ABSTRACT
Psoriasis is an autoimmune skin disorder influenced by genetic, epigenetic and environmental factors. We previously found CYP2S1 intragenic DNA methylation cg19430423 site strongly hypomethylated in psoriatic skin tissues. In this study, we performed methylation loci fine-mapping to search the top signals in the entire CYP2S1 gene region, and further carried out gene expression assay, cell proliferation, apoptosis, differentiation and migration in CYP2S1 overexpressed (CYP2S1over) and silenced (siRNA) human keratinocytes. Target bisulphite conversion sequencing revealed cg19430423 and nearby two loci were the top differentially methylated loci. These three loci located within active enhancer region marked by H3K4Me1 and H3K27AC peaks. Cg19430423 might not bind with ATF1 directly. CYP2S1over repressed NHEK cell proliferation, but have no confirmed evidence on affecting migration, apoptosis and differentiation. Real-time PCR showed that CYP2S1 inhibited expression of IL1β, IL8, IL33, IL36, LL37, CXCL10 and CCL20 gene. In summary, CYP2S1 might inhibit keratinocyte proliferation, and modulate immune response through IL-8, IL-33, IL-36, CXCL-10, CCL20, thus contribute to the development of psoriasis.
Introduction
Psoriasis is an autoimmune skin disease characterized by typical erythematous plaques covered with thick and silvery scales. The exact aetiology of psoriasis remains essentially undetermined, but it is commonly considered to be mediated by T lymphocytes and various cytokines. The factors triggered psoriasis include genetic susceptibility, nutrition, high mental stress, infections, psychological stress, alcohol, smoking and some other environmental components [Citation1–3]. Recently, studies found epigenetic factors might be involved in disease development [Citation4,Citation5]. Epigenetic refers to inherited expression changes without accompanying DNA sequence changes. Although most of these epigenetic studies revealed relationship between disease and epigenetic makers, it is hard to judge whether these epigenetic modifications are the causes or results of disease symptom [Citation6]. To reveal mechanisms of target epigenetics locus, additional function analysis is needed.
DNA methylation (DNAm) is a type of epigenetic modification adding a methyl group to the fifth position of cytosine by DNA methyltransferase. Reversely, DNAm can be removed through the enzymes of TET family, making it balanced under specific biological situation [Citation7]. It is commonly believed that DNAm, especially those located at gene promoter regions, would modulate gene expression by affecting the binding of transcription factors [Citation8]. Even great progress has been made in epigenetic community, the role of intragenic DNAm is poorly addressed [Citation9,Citation10].
Cytochromes P450 (CYPs) enzymes play an important function in metabolizing toxic compounds, including exogenous drugs and endogenous metabolisms such as steroids, fatty acids, bilirubin, retinoic acid and prostaglandins (PGE2) [Citation11]. PGE2 is one of the most well-known cyclooxygenase-derived prostanoids, which activates migration and proliferation while inhibits apoptosis in human epithelial cells [Citation12]. CYP2S1 is one member of CYPs family. CYP2S1 mRNAs can be induced by dioxin in the Hepa-1 mouse hepatoma cell line, or by ultraviolet irradiation and all-trans retinoic acid in human skin tissue [Citation13]. Smith et al revealed CYP2S1 expression was 3.3 times higher in psoriatic skins when compared to paired non-lesional skins [Citation14]. This line of evidence supports the argument that CYP2S1 is critical for psoriasis development.
We previously characterized the genome-wide DNAm profiles for psoriatic and normal skin tissues [Citation15]. By globally comparing methylation levels of case and control groups, we found cg19430423, an intragenic DNAm locus within CYP2S1, was significantly demethylated in psoriatic groups. In the current study, we firstly fine-mapped DNAm loci across the entire gene region by target bisulphite methylation sequencing. To make further exploration, we overexpressed and silenced CYP2S1 in human normal keratinocyte cells (HNEKs), and revealed their function on cell proliferation, migration, differentiation and apoptosis. We also noticed expression changes for some psoriasis-associated marker genes in silenced- and overexpressed-CYP2S1 NHEKs. This study illustrated CYP2S1 might regulate proliferation and immune response thus be involved in psoriasis development.
Materials and methods
Target bisulphite sequencing
Overall, 43 psoriatic skin (PP) and 47 controls (NN) were collected from the Department of Dermatology, the First Affiliated Hospital, Anhui Medical University, Anhui Province, China. All patients were diagnosed by at least two experienced clinicians. Histopathological examination for each PP tissue further provided clinical assistant diagnosis. The study was approved by the research ethics committees of Anhui Medical University (Number: 2015007). Informed consent forms were signed by all participants.
Genomic DNA were extracted by DNeasy Blood and Tissue Kit (Qiagen, Duesseldorf, Germany) according to the manufacturer’s protocol. DNA methylation levels of target loci were quantified by MethylTarget sequencing (Genesky Biotechnologies Inc., Shanghai, China). Briefly, 500ng genomic DNA was subjected to bisulphite conversion using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, United States). A multiplex PCR was performed with optimized combination of primer sets (Supplementary Table 1). Sample libraries were polled together and sequenced by Illumina Hiseq 2000. BSseeker2 was utilized for analysing raw sequence data. For data filtering, we excluded methylation loci with < 98% bisulphite conversion efficacy and average sequence coverage < 20 ×. Student t-test was used to calculate methylation differences between groups.
Cell culture and reagents
293 T cells were maintained in DMEM (Gibco, Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS). NHEKs were cultured in DMEM supplemented with 10% FBS (1.5 mg/L Glutamine, 100 U/ml Penicillin, 100 ug/ml Streptomycin). All cells were cultured in humidified incubator at 37°C with 5% CO2. The lentivirus for overexpressing CYP2S1 (LV-Cyp2s1) and the control lentivirus were obtained commercially from GenePharma Corporation (Shanghai, China). Lentivirus carrying green fluorescent protein (GFP) were used as the control. 293 T cells were cultured in 15 cm dish with 18 ml DMEM with 10% FBS until 50–60% confluence. Next, 3 ml DMEM containing recombinant lentiviruses, control lentiviruses and RNAi Mate were added into plates (GenePharma Corporation, Shanghai, China). Each transfection system contains 6 μg/ml polybrene (Sigma). The virus-containing medium was replaced with fresh complete medium 48 hours later. The expression level of GFP was observed under a microscope after 3 days’ culture. Medium containing 2 μg/ml puromycin (Thermofisher Scientific, United States) was added every 3 days to screen the stable infected cell.
Cell proliferation, scratch wound healing, cell apoptosis and differentiation
For proliferation assays, 3,000 cells were plated in each well of 96-well plate. And 10ul Cell Counting Kit mix was added, then optical density was measured to determine cell activity at a wavelength of 450 nm at 0, 24, 48 and 72 h with microplate reader (PerkinElmer EnSpire).
Scratch wound assays were performed for wild type NHEKs and CYP2S1 overexpressed NHEKs when they reached 70–80% confluence. A scratch was created on monolayer cells by a 200ul tip, then cells were washed twice with PBS. Image were taken after 24 h and 48 h with ImageJ software. For cell apoptosis detection, cells were cultured for 24 h, then harvested and tested using Annexin V-PE/7AAD (Kaiji Biotechnology, Shanghai, China) according to the manufacturer’s instructions. Accuri C6 flow cytometer (BD Biosciences, San Jose, CA) was used to detect the apoptosis. For keratinocyte differentiation, we quantified expression level of KRT6 in CYP2S1 overexpressed- and silenced-cells.
Real-time PCR
Total RNA of NHEK cells was extracted using RNeasy Protect Mini Kit (Qiagen) according to the manufacturer’s protocols. Five micrograms of total RNA were transcribed into cDNA using the PrimeScriptTM RT reagent Kit (Takara) according to the manufacturer’s instructions. The following primers were used for the quantitative real time PCR (RT-PCR): CYP2S1-F: CTCTTCTTCACCACCATCCTACA; CYP2S1-R: ACCACTTCCAAGTTGCCTTC. GAPDH-F: CATGAGAAGTATGACAACAGCCT; GAPDH-R: AGTCCTTCCACGATACCAAAGT. IL1-F: CATTGAGCCTCATGCTCTGTT; IL1-R: CGCTGTCTGAGCGGATGAA. IL6-F: TACATCCTCGACGGCATCTCAG. IL6-R: TGCACAGCTCTGGCTTGTTCC. IL8-F: CTGTGTGAAGGTGCAGTTTTGCC. IL8-R: CGCAGTGTGGTCCACTCTCAATC. IL23A-F: GGTGAACAACTGAGGGAACCAAA. IL23A-R: GCAGCAGCAACAGCAGCATTAC. IL33-F: GCCTGTCAACAGCAGTCTACTG. IL33-R: TGTGCTTAGAGAAGCAAGATACTC. IL36-F: CTGGAGCCACGATTCAGTCC. IL36-R: AGGGTCCACACTTGCTGATTC. LL37-F: AGGTCCTCAGCTACAAGGAAG. LL37-R: TCTTGAAGTCACAATCCTCTGGT. CXCL1-F: CTCACTGGTGGCTGTTCCTG; CXCL1-R: TCTCCTAAGCGATGCTCAAA. CXCL10-F: GTGGCATTCAAGGAGTACCTC. CXCL10-R: TGATGGCCTTCGATTCTGGATT. CCL5-F: CCAGCAGTCGTCTTTGTCAC. CCL5-R: CTCTGGGTTGGCACACACTT. CCL20-F: TGTCAGTGCTGCTACTCCAC. CCL20-R: GATTTGCGCACACAGACAAC. CK17-F: GGAGATTGCCACCTACCGC; CK17-R: TTGCCATCCTGGACCTCTT. TNF-α-F: CCGAGTCTGGGCAGGTCTA; TNF-α-R: GCGTTTGGGAAGGTTGGAT. S100A8-F: GTTGACCGAGCTGGAGAAAG; S100A8-R: CCTGTAGACGGCATGGAAA. Quantitative real time PCRs were performed using SYBR Premix Ex Taq II (Takara) according to the manufacturer’s instructions on ABI 7900 PCR system (Applied Biosystem). The thermal profile included 95°C for 3 min, 40 cycles of denaturation at 95°C for 30s and annealing at 62°C for 40s.
Immunofluorescence staining of CYP2S1
Skin biopsies were obtained from psoriatic skin, and then fixed in formaldehyde and embedded in paraffin to obtain serial 4-μm thick sections by pathology slides scanners (No. 1802642S, Pannoramic MiDi, 3DHISTECH Ltd.). Sections were then deparaffinized through xylene and graded ethanol to distilled water. Slides were undergone antigen retrieval in boiling 10 mM sodium citrate buffer (pH 6) in a microwave oven for 10 min. Non-Specific proteins were blocked with 100 μl of 2% bovine serum albumin (BSA)/phosphate buffered saline (PBS) (pH 7.6) for 1 h. Then sections were incubated overnight in a 1: 500 dilution of rabbit monoclonal anti-CYP2S1 antibody (Thermo-scientific). On the next day, sections were washed in PBS and stained with Alexa Fluor 488–conjugated anti–rabbit IgG (Thermo-scientific, United States) at 1:1000 for 2 h in the dark at room temperature. Sections were washed with PBS and Nuclei were stained with DAPI (1:1000) and mounted for imaging. The images were examined using an Olympus–Ix Microscope and merged using Image J Software.
Results
Fine-mapping of DNAm across entire CYP2S1 gene region
Based on Illumina 450 k Chip array, we found the average methylation level of cg19430423 in psoriatic skins was 36% lower than that in controls. This intragenic DNAm locus resides at about 2.5kb downstream of CYP2S1 promoter. To test whether significance of other loci would exceed cg19430423, we selected eight target regions, covering promoter, introns, exons with an average distance 1.5kb between each other, for further validation ().
Figure 1. Target methylation sequencing for entire CYP2S1 gene region. (a) All CpG loci covered by Illumina 450 k bead array. Cases and controls are presented by two different colour of lines. Solid lines represent Mean Value plot, and the dash lines represent the Loess line. (b) CYP2S1 gene context, position of array-covered loci and bisulphite target regions are depicted in the scheme. (c) Average methylation level for 46 loci are shown separately in case and control groups
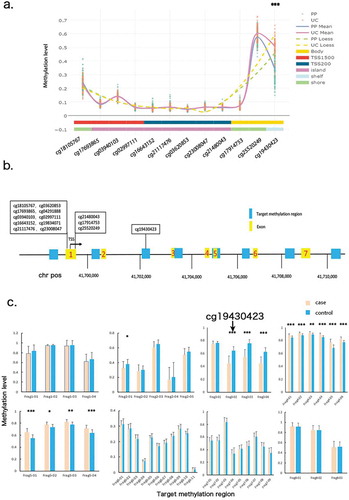
Forty-three cases and forty-seven sex and age matched controls were enrolled for target bisulphite convert sequencing, with an average of 36,850 clean reads and 99% methylation transferring efficiency for each sample. A total of 46 methylation loci were detected for the eight selected regions thus entered into final investigation. At a Bonferroni-corrected logistic regression test P-value < 0.01, twelve loci differed significantly between the two groups (Supplementary Table 1). We confirmed cg19430423 was the differentially hypomethylated locus in case group (Pcorrected = 5.6E-05). Meanwhile, the other two loci, located at 29bp and 35bp downstream of cg19430423, also showed strong hypomethylation, suggesting these loci might execute functions coordinately (). We term the three loci as cg19430423-block. Even some loci exceeded the significant threshold in the target regions, we reckoned cg19430423-block was the most top signal for two reasons. First, the mean methylation difference in cg19430423-block loci was much greater than that in the other loci. Consistent with our previous findings, cg19430423 methylation level of psoriatic tissue was 31% lower than that of control group. Second, when Area Under Curve (AUC) was used to predict sensitivity, specificity and accuracy of the model, we observed block loci performed better than others (Supplementary Table 1). In summary, through target methylation sequencing, we validated that cg19430423 and nearby loci were significantly hypomethylated.
Cg19430423-block might not directly regulate gene expression
We performed real time PCR to detect mRNA expression in 20 psoriatic (PP) and 20 paired non-lesional (PN) skin tissues. CYP2S1 were strongly upregulated in psoriatic tissues, consistent with published transcriptome microarray data from Elder JT’s team [Citation16] (). CYP2S1 upregulation in PP samples were confirmed in protein level assayed by immunohistochemistry (). We also noticed strong negative correlation between gene expression and cg19430423 (ρ = −0.56, Pcorrelation = 3.4E-06), suggesting cg19430423 might be a regulator for gene expression [Citation15]. Furthermore, cg19430423-block located in a transcriptional enhancer region marked by histone modification H3K4Me1 and H3K27AC peaks, further suggesting cg19430423 regulates enhancer activities ().
Figure 2. CYP2S1 gene expression in psoriatic tissues. (a)Validation of CYP2S1 gene expression with RT-PCR and public datasets. (b) Immunohistochemistry (IHC) revealed upregulation in psoriatic tissues. (c) Cg19430423 located within binding motif of ATF1 and ARG80 transcription factor. (d) Cg19430423 within active enhancer marked by H3K4Me1 and H3K27AC peaks. (e) Immunofluorescence (IF) indicated CYP2S1 expressed in cytoplasma of both basal and suprabasal keratinocytes
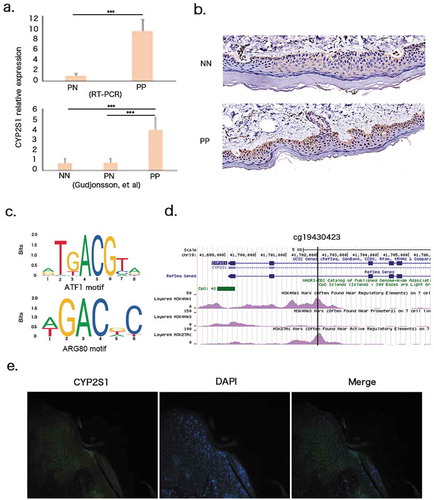
These clues triggered us to wander whether cg19430423-block directly control CYP2S1 expression. Cg19430423-block DNA sequence should be bound by transcription factor or proteins so as to be recognized to modulate RNA synthesis. We queried JASPAR transcription factor DNA binding databases and found cg19430423 lay on the core of binding motif for ATF1 and ARG80 (). ATF1 has been implicated in inducing chemokines production in keratinocytes, indicating a potential role in modulating immune response [Citation17]. We selected ATF1 for luciferase report assay in order to check whether DNA methylation of cg19430423-block affecting binding efficacy of ATF1. 293 T cells were cotransfected with ATF1 expression vector and pGL3-promoter vector fused to luciferase mRNAs. We set two different scenarios in which pGL3-promoter vector contains cg19430423-block wildtype/mutant sequence or treated with decitabine or not. No significant differences of luciferase signals were found in both situations when compared with normal controls, suggesting cg19430423-block might indirectly influencing binding of ATF1 (data not shown). Further investigations were needed to evaluate other transcription factors that might be involved in motif binding.
To find out whether CYP2S1 gene expressed in epidermal basal or suprabasal keratinocytes, we performed immunofluorescence staining in psoriatic tissue, and noticed CYP2S1 mainly expressed in cytoplasma of both basal and suprabasal layers (). We also queried mouse cell atlas database in which about 400,000 single cells were extensively portraited by single cell transcriptome analysis. In that study, nearly 4,000 skin cells were classified into 24 subgroups. It has been reported that CYP2S1 gene mainly expressed in keratinocyte subgroup expressing KRT5, KRT10, KRT15, consistent with our findings [Citation18]. In summary, cg19430423 was negatively correlated with CYP2S1 expression in human skin cells.
CYP2S1 might inhibit cell proliferation
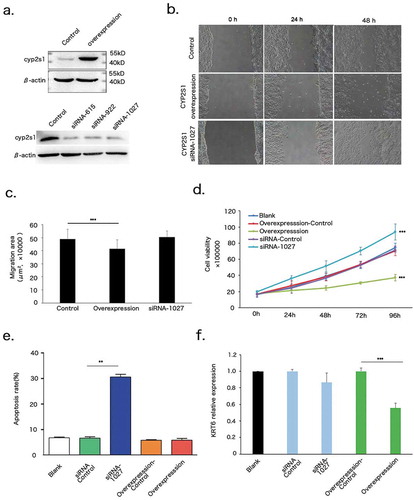
CYP2S1 gene activity has been implicated in Bronchial Epithelial BEAS-2B and Human colorectal cancer HCT116 cell line [Citation19,Citation20]. However, the specific function of CYP2S1 in human keratinocytes remains undefined. To test whether CYP2S1 influence morphology of keratinocytes, we constructed CYP2S1 overexpression (CYP2S1over) and silencing NHEK cells by using lentivirus system. CYP2S1over slightly inhibit cell proliferation and migration when compared with control group (). When CYP2S1 was silenced by siRNA-1027, with the strongest silencing effect of the three tested siRNAs, keratinocyte migration was not altered significantly, but proliferation was dramatically increased (). Meanwhile, cell apoptosis was increased in siRNA-1027 transfected- but not in CYP2S1over cells (). Krt6 is a key differentiation maker for keratinocyte. We found decreased expression of Krt6 in CYP2S1over cell lines, but no significant alteration was observed in siRNA-1027 cells (). These data suggested CYP2S1 inhibited keratinocyte proliferation, supported by both overexpressed- and silenced-CYP2S1 cells.
Figure 3. CYP2S1 repressed proliferation of NHEKs. (a) Overexpression and silencing model of CYP2S1 gene in NHEKs by using lentivirus system. (b) Migration assay for control, CYP2S1over and siRNA-1027. (c), Migration assay showed decreased area of confluent cell migration at 48 h in CYP2S1over NHEKs. (d) Proliferation analysis indicated significantly decreased proliferation for CYP2S1over, increased proliferation for siRNA-1027 induced cells when compared with normal controls at 96 h. (e) Apoptosis assay suggested siRNA-1027 cells promote cell apoptosis. (f) KRT6 expression was decreased in CYP2S1over cells. (n = 3, bar represents mean ± sem, ***P < 0.001)
CYP2S1 induce aberrant immune response
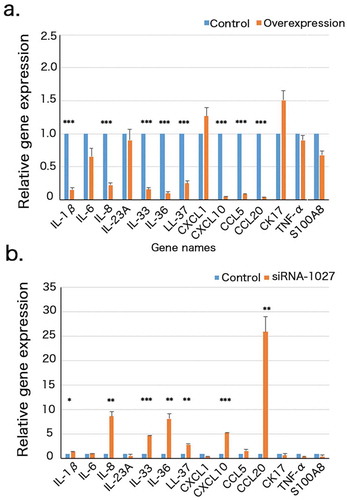
Given its upregulation in psoriatic tissues, we reasoned that CYP2S1 might be involved in cutaneous immune response. Psoriasis is considered to be a typical Th17-induced inflammation skin disease. Several lines of evidence suggested transcriptional or aberrant cellular signalling pathway of keratinocyte might trigger psoriasis [Citation21]. We mainly focused on chemokines, cytokines or some other psoriasis-related genes that have been confirmed by many groups. Fourteen genes (IL1β, IL6, IL8, IL33, IL36, CXCL10, CCL5, CCL20, LL37, CK17, CXCL1, S100A8, TNF-α, IL23A) were selected for RT-PCR analysis (). We found expression of IL1β, IL8, IL33, IL36, LL37, CXCL10, CCL20 were upregulated in siRNA-1027 silenced but downregulated in overexpressed keratinocytes (Pcorrected < 0.05, two tailed student t-test). TNF-α was decreased in siRNA-1027 silenced cells but did not altered significantly in overexpressed cells. These data suggested CYP2S1 might modulate expression of psoriasis-associated chemokines or cytokines.
Figure 4. Gene expression of 14 psoriatic-associated chemokines or cytokines in CYP2S1 overexpressed (CYP2S1over) and silenced (siRNA) NHEKs. (a). RT-PCR revealed significant downregulation of IL1β, IL6, IL8, IL33, IL36, CXCL10, CCL5, CCL20, LL37, CK17, IL23A and upregulation of CK17 and IL23A in CYP2S1over NHEKs. (b). RT-PCR suggested upregulation of IL1β, IL8, IL33, IL36, CXCL10, CCL20, LL37 siRNA NHEKs. (n = 3, bar represents mean ± sem, *P < 0.05, **P < 0.01, ***P < 0.001)
Discussion
In this study, we first validated our previous finding that cg19430423 was significantly hypomethylated in psoriatic skin tissue. Using fine-mapping strategy, we found cg19430423 and 35bp nearby loci were the strongest signals that deserved further investigation. Furthermore, cg19430423 lies at a keratinocyte-specific enhancer marked by H3K4me1 and H3K27AC peaks. CYP2S1 represses the proliferation of NHEK, and might cause aberrant expression of certain immune-related genes. This study provided some evidences that CYP2S1 might participated in the progress of psoriasis.
Promoter DNAm has been extensively studied in various cell lines, however, intragenic DNAm is less addressed. In this study, we used eight target sequencing primers covering the entire CYP2S1 region to evaluate methylation difference between cases and controls. Three nearby loci at the intragenic region, termed cg19430423-block, were discovered, which resided within the predicted ATF1 and ARG80 binding sites. ATF1, also known as activating transcription activator 1, has been implicated in tumour immune surveillance, cell proliferation and migration [Citation22]. Luciferase report assay showed that ATF1 regulate CYP2S1 expression by binding cg19430423-block indirectly. However, due to strong negative correlation between methylation and CYP2S1 gene expression, we could not exclude other possibilities that cg19430423-block modulated expression level. For example, ATF1 or some other transcription factors interacted with methylation binding proteins and formed a complex, whose activities might largely depend on the expression level of the methylation sensitive protein. In another case, ATF1 has the capacity to bind methylation block, and this intermediate state needs further cofactors, in order to interact with CYP2S1 promoter. Whether cg19430423-block mediate expression of CYP2S1 need further investigation.
DNAm may act as a cause or arise as a consequence of the disease. Thus, it is important to figure out the real function of DNAm in regulating psoriasis. Psoriasis is characterized by abnormal and excessive proliferation of basal keratinocytes. However, our in vitro study showed that CYP2S1 inhibited NHEK proliferation, suggesting CYP2S1 upregulation might be the consequence rather than the cause of disease manifestation. We suspected cg19430423 demethylation might be a kind of self-protection against keratinocyte hyperproliferation. In line with this hypothesis, we previously performed causal inference test for all the psoriasis-related DNAm loci, but failed to detect any causal effect for cg19430423 [Citation6].
CYP2S1 gene is an extrahepatic cytochrome P450 that constitutively or inducibly expressed in lung, spleen, skin and some other tissues [Citation23]. CYP2S1 expression can be induced by UV radiation (UVR) and topical drugs, including coal tar and all-trans retinoic acid (atRA). The precise mechanisms through which UVR induces CYP2S1 expression are still unknown. It is believed that UV-induced derivatives act as aryl hydrocarbon receptor (AhR) agonists, suggesting that induction may be mediated via the AhR pathway [Citation20]. The AhR is a ligand-activated transcription factor involved in the regulation of CYP2S1 enzyme induction. We are not sure whether AhR execute its function through binding demethylated cg19430423, however, some evidence supported the notion that AhR can bind to CYP gene enhancer in a methylation sensitive manner. Oesch-Bartlomowicz et al reported CpG methylation patterns within the dioxin responsive element (DRE) of the CYP1A1 enhancer would affect the recognition and binding efficiency of TCDD-liganded AhR complex. They carried out Electrophoretic Mobility Shift Assay (EMSA) and found methylated guanine strongly interfered with the binding of TCDD-liganded AhR to its binding motif [Citation24].
There are clear evidences that treatment with AtRA has a positive effect on alleviating psoriasis severity. Meanwhile, individual difference of clinical therapeutic effect is observed. We suspected one possible reason lay in the individual variation of CYP2S1 level. AtRA is not only the source to induce CYP2S1, but its synthesis and metabolism can also be mediated by CYP2S1, suggesting CYP2S1 plays an important function in both metabolism and disease development. We surmised that upregulated CYP2S1 might influence psoriasis mainly in two possible ways. Firstly, CYP2S1 might inhibit immune response as indicated by repressing the expression of IL1β, IL8, IL33, IL36, LL37, CXCL10, CCL20 in NHEK. These chemokines or cytokines are key mediator for psoriasis development. CCL20, with the most significant change in both overexpressed and silenced cells, is the only receptor for CCR6. Several studies revealed that CCL20/CCR6 axis played a critical role in recruiting Th17 cells to the epidermis, which was involved in the maintenance of IL23/Th17 signalling pathway [Citation25,Citation26]. Secondly, CYP2S1 might inhibit cell proliferation. CYP2S1 metabolizes cyclooxygenase (COX)-derived prostaglandin G2 (PGG2) in a NADPH P450-reductase-independent manner [Citation19]. PGGs can be used to synthesize numerous prostanoid, including PGE2. PGE2 further promotes proliferation and migration of many cell types, like fibroblasts, keratinocyte, cutaneous melanoma cells and among others [Citation12,Citation27,Citation28].
In this study, we found that CYP2S1 intragenic methylation locus cg19430423 was hypomethylated in psoriatic tissues. CYP2S1 might contributed to psoriasis by both inhibit proliferation and immune response of keratinocyte.
Supplemental Material
Download MS Excel (13.4 KB)Acknowledgments
The authors wish to thank all the subjects and their families for their participation in this study and the Center for Scientific Research of Anhui Medical University for valuable help in our experiment.
Disclosure statement
The authors have no conflict of interest to declare.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Prieto-Perez R, Cabaleiro T, Dauden E, et al. Genetics of psoriasis and pharmacogenetics of biological drugs. Autoimmune Dis. 2013;2013:613086.
- Yang EJ, Beck KM, Sanchez IM, et al. The impact of genital psoriasis on quality of life: a systematic review. Psoriasis (Auckl). 2018;8:41–47.
- Takeshita J, Grewal S, Langan SM, et al. Psoriasis and comorbid diseases: epidemiology. J Am Acad Dermatol. 2017;76:377–390.
- Chandra A, Senapati S, Roy S, et al. Epigenome-wide DNA methylation regulates cardinal pathological features of psoriasis. Clin Epigenetics. 2018;10:108.
- Zhang P, Zhao M, Liang G, et al. Whole-genome DNA methylation in skin lesions from patients with psoriasis vulgaris. J Autoimmun. 2013;41:17–24.
- Zhou F, Shen C, Xu J, et al. Epigenome-wide association data implicates DNA methylation-mediated genetic risk in psoriasis. Clin Epigenetics. 2016;8:131.
- Luo C, Hajkova P, Ecker JR. Dynamic DNA methylation: in the right place at the right time. Science. 2018;361:1336–1340.
- Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326.
- Kulis M, Queiros AC, Beekman R, et al. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. 2013;1829:1161–1174.
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492.
- Deb S, Bandiera SM. Characterization and expression of extrahepatic CYP2S1. Expert Opin Drug Metab Toxicol. 2009;5:367–380.
- Sawada Y, Honda T, Nakamizo S, et al. Prostaglandin E2 (PGE2)-EP2 signaling negatively regulates murine atopic dermatitis-like skin inflammation by suppressing thymic stromal lymphopoietin expression. J Allergy Clin Immunol. 2019;144:1265–1273 e1269.
- Bebenek IG, Solaimani P, Bui P, et al. CYP2S1 is negatively regulated by corticosteroids in human cell lines. Toxicol Lett. 2012;209:30–34.
- Smith G, Wolf CR, Deeni YY, et al. Cutaneous expression of cytochrome P450 CYP2S1: individuality in regulation by therapeutic agents for psoriasis and other skin diseases. Lancet. 2003;361:1336–1343.
- Zhou F, Wang W, Shen C, et al. Epigenome-wide association analysis identified nine skin DNA methylation loci for psoriasis. J Invest Dermatol. 2016;136:779–787.
- Gudjonsson JE, Ding J, Johnston A, et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol. 2010;130:1829–1840.
- Nijnik A, Pistolic J, Filewod NC, et al. Signaling pathways mediating chemokine induction in keratinocytes by cathelicidin LL-37 and flagellin. J Innate Immun. 2012;4:377–386.
- Han X, Wang R, Zhou Y, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–1107 e1017.
- Madanayake TW, Fidler TP, Fresquez TM, et al. Cytochrome P450 2S1 depletion enhances cell proliferation and migration in bronchial epithelial cells, in part, through modulation of prostaglandin E(2) synthesis. Drug Metab Dispos. 2012;40:2119–2125.
- McNeilly AD, Woods JA, Ibbotson SH, et al. Characterization of a human keratinocyte HaCaT cell line model to study the regulation of CYP2S1. Drug Metab Dispos. 2012;40:283–289.
- Dainichi T, Kitoh A, Otsuka A, et al. The epithelial immune microenvironment (EIME) in atopic dermatitis and psoriasis. Nat Immunol. 2018;19:1286–1298.
- Tian J, Chang J, Gong J, et al. Systematic functional interrogation of genes in GWAS loci identified ATF1 as a key driver in colorectal cancer modulated by a promoter-enhancer interaction. Am J Hum Genet. 2019;105:29–47.
- Yan P, Eng OC, Yu CJA. Review on the expression and metabolic features of orphan human cytochrome P450 2S1 (CYP2S1). Curr Drug Metab. 2018;19:917–929.
- Oesch-Bartlomowicz B, Janssen K, Wiss O, et al. Guanine 6-O-methylation pattern within the dioxin responsive element of the CYP1A1 enhancer shows two critical guanines for AhR/ARNT binding. Chem Biodivers. 2004;1:473–480.
- Kim TG, Jee H, Fuentes-Duculan J, et al. Dermal clusters of mature dendritic cells and T cells are associated with the CCL20/CCR6 chemokine system in chronic psoriasis. J Invest Dermatol. 2014;134:1462–1465.
- Hedrick MN, Lonsdorf AS, Shirakawa A-K, et al. CCR6 is required for IL-23-induced psoriasis-like inflammation in mice. J Clin Invest. 2009;119:2317–2329.
- Saalbach A, Janik T, Busch M, et al. Fibroblasts support migration of monocyte-derived dendritic cells by secretion of PGE2 and MMP-1. Exp Dermatol. 2015;24:598–604.
- Kim SH, Roszik J, Cho S-N, et al. The COX2 effector microsomal PGE2 synthase 1 is a regulator of immunosuppression in cutaneous melanoma. Clin Cancer Res. 2019;25:1650–1663.
