21). DOI:10.1128/JVI.01208-18.
- Whitworth KM, Lee K, Benne JA, et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91(3):78.
- Santos F, Hyslop L, Stojkovic P, et al. Evaluation of epigenetic marks in human embryos derived from IVF and ICSI. Hum Reprod. 2010;25(9):2387–2395.
- Deshmukh RS, Ostrup O, Ostrup E, et al. DNA methylation in porcine preimplantation embryos developed in vivo and produced by in vitro fertilization, parthenogenetic activation and somatic cell nuclear transfer. Epigenetics. 2011;6(2):177–187.
- Fulka J, Fulka H, Slavik T, et al. DNA methylation pattern in pig in vivo produced embryos. Histochem Cell Biol. 2006;126(2):213–217.
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–461.
- Prather RS. Nuclear control of early embryonic development in domestic pigs. J Reprod Fertil Suppl. 1993;48:17–29.
- Yoshioka K, Suzuki C, Tanaka A, et al. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod. 2002;66(1):112–119.
