ABSTRACT
Prenatal exposure to endocrine disrupting chemicals can interfere with development, and has been associated with social-cognitive functioning and adverse health outcomes later in life. Exposure-associated changes of DNA methylation (DNAm) patterns have been suggested as a possible mediator of this relationship. This study investigated whether prenatal low-dose exposure to polychlorinated biphenyls (PCBs) and polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs) is associated with altered DNAm patterns across the genome in a Western urban-industrial population. In 142 mother-infant pairs from the Duisburg Birth Cohort Study, PCBs and PCDD/Fs levels were quantified from maternal blood during late pregnancy and associated with DNAm levels in cord blood using the Illumina EPIC beadchip. The epigenome-wide association studies (EWAS) identified 32 significantly differentially methylated positions (DMPs) and eight differentially methylated regions (DMRs) associated with six congeners of PCB and PCDD in females or males (FDRs < 0.05). DMPs and DMRs mapped to genes involved in neurodevelopment, gene regulation, and immune functioning. Weighted gene correlation network analysis (WGCNA) showed 31 co-methylated modules (FDRs < 0.05) associated with one congener of PCDF levels in females. Results of both analytical strategies indicate that prenatal exposure to PCBs and PCDD/Fs is associated with altered DNAm of genes involved in neurodevelopment, gene expression and immune functioning. DNAm and gene expression levels of several of these genes were previously associated with EDC exposure in rodent models. Follow-up studies will clarify whether these epigenetic changes might contribute to the origin for adverse mental and health outcomes.
Background
The developing foetus is highly sensitive to perturbations in the internal and external environment [Citation1]. Foetal exposure to a range of environmental influences, including maternal subnutrition [Citation2,Citation3], maternal psychosocial stress [Citation4,Citation5], or exposure to chemicals [Citation6] has been associated with lifelong consequences for the developing organism [Citation7–9]. There is a large body of literature investigating the long-term effects of prenatal exposure to endocrine disrupting chemicals (EDCs) on physiological and psychological functioning [Citation10]. EDCs, including polychlorinated biphenyls (PCBs), polychlorinated dibenzo-p-dioxins and dibenzofurans (PCDD/Fs), are widely dispersed, environmentally persistent organic pollutants (POPs) that can act as exogenous agents interfering with endocrine processes [Citation11]. PCDD/Fs, also known as dioxins, and PCBs are complex mixtures of congeners of polychlorinated organic compounds that share a common molecule origin and structure but vary in the number and location of chlorine atoms introducing differential functioning of each congener [Citation12,Citation13]. Since 1929, PCBs have been synthesized and used as synthetic oils for various industrial deployments such as in surface coating or in electrical apparatus as dielectric and coolant fluids until the late 1970s. PCDD/Fs are inadvertent by-products of synthetic processes such as smelting, chlorine bleaching of paper, or manufacturing of several herbicides and pesticides. Due to their chemical lipophilic characteristics and a generally long half-life, PCBs and PCDD/Fs persist in widely geographically dispersed environments over long periods of time and are able to bioaccumulate in the fatty tissue of humans and wildlife [Citation11].
The exposure to EDCs has decreased in the general population over the last years [Citation14]. Nevertheless, exposure to PCBs and PCDD/Fs below toxic level – so called low-dose exposure – is still of great concern, since exposure to even low doses can be harmful for the developing organism [Citation15]. There is empirical evidence for adverse effects of prenatal low-dose exposure to PCBs and PCDD/Fs on cognitive [Citation16–18] and motor development [Citation18–20], pubertal development [Citation21,Citation22] as well as gender-typical play behaviour [Citation23], attention deficits [Citation24,Citation25] and autistic behaviour [Citation26,Citation27].
The observation of such long-term influences of foetal exposures has led to the hypothesis that epigenetic mechanisms might explain how these effects retain their stability [Citation28]. The most extensively studied epigenetic mark in humans is DNA methylation (DNAm), a chemical modification of the DNA with gene regulatory function [Citation29]. During foetal development, DNAm is involved in the orchestration of developmental programming processes and is essential for normal cellular development and differentiation [Citation30]. Perturbations of DNAm processes through exogenous factors such as chemicals are thought to underlie structural and functional changes in cells, tissues, and organ systems, increasing disease risk across the lifespan (developmental origin of health and disease; DOHaD) [Citation7,Citation31,Citation32].
So far, it has rarely been investigated in humans whether prenatal exposure to PCBs or PCDD/Fs might be associated with altered DNAm [Citation28,Citation33,Citation34]. First empirical insights into this question come from animal models that have shown global DNAm changes after prenatal exposure to PCBs or PCDD/Fs [e.g., Citation35,Citation36]. Findings from animal models have been partly confirmed in humans by very few studies, reporting associations between exposure to PCBs or PCDD/Fs and global DNAm, which was estimated via the Alu and LINE1 assay [Citation37,Citation38].
The first epigenome-wide association study (EWAS) of prenatal exposure to various PCB congeners was realized in a cohort of Faroes individuals (N = 72) using the Infinium Methylation 450k chip [Citation33]. These individuals were exposed to high levels of PCBs, much higher than other Western populations, due to Faroes traditional dietary habits including whale meat and blubber. Leung et al. [Citation33] showed that higher doses of single PCB congeners were significantly associated with DNAm changes in cord blood in males and females. Among the investigated PCB congeners, mainly PCB congener 105 (PCB #105) was significantly associated with differential DNAm in sex-combined analysis. Notably, in a sex-stratified analysis, DNAm changes were significantly associated with PCB #105 exposure in females only, whereas in males, literally no single PCB congener remained significantly associated with DNAm changes.
Lastly, studies investigating other EDCs showed gene-specific or global DNAm changes after prenatal exposure to EDCs. For instance, significant associations were found between phthalates and gene-specific DNAm changes in Mexican-American newborns [Citation39] as well as between bisphenol A (BPA) and gene-specific DNAm changes in the Michigan Mother-Infant Pairs birth cohort [Citation40]. Also, prenatal exposure to phthalates revealed significant associations with epigenome-wide DNAm marks in placental tissues [Citation41].
Taken together, since prenatal low-dose exposure to EDCs including PCBs and PCDD/Fs seems to be especially harmful for the developing organism [Citation11], and has been associated with developmental sequelae, we aimed to explore whether prenatal low-dose exposure to PCBs and PCDD/Fs is associated with specific DNAm patterns in a Western urban-industrial population. In the Duisburg Birth Cohort, where maternal blood levels of PCBs and PCDD/Fs were objectively quantified during late pregnancy, DNAm in cord blood was quantified at around 850,000 sites across the genome with the Illumina Methylation EPIC beadchip. Based on previous results reporting sex-specific associations between EDC exposure and DNAm patterns, we hypothesized sex-specific effects and conducted sex stratified analyses.
Methods
Study area and participants
The Duisburg Birth Cohort study was conducted in Duisburg, Germany after one ton of dioxinated dust leaked from a recycling firm in 1999. Duisburg belongs to the Ruhr District, an industrial conurbation in Western Germany. A detailed description of the study area and study population is given in Wilhelm et al. [Citation42]. Initially, 232 healthy mothers from Duisburg aged between 18 and 42 years who became pregnant after the leakage were recruited between 2000 and 2002. During the third trimester of pregnancy, the concentration levels of PCBs and PCDD/Fs (reflecting cumulative exposure from various potential EDC sources) were assessed from maternal blood. The mother´s children have been followed from birth until today almost annually. Healthy mother-child pairs were included if they met the following criteria: German or Turkish as a first language, born at term (weeks 38–42 of pregnancy), with an APGAR score of at least 8, of parity 1–3, without serious complications or illnesses during pregnancy or parturition and without congenital anomalies. Since 2007, regular follow-up studies have been conducted to examine child development. See additional file 1 supplementary table (ST) 1 and 2 for descriptive data on neonate, mother and pregnancy characteristics.
Ethics approval and consent to participate
The Duisburg Birth Cohort study was approved by the Ethics Committee of the Medical Faculty of the Ruhr-University Bochum (registry no. 1478); follow-up was approved by the Ethics Committees of the Faculty of Psychology of the Ruhr-University Bochum in 2016 (No. 20160126). The study was conducted in accordance with the ethical principles for medical research involving human subjects as defined by the Declaration of Helsinki. All parents gave written informed consent.
Collection and analysis of maternal blood samples
To assess the concentration levels of the congeners of PCB and PCDD/F, maternal blood samples (50 mL, N = 226) were taken between the 28th and 42nd week of pregnancy (median: 32 weeks). In seven cases, blood samples were taken during the first 38 days after birth. Blood samples were stored at −80°C until analysis. Analysis of PCBs and PCDD/Fs in maternal blood was performed at the Department of Hygiene of the Ruhr-University Bochum by standard procedure with capillary gas chromatography and high-resolution mass spectrometry described in Wittsiepe et al. [Citation43]. Measured concentrations of single congeners of PCBs and PCDD/Fs were relativized on 0.5 limit of detection (<1 pg/g on lipid base). The measured levels of PCBs and PCDD/Fs have been classified as low-dose by comparison to reference levels of the human biomonitoring data from Germany matching sampling years and participant’s age [Citation43].
Sixteen congeners of PCBs and PCDD/Fs with concentrations higher than the limit of detection in at least 75% of the samples were included in the data analysis (congener list in additional file 1 ST1). Concentration of single congeners of PCBs and PCDD/Fs were measured in natural logarithm transformed (nat.log.) pg/g lipid base.
DNA isolation and genome-wide methylation assessment
Genomic DNA was isolated from umbilical cord blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden; Germany). Cord blood was taken directly after birth in the obstetrical clinic and stored at −80°C until DNA extraction and further analysis. DNA bisulphite conversion and assessment of genome-wide DNAm levels at 850,000 CpG sites using the Infinium HumanMethylationEPIC BeadChip array (Illumina, San Diego, CA, USA) were performed according to the manufacturer’s protocol at the Life&Brain Core Facility (University of Bonn, Germany).
Data analysis
All data analyses were performed with R [Citation44] using RStudio 3.6.1 (except for WGCNA using RStudio 3.3.4). The supporting R-code of all data analyses is available at https://github.com/kattamatta/duisburgbirthcohortstudy.
DNA methylation data preprocessing
Raw Infinium EPIC data was preprocessed using the pipeline of the RnBeads package [Citation45,Citation46] for intensity summarization and calculation of methylation ratio (beta values). The preprocessing steps included primary quality control as well as a filtering step before and after normalization. Seven samples with low bisulphite conversion efficiency compared to control probes were removed, as well as one of two samples with identical SNP methylation patterns. Moreover, six samples that did not match a priori criteria (Caucasian ethnicity as stated by the participant’s country of origin, no missing data regarding congers and sex of neonate) were excluded. During the first filtering step, cross-reactive and SNP-enriched probes overlapping with at least three bases in the target sequence with annotated SNPs were eliminated. Low-quality probes were removed using RnBeads´ Greedycut algorithm with default settings. Normalization was conducted using the dasen method from the wateRmelon R-package [Citation47] implemented in RnBeads. For the second filtering step, non-CpG probes (i.e., CC, CAG, CAH, CTG and CTH) and CpGs containing missing beta values were excluded. CpGs on the X chromosome for male and female neonates and Y chromosomes for male neonates were not filtered to conduct sex-stratified analyses, since recent research [Citation13,Citation33] has indicated that epigenetic marks due to prenatal exposure to EDCs might be sex-dependent. The processing procedure lead to a final set of 760,403 CpGs for female samples and 760,464 CpGs for male samples. Final quality control revealed three samples with distinct genome-wide DNAm patterns using multidimensional scaling plots of M values (related to beta values by logit transformation) and three samples mismatching known and predicted sex and were removed. This led to a total of 142 samples (73 females, 69 males).
Assessment of cell-type heterogeneity
To account for cell-type heterogeneity in the DNAm profiles, the reference-based approach by Houseman et al. [Citation48] implemented in RnBeads was used with default settings to estimate cell-type compositions from the assessed DNAm profiles. There has been only one reference panel available for cord blood assessed with the EPIC Infinium array from 83 Asian neonates including CD4T, CD8T, natural killer cells, B cells, granulocytes, and monocytes (nucleated red blood cells were not available) [Citation49]. Thus, DNAm profiles of this reference panel were used and processed together with the target DNAm profiles to estimate the relative contribution of six cell types in the target profiles. The six estimated cell-type proportions of CD4T, CD8T, natural killer cells, B cells, granulocytes, and monocytes were included as covariates in the inferential analyses.
Epigenome-Wide Association Studies (EWAS)
A series of sex-stratified EWAS was conducted to identify DNAm sites and regions significantly associated with the single PCB and PCDD/F congeners. In a first set of EWAS, differentially methylated positions (DMPs) were assessed using limma [Citation50] by fitting multiple regression models in combination with outlier-robust empirical Bayes moderation for each retrieved CpG in M values as criterion and single PCB and PCDD/F congeners (nat.log. pg/g lipid base in maternal blood) as continuous predictor. For each congener an EWAS was conducted, since in general individual congeners are discussed to differ in the effects on health [Citation13]. Following this argument assuming independency of congener’s effects, the single congener might also have differential effects on DNAm. Additionally, a set of possible confounders (smoking status of mother during pregnancy [yes or no], age of mother at birth [years], gestational age [weeks], weight of neonate [g]), technical batch (EPIC assay plate [plate 1 or plate 2]) and the six estimated cell type contributions to account for cell-type heterogeneity were included in each regression model simultaneously. Since PCBs and PCDD/Fs assessed during pregnancy are generally known to be associated with pregnancy parameters, such as gestational length [Citation51] and birth weight [Citation52], interaction terms, were included in a model, when the regarding predictor was significantly (p < 0.01, additional file 1 ST3-ST5) correlated with an above listed covariate. For each regression model separately, outlier CpGs indicating hemimethylated sites were excluded to avoid a bias regarding false-positive associations due to outliers [Citation53].
Following EWAS that identified DMPs significantly associated with a particular congener of PCB or PCDD/F for the given sex, a second set of EWAS was implemented to identify differentially methylated regions (DMRs) associated with that same single PCB and PCDD/F congener (nat.log. pg/g lipid base from maternal blood). Thus, a separate model for each congener of PCB or PCDD/F significantly associated with a DMP in the given sex was conducted for this sex including the same confounders as in DMPs-models to identify DMRs. Each model to identify DMRs was separately implemented using dmrCate [Citation54] with default parameters. Significant DMRs were defined by a minimum of two significant CpGs in a genomic bin smaller than 1,000 nucleotides.
To gain insight into possible over-represented gene pathways in the identified significant DMPs and DMRs, gene ontology (GO) enrichment analyses were performed for these DMPs and DMRs in a sex-stratified manner using the R-package missMethyl [Citation55]. By taking into account all tested CpGs to correct for biases introduced due to differing numbers of probes per gene present on the EPIC array, the significant DMPs and DMRs were tested for GO term enrichment.
Weighted gene correlation network analysis (WGCNA)
To undertake a systems-level view of the DNAm data, weighted gene correlation network analysis (WGCNA) [Citation56] was applied to the top 50% of the ranked by median absolute deviation (MAD) preprocessed CpGs in a sex-stratified manner. WGCNA offers an alternative clustering strategy to the grouping approach of DMRs by employing network analysis to the beta values of CpGs. Using the R-package WGCNA [Citation56], scale-free correlation-based hierarchical clusters of the methylation profiles of CpGs were used to construct weighted co-methylation modules. These modules represent discrete networks of co-methylated CpGs. An unsigned network with outlier-robust bidweight midcorrelation and a soft-threshold for power adjacency transformation was chosen. For reasons of computational time, block-wise module detection of pre-clustered blocks and soft-threshold was applied on the preprocessed beta-values of the top 50% of CpGs ranked by MAD, including those on sex chromosomes for samples of each sex separately (female: 380,202 CpGs; male: 380,232 CpGs). The default procedure was used to estimate the soft-threshold power (female: ß = 5, male: ß = 2) for network construction based on scale-free topology analysis, followed by the topological overlap approach to calculate a dissimilarity between MAD filtered CpGs to reflect interconnectedness. The approach of average linkage hierarchical clustering was then used to divide CpGs with high topological overlap into modules. Modules were summarized by intramodular connectivity as well as modules EigenCpGs. The intramodular connectivity was assessed by the so-called hub-CpG, which represents the most highly connected CpG within a module. To assess whole-module behaviour, a single EigenCpG reflecting the first principal component of variance was calculated for each module separately and for distinction between EigenCpG labelled by colours. These EigenCpGs were linearly regressed onto the single PCB and PCDD/F congeners in a sex-stratified manner, including the same confounders used in the EWAS described above. GO enrichment analyses using the R-package missMethyl [Citation55] were performed for significant modules´ CpGs, to gain insight into possible over-represented gene pathways using the same approach used in EWAS described above.
For all analyses of EWAS and WGCNA, p-values were adjusted for multiple testing by the Benjamini-Hochberg approach and the p-value cut-off was determined by a FDR less than 0.05.
Results
Descriptive data of participants, PCBs and PCDD/Fs
One hundred and forty-two mother-infant pairs were included in all data analyses; 73 (51.41%) neonates were female and 69 (48.59%) were male (additional file 1 ST1 & ST2). Levels of concentration for the individual congeners of PCBs and PCDD/Fs did not significantly differ between sexes (p > 0.05; additional file 1 ST6 & ST7). Their average birth weight was M = 3408.98 g (SD = 569.50 g). On average, the mothers were M = 31.87 years (SD = 4.67 years) old at giving birth and their BMI before pregnancy was M = 24.02 kg/m2 (SD = 4.74 kg/m2). Thirty-eight (26.76%) mothers indicated that they had consumed at least one cigarette during the pregnancy. Mean gestation length was M = 39.39 weeks (SD = 1.60 weeks). Fifty (35.21%) mothers left school with University Entrance Exam (German Abitur), 13 (9.15%) with an advanced technical college certificate, 49 (34.51%) with a secondary school certificate, 25 (17.61%) with basic school qualification and five (3.52%) mothers had no or another school-leaving qualification.
Epigenome-Wide Association Studies (EWAS)
Differentially methylated positions (DMPs)
Results of the limma models showed that DNAm of 32 CpG sites were significantly associated with levels of six of the 16 PCB and PCDD/F congener as continuous predictors at FDR-corrected threshold of less than 0.05 (, ; additional file 1 ST8 & ST9). The various significant DMPs differed between female and male neonates and were found for PCB and PCDD but not PCDF congener predictors ().
Table 1. Differentially methylated positions associated with PCB and PCDD congeners in A) female or B) male subsample.
Figure 1. Stacked manhattan plot of DMPs from six EWAS for each sex. Stacked manhattan plot of results from the six EWAS for each sex that indicates significant DMPs. -log10 transformed FDR of each CpG included in the EWAS are plotted in order of its location on the chromosome. The dashed line indicates the array-wide threshold of -log10(0.05). Each DMP is labelled by the associated congener predictor and sex, i.e., which congener was significantly associated with DNAm in the male or female subsample. DMPs that are associated with more than one congener are additionally labelled with the gene the DMP maps to (genomic location in ).
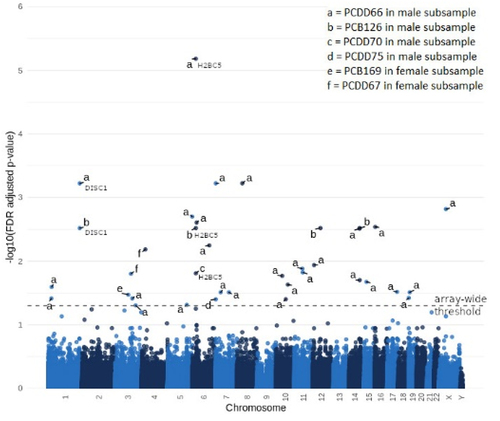
Figure 2. Significant DMPs common between different congener predictors. Euler diagram of the number of significant DMPs that were common between different congener predictors and A) males or B) females.
In the female sample, a total of three significant DMPs were identified (; additional file 1 ST9). An increasing level of PCB169 was associated with increased DNAm of cg08931983 (FDR = 0.03). An increase of the PCDD67-level was associated with two DMPs (cg22358250, FDR = 0.01; cg07612923, FDR = 0.02).
In males, four out of 29 DMPs were significantly associated with levels of more than one congener (; additional file 1 ST9). Cg02887499 annotated for gene H2B Clustered Histone 5 (H2BC5) was positively associated with PCB126 (FDR = 0.003), PCDD66 (FDR = 6.58×10−6) as well as PCDD70 (FDR = 0.02). In addition, cg05889395, annotated for gene Disrupted In Schizophrenia 1 (DISC1) was positively associated with PCB126 (FDR = 0.003) and PCDD66 (FDR = 0.0006). Cg04423493, not annotated for a gene, was positively associated with PCB126 (FDR = 0.003) and PCDD66 (FDR = 0.02). Cg17130815, annotated for LFNG O-Fucosylpeptide 3-Beta-N-Acetylglucosaminyltransferase (LFNG), was positively associated with PCCD75 (FDR = 0.04) and PCDD66 (FDR = 0.0006). GO analyses of the identified DMPs revealed no significant enriched pathways (FDR > 0.05; additional file 1 ST19-ST24).
Differentially methylated regions (DMRs)
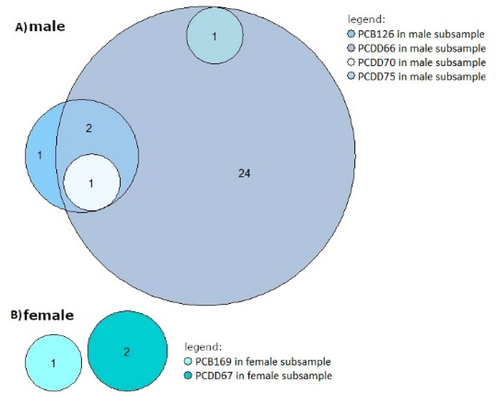
Eight significantly differentially methylated regions (DMRs) were identified in sex-stratified EWAS of three different PCB and PCDD congeners (FDR-adjusted p < 0.05; mean beta-fold-changes are presented in ; additional file 1 ST8 & ST10). In females, one DMR was identified. An increasing level of exposure to PCDD67 was associated with increased DNAm levels (mean beta-fold-change = 0.20, FDR = 6.52×10−9; , additional file 1 ST10) across three CpGs located in a DMR which overlaps the promoter region (± 2,000 base pairs [bp] from the transcription start site [TSS]) of the gene coding for phosphodiesterase 6B (PDE6B; .
Table 2. Differentially methylated regions associated with PCB and PCDD congeners in A) female or male B) subsample.
Figure 3. DMR associated with PCDD67 in female subsample. A) Differentially methylated region (DMR) associated with PCDD67 in female neonates. In green, the position of each assayed CpG in the DMR is shown with its -log10 transformed FDR above the FDR-corrected significance threshold (blue horizontal line). The DMR´s location related to NCBI RefSeq gene annotation (GRCh37/hg19), in a range of ± 2,000 bp from TSS is shown at the bottom of the plot. Here, it displays three PDE6B isoforms (NM_001350155.2, NM_001350154.2, & NM_001145292.1), and in addition two other PDE6B isoforms (NM_001145291.1 & NM_000283.3), that are located some 25kbp upstream of the chromosomal locus. B) For purpose of illustration only, the mean beta values of the three correlated (all p < 0.01; additional file 1 ST11) CpGs located in the DMR in chromosome 4 are linearly regressed on PCDD67 in female subsample without confounder adjustment and robustness for outliers in a scatterplot.
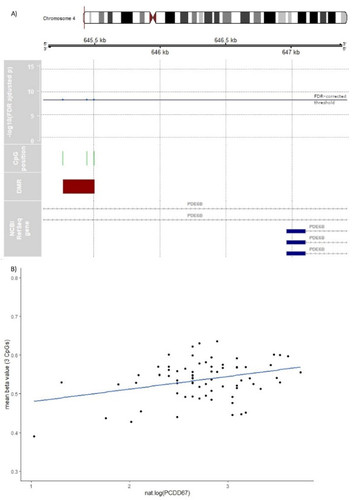
In males, seven DMRs were identified (; additional file 1 ST10; additional file 2 supplementary figure (SF) 1A-SF5B). One DMR is located in the promoter region of H2B Clustered Histone 5 (H2BC5; ). It comprises seven adjacent CpGs, including cg02887499, which was also identified as a DMP (see above). Cg02887499 [chr6:26,158,339] is located within the transcription factor-binding site motifs for YY1 (chr6:26,158,329–26,158,343), and TBP (chr6:26,158,320–26,158,339). Furthermore, this DMR includes the transcription-binding site of NFYA (chr6:26,158,003–26,158,322) and NFYB (chr6:26,158,058–26,158,316). GO analyses of the identified DMRs revealed no significant enriched pathways (FDR > 0.05; additional file 1 ST25-ST27).
Figure 4. DMR associated with PCDD66 in male subsample. A) Differentially methylated region (DMR) in chromosome 6 associated with PCDD66 in male neonates. In green, the position of each assayed CpG in the DMR is shown with its -log10 transformed FDR above the FDR-corrected significance threshold (blue horizontal line). The light red marked CpG position also represents a significant DMP (cg02887499). The DMR´s location related to NCBI RefSeq gene annotation (GRCh37/hg19) shows two H2BC5 isoforms (NM_138720 & NM_021063). B) For purpose of illustration only, the beta values of the seven uncorrelated (all p > 0.05; additional file 1 ST18) CpGs located in the DMR in chromosome 6 are linearly regressed on PCDD66 in male subsample without confounder adjustment and robustness for outliers in a scatterplot.
Weighted gene co-methylation network analysis (WGCNA)
Two weighted gene co-methylation networks were constructed separately for females and males. In female samples, 174 modules were identified from the 50% most variable CpG sites (380,202 CpGs). In males, 113 modules were identified from the 50% most variable CpG sites (380,232 CpGs).
EigenCpG-values, reflecting the first principal component of each module, were further used as indicator of the whole module and linearly regressed on the individual congeners and confounders in same order as in the EWAS-analyses in a sex-stratified manner. The first principal component in each module explained approximately half of the total variance, while the other components of a module explained minimal variance (selected scree plots in additional file 2 SF6-SF11). We did not observe any significant associations between EigenCpG-values and any PCB or PCDD/F predictor in male neonates. In females, 31 modules were identified to be significantly associated with exposure levels of PCDF130 after adjustment for multiple testing (FDR < 0.05; ). For 20 of these modules, there was a negative association between the module’s EigenCpG-value and the PCDF130-congener, indicating that with increasing level of PCDF130 the weighted average DNAm profile of a module decreased.
Table 3. 31 WGCNA-identified modules significantly associated with PCDF130-congener in female subsample adjustment for confoundersa.
For each module significantly associated with PCDF130 in female neonates, the most connected intramodular CpG, the so-called hub-CpG, was identified (). None of these 31 hub-CpGs were identical to any DMPs or CpGs in DMRs identified in EWAS with female neonates.
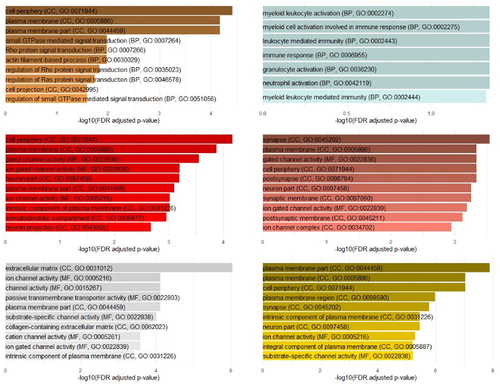
Pathway analysis showed that CpGs from six (brown, light cyan, red, salmon, white, & yellow) modules of the 31 modules identified as significantly associated with PCDF130 exposure-levels in female samples were significantly enriched in gene pathways after adjustment for multiple testing (FDR < 0.05; ; additional file 1 ST28-ST33; additional file 2 SF12-SF25). Analysis of the relationship between significantly enriched GO-terms across modules identified GO-terms related to, among others, neurotransmission, synapse and neuron parts, synapse organizations, and immune response.
Figure 5. Top enriched pathways in six modules associated with PCDF130 in female subsample. Top enriched pathways in the brown, light cyan, red, salmon, white, & yellow module significantly associated with PCDF130 in female neonates. Biological pathway terms are ranked by -log10 FDR. GO aspect and GO ID are shown in parentheses. Note. GO aspects are CC = cellular component, MF = molecular function, BP = biological process.
Discussion
Prenatal exposure to endocrine disrupting chemicals (EDCs) can interfere with developmental processes, and it has been suggested that long-term effects of in utero EDC exposure might be caused by the influence of EDCs on basic gene regulatory mechanism. Altered DNAm levels might be functionally involved in these processes, or serve as biomarkers of exposure. Here, using an epigenome-wide screen, we investigated whether prenatal low-dose exposure to PCBs and PCCD/Fs was associated with altered DNAm patterns in cord blood in a Western urban-industrial population in a sex-stratified manner. Our EWAS showed that six congeners of PCB and PCDD were significantly associated with DNAm levels at 32 CpG sites in female or male neonates. In addition to these DMPs, we identified eight differentially methylated regions (DMRs), one of those in females. The implicated genes are involved in neurodevelopment processes (DISC1, LFNG, TPD52L1, PAQR4), gene regulation (SMC1A, H2BC5), and immune functioning (ALOX5, HSPA1B), and for several of these genes, there is evidence for effects of EDC exposure on gene expression and differential DNAm in rodent models.
Of note, we found one DMP annotated to DISC1 that was associated with PCB126 and PCDD66 levels in male neonates. DISC1 codes for disrupted in schizophrenia 1 and is involved in the regulation of various processes of embryonic neurogenesis. It is especially known for its role in the pathophysiology of schizophrenia and other mental health problems due to perturbation of DISC1 expression [Citation57]. In relation to our finding, a study on mutant Disc1 (mDisc1) mice exposed to lead observed neurobehavior consistent with schizophrenia regarding locomotive activity and other mental disorders such as anxiety [Citation58]. In human studies, it has been shown that prenatal exposure to lead or BPA is associated with increased likelihood of schizophrenia later in life [Citation59–62].
PCDD66 and PCDD75 exposure in male neonates was also associated with DNAm levels of a CpG site and a DMR which mapped to LFNG, also involved in neurodevelopment. It codes for LFNG O-Fucosylpeptide 3-Beta-N-Acetylglucosaminyltransferase, which is involved in neuronal differentiation by acting in the Notch signalling pathway to define boundaries during embryonic development [Citation63]. Altered expression levels of LFNG have also been reported before to be associated with ultrafine particles (UFP) exposed onto COPD-diseased human bronchial epithelial cells [Citation64]. UFP are able to act as endocrine disruptor on the developing organism [Citation65].
An additional DMP associated with PCDD66 in the male samples was located in the gene TPD52L1, coding for Tumour protein D52-like 1, involved in carcinogenesis and cell proliferation. PCDD66 in mixture with other PCDD/Fs, PCBs and pesticides fed to mother rats during pregnancy and nursing, led to differentially expressed Tpd52l1 in liver tissue of the offspring [Citation66]. Furthermore, Tpd52l1 was reported to be differentially expressed in zebrafish exposed to low doses of atrazine, showing endocrine disrupting effects [e.g., Citation67], during early development [Citation68].
Finally, one DMP related to neurodevelopment that was associated with PCDD66 in male neonates of our study, was annotated for the gene PAQR4. PAQR4 is coding for Progestin And AdipoQ Receptor Family Member 4 and is associated with Isolated Growth Hormone Deficiency (Type Ib), a neurodevelopmental disease that is primarily characterized by short stature and is also associated with cognitive impairment [Citation69–71]. In male rodent neonates, which were exposed to low-dose BPA and 17β-oestradiol-3-benzoate during neonatal development, Paqr4 was differentially methylated in dorsal prostate tissue [Citation72].
Furthermore, we found two DMPs and one DMR associated with genes involved in gene regulation. Most notably, a strong association was found between PCB126, PCDD66 or PCDD70 exposure in males with DNAm for H2BC5. In addition to one DMP in H2BC5, we identified a DMR harbouring seven CpGs in the promoter region of H2BC5. The gene codes for H2B Clustered Histone 5 and is one of the four core histones responsible for the nucleosome structure of the chromosomal fibre in eukaryotes and thus involved in modulating DNA accessibility. The DMR includes several transcription factor-binding site motifs, and cg02887499 overlaps or directly neighbours those for YY1 (Ying-Yang 1) and TBP (TATA-box binding protein).
Lastly, we found DMPs and DMRs annotated to genes involved in immune functioning. In our study, one DMP was associated with PCDD66 in male neonates, which was annotated for the gene ALOX5. ALOX5 is coding for arachidonate 5-Lipoxygenase and plays an important role in the synthesis of leukotrienes from arachidonic acid. Leukotrienes are involved in various inflammatory and allergic processes, linking ALOX5 indirectly to immune processes. In embryo-larvae of zebrafish exposed to chiral PCB149, Alox5 expression was found to be dysregulated [Citation73]. Additionally, we identified one DMP annotated for gene HSPA1B that was associated with PCDD66 in male neonates. HSPA1B is coding for Heat Shock Protein Family A (Hsp70) Member 1B, that is involved in cellular stress regulation by maintaining the proteostasis [Citation74] and plays a mediating role between transcription factor Foxp3 and ubiquitin ligase Sub1 in regulatory T-cells during inflammation [Citation75]. In male offsprings of rats, fed to mixture of PCBs, PCDD/Fs (including PCDD66) and pesticides during pregnancy and nursing, Hspa1b was differentially expressed compared to male offspring of rats that were fed with corn oil [Citation66].
Overall, DNA methylation levels associated with PCB and PCDD exposure have been previously reported as associated with exposure to other EDCs (e.g., BPA), which might suggest a set of genes with broad sensitivity to chemical exposure.
We additionally examined epigenome-wide DNAm data from a systems-level view by conducting WGCNA to identify clusters of highly connected CpGs – so-called modules. Controlling for biological and technical confounders, six modules were significantly associated with PCDF130 in female samples and significantly enriched for gene pathways of neurodevelopment and immune functioning, which converges on the results discussed above.
The following limitations need mention. First, DNAm was assessed in peripheral cord blood cells although – given the hypotheses that prenatal EDC exposure can programme neurodevelopment resulting in long-term changes in behaviour and risk for mental health problems – the brain is the primary tissue of interest. However, DNAm profiles from cord blood might be understood as proxy or marker of epigenetic patterns in neuronal tissue cells, since epigenetic modifications established during early development might become propagated soma-wide [Citation76]. Furthermore, systematic research on tissue concordance showed that blood-based EWAS are suitable to identify DNAm patterns as biomarker for mental phenotypes manifesting in the brain [Citation77]. Second, in our study, DNAm was measured from cord blood tissue, which compromises multiple cell types having distinct epigenetic profiles, including DNAm patterns [Citation78]. Potential confounding effects in the epigenetic profile due to cell-type heterogeneity in cord blood were, however, accounted for [Citation48,Citation78,Citation79]. Third, it is known that a large part of the variance of DNA methylation levels is accounted for by genetic variation. Whereas we accounted for SNPs overlapping probes on the array, we cannot rule out potential effects of methylation quantitative trait loci (methQTLs) as potential confounders. Fourth, the identified differentially methylated CpGs were not validated. Local deep bisulphite sequencing of the CpGs on MiSeq (Illumina, San Diego, CA, USA) would be an adequate approach to evaluate the results further. Lastly, it has to be noted that our EWAS mainly identified associations between exposure for PCB and PCDD congeners and DNAm in the male sample, whereas significant associations between exposure for PCDF congener and DNAm of co-methylated modules was limited to females. These results might indicate that there is a sex-dependent susceptibility to the different congeners of PCB and PCDD/F [Citation13]. However, it also might be reasonable that the small sample size of each sex introduced an issue of statistical power of unidentified false-negative results.
Little is known about sex-specific effects of EDC exposure on DNAm levels. Potential mechanisms underlying the sex-dependent susceptibility and effects of the different congeners of PCB and PCDD/F might be that these chemicals are known to interact with the sex-hormonal system [Citation28]. Different congeners of PCB and PCDD/F seem to have different binding affinities to sex-hormone nuclear receptors involved in the gene expression machinery [Citation80,Citation81].
A strength of our study design is the temporal proximity as well as the chronological order of measurements of levels of PCBs and PCDD/F from maternal blood during late pregnancy and DNAm in cord blood after birth, which reduces threat of reverse causation. Furthermore, EDCs levels were objectively quantified using mass spectrometry in maternal blood. Future confirmatory studies will be required to follow up on these results, and the present findings should be considered hypothesis generating rather than definitive. For instance, to further understand the regulatory effects of our identified differentially DNAm patterns associated with prenatal exposure to PCBs and PCDD/Fs, cell culture-based experiments with varying exposure levels followed by downstream analyses of gene expression are warranted. Regarding possible long-term effects, it will be of interest whether the identified differentially methylated marks in cord blood are associated with behavioural outcomes in general and in specific with developmental outcomes that have been shown to be altered by PCBs and PCDD/Fs. The longitudinal nature of our study will enable causal mediation analyses to investigate the potential role of DNAm in mediating any exposure-outcome associations [Citation82]. Promising future studies are systematic comparisons how the measured levels of this cohort are in line with levels from other populations than the German population. The measured levels of PCBs and PCDD/Fs of the samples in the present study are representative for the German population as qualified by a German reference panel matching for sampling year and sample’s age [Citation43]. A systematic comparison of the exposure levels between different populations would help to get insight into how representative the results of this study might be for other populations.
To conclude, a better understanding of the mechanisms involved in the exposure-outcome relationship between EDCs and mental disorders continues to be an important research aim. Our findings on differentially methylated DNAm levels of single CpG sites, regions and modules indicate that PCBs and PCDD/Fs induce epigenetic changes that are associated with neurodevelopment, gene expression and immune functioning and thereby might set origin for adverse mental and health outcomes over the life span.
Discloser of interest
The authors report no conflict of interest.
Authors’ contributions
RK, AS, & MW designed the study. Data was acquired by MKS & MW. Labor analyses were conducted by MKS, MW, & DM. KM, RK, & AS conceived of the analyses. KM & VV performed all data analyses. ST & MS reviewed the data analyses. KM, VV, & NNW drafted the manuscript. DM, ST, MS, JW, JW, JGH, & AS substantive reviewed. RK, AS, JW, & JGH supervised the research project. All authors have read and approved the manuscript.
Consent
All parents gave written informed consent about the inclusion of material pertaining to themselves and their child, about their acknowledgement that they cannot be identified via the paper, and about the full anonymization of their data. We confirm that all mandatory laboratory health and safety procedures have been complied with in the course of conducting the research reported in our paper.
Supplemental Material
Download MS Word (182.4 KB)Supplementary Material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Zhang X, Tilling K, Martin RM, et al. Analysis of ‘sensitive’ periods of fetal and child growth. Int J Epidemiol. 2019;48(1):116–123.
- Lumey LH, Stein AD, Kahn HS, et al. Cohort Profile: the Dutch Hunger Winter Families Study. Int J Epidemiol. 2007;36(6):1196–1204.
- Lumey LH, Stein AD, Susser SE. Prenatal Famine and Adult Health. Annual Review of Public Health. 2011;32(1):237–262.
- Entringer S, Buss C, Kumsta R, et al. Prenatal psychosocial stress exposure is associated with subsequent working memory performance in young women. Behavioral Neuroscience. 2009;123(4):886–893.
- Entringer S, Buss C, Wadhwa PD. Prenatal stress and developmental programming of human health and disease risk: concepts and integration of empirical findings: current Opinion in Endocrinology. Diabetes and Obesity. 2010;17:507–516.
- Vandenberg LN, Colborn T, Hayes TB, et al. Hormones and Endocrine-Disrupting Chemicals: low-Dose Effects and Nonmonotonic Dose Responses. Endocr Rev. 2012;33:378–455.
- Barker DJP. Developmental origins of adult health and disease. J Epidemiol Community Health. 2004;58(2):114–115.
- Barker DJP. The origins of the developmental origins theory. J Intern Med. 2007;261(5):412–417.
- Wadhwa P, Buss C, Entringer S, et al. Developmental Origins of Health and Disease: brief History of the Approach and Current Focus on Epigenetic Mechanisms. Seminars in Reproductive Medicine © Thieme Medical Publishers 2009;27:358–368.
- Walker DM, Gore AC. Endocrine-Disrupting Chemicals and the Brain [Internet]. In: Gore AC, editor. Endocrine-Disrupting Chemicals Totowa NJ: Humana Press. 2007; cited 2019 May 20]. 63–109. Available from. http://link.springer.com/10.1007/1-59745-107-X_4
- Diamanti-Kandarakis E, Bourguignon J-P, Giudice LC, et al. Endocrine-Disrupting Chemicals: an Endocrine Society Scientific Statement. Endocr Rev. 2009;30:293–342.
- Cao Y, Winneke G, Wilhelm M, et al. Environmental exposure to dioxins and polychlorinated biphenyls reduce levels of gonadal hormones in newborns: results from the Duisburg cohort study. Int J Hyg Environ Health. 2008;211:30–39.
- Pessah IN, Lein PJ, Seegal RF, et al. Neurotoxicity of polychlorinated biphenyls and related organohalogens. Acta Neuropathol. 2019;138:363–387.
- Wittsiepe J, Schrey P, Ewers U, et al. Decrease of PCDD/F levels in human blood from Germany over the past ten years (1989–1998). Chemosphere. 2000;40(9–11):1103–1109.
- Myers JP, Guillette LJ, Swan SH, et al. Endocrine Disruptor Chemicals: overview [Internet]. In: Jorgensen SE, Fath B, editors. Encyclopedia of Ecology. Elsevier 2008; cited 2018 Jun 11]. 1265–1269. Available from http://linkinghub.elsevier.com/retrieve/pii/B9780080454054002457
- Jacobson JL, Jacobson SW, Humphrey HEB. Effects of in utero exposure to polychlorinated biphenyls and related contaminants on cognitive functioning in young children. The Journal of Pediatrics. 1990;116(1):38–45.
- Patandin S, Lanting CI, Mulder PGH, et al. Effects of environmental exposure to polychlorinated biphenyls and dioxins on cognitive abilities in Dutch children at 42 months of age. J Pediatr. 1999;134(1):33–41.
- Walkowiak J, Wiener J-A, Fastabend A, et al. Environmental exposure to polychlorinated biphenyls and quality of the home environment: effects on psychodevelopment in early childhood. Lancet. 2001;358(9293):1602–1607.
- Gladen BC, Rogan WJ, Hardy P, et al. Development after exposure to polychlorinated biphenyls and dichlorodiphenyl dichloroethene transplacentally and through human milk. J Pediatr. 1988;113(6):991–995.
- Koopman-Esseboom C, Weisglas-Kuperus N, Ridder MAJ, et al. Effects of Polychlorinated Biphenyl/Dioxin Exposure and Feeding Type on Infants’ Mental and Psychomotor Development. Pediatrics. 1996;97:700–706.
- Leijs MM, Koppe JG, Olie K, et al. Delayed initiation of breast development in girls with higher prenatal dioxin exposure; a longitudinal cohort study. Chemosphere. 2008;73(6):999–1004.
- Rennert A, Wittsiepe J, Kasper-Sonnenberg M, et al. Prenatal and Early Life Exposure to Polychlorinated Dibenzo-p-Dioxins, Dibenzofurans and Biphenyls May Influence Dehydroepiandrosterone Sulfate Levels at Prepubertal Age: results from the Duisburg Birth Cohort Study. J Toxicol Environ Health Part A. 2012;75(19–20):1232–1240.
- Winneke G, Ranft U, Wittsiepe J, et al. Behavioral Sexual Dimorphism in School-Age Children and Early Developmental Exposure to Dioxins and PCBs: a Follow-Up Study of the Duisburg Cohort. Environ Health Perspect. 2014;122(3):292–298.
- Neugebauer J, Wittsiepe J, Kasper-Sonnenberg M, et al. The influence of low level pre- and perinatal exposure to PCDD/Fs, PCBs, and lead on attention performance and attention-related behavior among German school-aged children: results from the Duisburg Birth Cohort Study. Int J Hyg Environ Health. 2015;218(1):153–162.
- Verner M-A, Hart JE, Sagiv SK, et al. Measured Prenatal and Estimated Postnatal Levels of Polychlorinated Biphenyls (PCBs) and ADHD-Related Behaviors in 8-Year-Old Children. Environ Health Perspect. 2015;123(9):888–894.
- Braun JM, Kalkbrenner AE, Just AC, et al. Gestational Exposure to Endocrine-Disrupting Chemicals and Reciprocal Social, Repetitive, and Stereotypic Behaviors in 4- and 5-Year-Old Children: the HOME Study. Environ Health Perspect. 2014;122(5):513–520.
- Nowack N, Wittsiepe J, Kasper-Sonnenberg M, et al. Influence of Low-Level Prenatal Exposure to PCDD/Fs and PCBs on Empathizing, Systemizing and Autistic Traits: results from the Duisburg Birth Cohort Study. PLOS ONE. 2015;10(6):e0129906.
- Walker DM, Gore AC. Epigenetic impacts of endocrine disruptors in the brain. Front Neuroendocrinol. 2017;44:1–26.
- Barker ED. Epigenetics, early adversity and child and adolescent mental health. Psychopathology. 2018;51(2):71–75.
- Geiman TM, Muegge K, DNA methylation in early development. Molecular Reproduction and Development. Molecular Reproduction and Development: Incorporating Gamete Research, 20.
- El-Heis S, Godfrey K. Developmental origins of health and disease. Obstetrics, Gynaecology & Reproductive Medicine. 2015;25(8):236–238
- Rogers JM, Lau C, Ellis-Hutchings RG. 5.08 - Epigenetics and the Developmental Origins of Health and Disease☆ [Internet]. In: McQueen CA, editor. Comprehensive Toxicology Third Oxford: Elsevier. 2018; cited 2019 May 20]. 118–136. Available from http://www.sciencedirect.com/science/article/pii/B9780128012383994832
- Leung Y-K, Ouyang B, Niu L, et al. Identification of sex-specific DNA methylation changes driven by specific chemicals in cord blood in a Faroese birth cohort. Epigenetics. 2018;13(3):290–300.
- Su K-Y, Li M-C, Lee N-W, et al. Perinatal polychlorinated biphenyls and polychlorinated dibenzofurans exposure are associated with DNA methylation changes lasting to early adulthood: findings from Yucheng second generation. Environ Res. 2019;170:481–486.
- Desaulniers D, Cooke GM, Leingartner K, et al. Effects of Postnatal Exposure to a Mixture of Polychlorinated Biphenyls,p, p ′-dichlorodiphenyltrichloroethane, and p- p′-dichlorodiphenyldichloroethene in Prepubertal and Adult Female Sprague-Dawley Rats. Int J Toxicol. 2005;24(2):111–127.
- Walker DM, Goetz BM, Gore AC. Dynamic postnatal developmental and sex-specific neuroendocrine effects of prenatal polychlorinated biphenyls in rats. Molecular Endocrinology. 2014;28(1):99–115.
- Kim K-Y, Kim D-S, Lee S-K, et al. Association of Low-Dose Exposure to Persistent Organic Pollutants with Global DNA Hypomethylation in Healthy Koreans. Environ Health Perspect. 2010;118(3):370–374.
- Rusiecki JA, Baccarelli A, Bollati V, et al. Global DNA Hypomethylation Is Associated with High Serum-Persistent Organic Pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116(11):1547–1552.
- Tindula G, Murphy SK, Grenier C, et al. DNA methylation of imprinted genes in Mexican–American newborn children with prenatal phthalate exposure. Epigenomics. 2018;10(7):1011–1026.
- Montrose L, Padmanabhan V, Goodrich JM, et al. Maternal levels of endocrine disrupting chemicals in the first trimester of pregnancy are associated with infant cord blood DNA methylation. Epigenetics. 2018;13(3):301–309.
- Grindler NM, Vanderlinden L, Karthikraj R, et al. Exposure to Phthalate, an Endocrine Disrupting Chemical, Alters the First Trimester Placental Methylome and Transcriptome in Women. Scientific Reports [Internet] 2018 [ cited 2019 Feb 18]; 8. Available from: http://www.nature.com/articles/s41598-018-24505-w
- Wilhelm M, Wittsiepe J, Lemm F, et al. The Duisburg birth cohort study: influence of the prenatal exposure to PCDD/Fs and dioxin-like PCBs on thyroid hormone status in newborns and neurodevelopment of infants until the age of 24 months. Mutat Res/Rev Mutat Res. 2008;659(1–2):83–92.
- Wittsiepe J, Fürst P, Schrey P, et al. PCDD/F and dioxin-like PCB in human blood and milk from German mothers. Chemosphere. 2007;67(9):S286–94.
- R Core Team. R: the R Project for Statistical Computing [Internet]. 2019 [cited 2020 Aug 14];Available from: https://www.r-project.org/
- Assenov Y, Müller F, Lutsik P, et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11(11):1138–1140.
- Müller F, Scherer M, Assenov Y, et al. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol. 2019;20(1):55.
- Pidsley R,Y, Wong CC, Volta M, et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14(1):293.
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86.
- Lin X, Tan JYL, Teh AL, et al. Cell type-specific DNA methylation in neonatal cord tissue and cord blood: a 850K-reference panel and comparison of cell types. Epigenetics. 2018;13(9):941–958.
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47.
- Verner M-A, McDougall R, Glynn A, et al. Is the Relationship between Prenatal Exposure to PCB-153 and Decreased Birth Weight Attributable to Pharmacokinetics? Environ Health Perspect. 2013;121(10):1219–1224.
- Konishi K, Sasaki S, Kato S, et al. Prenatal exposure to PCDDs/PCDFs and dioxin-like PCBs in relation to birth weight. Environ Res. 2009;109(7):906–913.
- Mansell G, Gorrie-Stone TJ, Bao Y, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics. 2019;20(1):366.
- Peters TJ, Buckley MJ, Statham AL, et al. De novo identification of differentially methylated regions in the human genome De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015. 8(1):6.
- Phipson B, Maksimovic J missMethyl: analysing Illumina HumanMethylation BeadChip Data [Internet]. Bioconductor version: Release (3.10); 2019 [ cited 2019 Dec 18]. Available from: https://bioconductor.org/packages/missMethyl/
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559.
- Millar JK. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9(9):1415–1423.
- Abazyan B, Dziedzic J, Hua K, et al. Chronic Exposure of Mutant DISC1 Mice to Lead Produces Sex-Dependent Abnormalities Consistent With Schizophrenia and Related Mental Disorders: a Gene-Environment Interaction Study. Schizophr Bull. 2014;40(3):575–584.
- Brown JS. Effects of Bisphenol-A and Other Endocrine Disruptors Compared With Abnormalities of Schizophrenia: an Endocrine-Disruption Theory of Schizophrenia. Schizophr Bull. 2009;35:256–278.
- Opler MGA, Brown AS, Graziano J, et al. Prenatal lead exposure, delta-aminolevulinic acid, and schizophrenia. Environ Health Perspect. 2004;112(5):548–552.
- Opler MGA, Buka SL, Groeger J, et al. Prenatal Exposure to Lead, δ-Aminolevulinic Acid, and Schizophrenia: further Evidence. Environ Health Perspect. 2008;116(11):1586–1590.
- Schultheiss NW, McGlothan JL, Guilarte TR, et al. Chronic Lead Exposure Causes Theta and Gamma Hypersynchrony in the Hippocampus and Disrupts Sensorimotor Gating. bioRxiv. 2020;2020.06.30.181149.
- Johnston SH, Rauskolb C, Wilson R, et al. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124(11):2245–2254.
- Sotty J, Garçon G, Denayer F-O, et al. Toxicological effects of ambient fine (PM2.5-0.18) and ultrafine (PM0.18) particles in healthy and diseased 3D organo-typic mucocilary-phenotype models. Environ Res. 2019;176:108538.
- Sobolewski M, Anderson T, Conrad K, et al. Developmental exposures to ultrafine particle air pollution reduces early testosterone levels and adult male social novelty preference: risk for children’s sex-biased neurobehavioral disorders. Neurotoxicology. 2018;68:203–211.
- Desaulniers D, Khan N, Cummings-Lorbetskie C, et al. Effects of cross-fostering and developmental exposure to mixtures of environmental contaminants on hepatic gene expression in prepubertal 21 days old and adult male Sprague-Dawley rats. J Toxicol Environ Health Part A. 2019;82(1):1–27.
- Holloway AC, Anger DA, Crankshaw DJ, et al. Atrazine-induced changes in aromatase activity in estrogen sensitive target tissues. J Appl Toxicol. 2008;28(3):260–270.
- Weber GJ, Sepúlveda MS, Peterson SM, et al. Transcriptome Alterations Following Developmental Atrazine Exposure in Zebrafish Are Associated with Disruption of Neuroendocrine and Reproductive System Function, Cell Cycle, and Carcinogenesis. Toxicol Sci. 2013;132(2):458–466.
- Lagrou K, Xhrouet-Heinrichs D, Massa G, et al. Quality of life and retrospective perception of the effect of growth hormone treatment in adult patients with childhood growth hormone deficiency. J Pediatr Endocrinol Metab. 2001;14(Suppl 5):1249–1260. discussion 1261-1262.
- Lijffijt M, Van Dam PS, Kenemans JL, et al. Somatotropic-axis deficiency affects brain substrates of selective attention in childhood-onset growth hormone deficient patients. Neurosci Lett. 2003;353(2):123–126.
- Sytze van Dam P, De Winter CF, Vries R, et al. Childhood-onset growth hormone deficiency, cognitive function and brain N-acetylaspartate. Psychoneuroendocrinology. 2005;30(4):357–363.
- Cheong A, Zhang X, Cheung -Y-Y, et al. DNA methylome changes by estradiol benzoate and bisphenol A links early-life environmental exposures to prostate cancer risk. Epigenetics. 2016;11(9):674–689.
- Chai T, Cui F, Mu X, et al. Exploration of Stereoselectivity in Embryo-Larvae (Danio rerio) Induced by Chiral PCB149 at the Bioconcentration and Gene Expression Levels. PLoS ONE. 2016;11(5):e0155263.
- Seo JH, Park J-H, Lee EJ, et al. ARD1-mediated Hsp70 acetylation balances stress-induced protein refolding and degradation. Nat Commun. 2016;7(1):12882.
- Chen Z, Barbi J, Bu S, et al. The Ubiquitin Ligase Stub1 Negatively Modulates Regulatory T Cell Suppressive Activity by Promoting Degradation of the Transcription Factor Foxp3. Immunity. 2013;39(2):272–285.
- Waterland RA, Jirtle RL. Transposable Elements: targets for Early Nutritional Effects on Epigenetic Gene Regulation. Molecular and Cellular Biology. 2003;23(15):5293–5300.
- Hannon E, Lunnon K, Schalkwyk L, et al. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10(11):1024–1032.
- Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362.
- Gervin K, Salas LA, Bakulski KM, et al. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin Epigenet. 2019;11(1):125.
- Fang H, Tong W, Branham WS, et al. Study of 202 natural, synthetic, and environmental chemicals for binding to the androgen receptor. Chem Res Toxicol. 2003;16(10):1338–1358.
- Kuiper GGJM, Lemmen JG, Carlsson B, et al. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor β. Endocrinology. 1998;139(10):4252–4263.
- Barker ED, Walton E, Cecil CAM. Annual Research Review: DNA methylation as a mediator in the association between risk exposure and child and adolescent psychopathology. J Child Psychol Psychiatry. 2018;59(4):303–322.
