ABSTRACT
Transposable elements (TEs) have been seen as selfish genetic elements that can propagate in a host genome. Their propagation success is however hindered by a combination of mechanisms such as mutations, selection, and their epigenetic silencing by the host genome. As a result, most copies of TEs in a given genome are dead relics: their sequence is too degenerated to allow any transposition. Nevertheless, these TE relics often, but not always, remain epigenetically silenced, and if not to prevent transposition anymore, one can wonder the reason for this phenomenon. The mere self-perpetuating loop inherent to epigenetic silencing could alone explain that even when inactive, TE copies remain silenced. Beyond this process, nevertheless, antagonistic selective forces are likely to act on TE relic silencing. Especially, without the benefit of preventing transposition, TE relic silencing may prove deleterious to the host fitness, suggesting that the maintenance of TE relic silencing is the result of a fine, and perhaps case-by-case, evolutionary trade-off between beneficial and deleterious effects. Ultimately, the release of TE relics silencing may provide a ‘safe’ ground for adaptive epimutations to arise. In this review, we provide an overview of these questions in both plants and animals.
Introduction
“Any relic of the dead is precious, if they were valued living.” Emily Brontë
Transposable elements (TEs) and their hosts are involved in an ancient and complex evolutionary conflict. On one side, this conflict results from the pressure of TEs to propagate in the host genome; on the other, from the need of the host to protect its genome integrity from TEs mutagenic potential. The mutagenic potential of TEs and the host defensive adaptation thus triggered an evolutionary arms race that contributed to shaping genomic landscapes and function (reviewed in [Citation1] and [Citation2]). For instance, one potential outcome of this arms race is the huge diversity of TEs. TEs can be broadly divided into two classes: DNA transposons and retrotransposons [Citation3]. DNA transposons move either as double-stranded DNA (dsDNA) elements via a ‘cut-and-paste’ strategy, or as single-stranded DNA (ssDNA) via a ‘peel-and-paste’ method (i.e., helitrons). Instead, retrotransposons replicate through an RNA intermediate with a ‘copy-and-paste’ mechanism, thus resulting in an extensive increase in copy number for each transposition event. Retrotransposons include long terminal repeat (LTR) and non-LTR elements, distinguished by the presence or not of long terminal repeats (reviewed in [Citation4] and [Citation5]). LTR elements include, in particular, endogenous retroviruses (ERVs), which are remains from past viral infection of the host genome that invaded the germline [Citation6]. Both DNA transposons and retrotransposons can encode the enzymatic machinery required for transposition. Such machinery can rely on a few proteins (e.g., DNA transposons), or can include an elaborate set of molecular activities (e.g., LTR elements), all of which are required for mobilization.
The life-cycle of a TE includes invasion, proliferation, degeneration, and eventually senescence or extinction [Citation7]. An invading TE proliferates until its transposition rate is lowered by the host defence machinery, which is crucial to limit the deleterious effects associated with TEs activity. The defence machinery is conserved in most eukaryotes, and relies on small RNAs (sRNAs) and epigenetic silencing, such as histone modifications and DNA methylation [Citation8,Citation9]. In plants, such machinery relies on the RNA-directed DNA methylation (RdDM) pathway (reviewed in [Citation10]) leading to the methylation of CG, CHG, and CHH sequences (where H is A, T, or C), while in animals on PIWI-interacting RNAs (piRNAs), small RNAs produced from discrete genomic regions enriched in inactive or truncated TEs [Citation11,Citation12], leading to methylation of CG sequences among other repressive marks. Once ‘immobilized’ by the host epigenetic defence machinery, TEs will accumulate inactivating mutations, leading slowly to their definitive immobilization [Citation7]. Some elements may first lose the genes required for mobility, yet retain the ability to transpose non-autonomously thanks to intact sequences required for transposition, and which are recognized by the enzymatic machinery of full-length copies (autonomous elements). For example, Miniature Inverted Repeats (MITEs) are decayed DNA transposons [Citation13] and may still mobilize non-autonomously as they may retain the terminal inverted repeats (TIRs) used as substrate by transposases from other autonomous DNA transposons. Eventually, the mobility of most TEs will be impaired due to extensive sequence degeneration, insertion of new TEs within an existing one, or deletion by recombination, thus resulting in TE relics or ‘dead’ copies [Citation7]. A classic example of dead copies is solo LTRs, which arise as a product of non-homologous recombination of flanking LTRs of a retroelement, leading to the elimination of the inner sequence of this element. TE relics occupy a large fraction of the genome [Citation14], and as with other TEs, they are targeted by epigenetic processes [Citation15–18]. However, why TE relics are repressed by the host defence machinery appears counterintuitive if they are immobile.
TE silencing is a self-perpetuating phenomenon (reviewed in [Citation19]): once the initial silencing is established, the heterochromatin marks and/or sRNAs affecting a TE will be the signal for silencing maintenance through mitosis and meiosis. Relic repression might then result from the difficulty of the host to discriminate degenerated from full-length copies. In principle, after a removal of TE relic silencing, such a process would not be restored if these copies were not harmful to the host. In Arabidopsis thaliana, after a transient loss of epigenetic silencing, some TE loci show re-establishment of DNA methylation in all contexts (CG, CHG, and CHH), while others retain the unmethylated identity [Citation20]. This illustrates that some TEs are simply silenced by self-perpetuation, since a perturbation of their epigenetic state does not trigger the restoration of their methylation level. Once the silencing is lost, it even seems that unmethylated TEs are protected from resilencing: the irreversible loss of methylation seems to depend on an active DNA demethylation that prevents the re-establishment of the epigenetic memory ([Citation21]; reviewed in [Citation22]). TE relics may constitute a significant part of these non-remethylable TEs, as they may not be detected as active TEs by the host genome. However, since the biological basis of remethylable and non-remethylable sequences remains uncertain, as their analogy with relics does, this interpretation is as yet only hypothetical. Nevertheless, it is plausible that TE relics are mostly kept silenced by mere perpetuation, as a by-product rather than having a role in itself. In other words, a sensible null hypothesis would postulate that epigenetic silencing of TE relics has little effect on the fitness of the host [Citation23,Citation24].
Rather than having no function, the silencing of TE relics might impact the fitness of the host [Citation25]. For example, irrespective of the transposition process – which relics lack by definition -, TE sequences or transcripts can have profound effects on the genome: they may induce chromosome breaks and genomic rearrangements [Citation26], alt(er gene expression and chromatin accessibility [Citation27,Citation28], and regulate the activity of other TE copies [Citation29,Citation30]. These topics are central to this review, where we evaluate the impact of relic silencing on (1) ectopic homologous recombination, (2) interference with gene transcription, and (3) the surveillance of active copies. Indeed, as the transposition threat lacks in TE relics, the maintenance of relic epigenetic targeting may depend on a trade-off between its beneficial and deleterious effects involving one or more of these three processes.
What we mean by ‘TE relics’
We consider TE relics to be TEs that lost their capacity to transpose autonomously or non-autonomously and the capacity to facilitate the transposition of other elements (e.g., production of a functional transposase for DNA transposons). The lack of a functional protein may be due to a truncated TE sequence, for example. In this definition, we therefore do not consider non-autonomous mobile elements to be relics, as they still have the ability to transpose. However, in our definition, TE relics may still contain ‘active’ components, such as a transcription start site or transcription factor-binding motifs. A solo LTR would be a perfect example of a TE relic, as it cannot transpose on its own, and yet may contain regulatory elements.
The central goal of this review is thus to evaluate the biological implications of TE relics remaining epigenetically silenced, which cannot be related to the prevention of transposition. A major problem arises when asking this question: very few studies actually focused on the epigenetic targeting of TE relics per se [Citation17,Citation18,Citation31]. Therefore, we base this review on previous works that mostly did not explicitly study TE relic silencing and yet, because of certain properties associated with TE relics, these studies can indirectly help us infer the role of epigenetic targeting of TE relics. These properties are:
1 – As mentioned above, in most genomes, the majority of TEs are ‘dead.’ For example, 99.9% of L1 copies in the human genome are no longer mobile [Citation32]. Therefore, studies focusing on general epigenetic patterns of TEs largely reflect the pattern of epigenetic silencing of TE relics.
2 – In species such as the plant model Arabidopsis, TE relics are predominant in euchromatic regions, while active copies are mostly found in (heterochromatic) pericentromeric regions [Citation31,Citation33], a likely consequence of negative selection against the insertion of new TEs nearby genes. Therefore, we can assume that generally, studies focusing on the epigenetic silencing of TEs located close to genes [Citation34,Citation35] focus on TE relic silencing and similarly, studies comparing the silencing of euchromatic vs heterochromatic TEs [Citation15,Citation31] generally compare the silencing of TE relics and active TEs. Please note, however, that this contradicts conclusions of another work [Citation25], which suggested that younger TEs are found closer to genes compared to older TEs. However, this work used polymorphic/fixed TE insertions as a proxy for young/old age of TEs, respectively, and found that fixed insertions are located further away from genes compared to polymorphic insertions. This may be a too indirect proxy, as the fixation of TE insertions may be influenced by drift and selection [Citation36], especially nearby genes (see further sections of this review). Instead, measuring the sequence identity of TE copies compared to consensus sequences of respective elements appears as a more direct method to infer the age of TE copies [Citation37], and with this method, authors found that young TEs are mostly found in pericentromeric regions, while gene-rich regions mostly contain old TEs [Citation31,Citation33]. Finally, this pattern concerns the model species A. thaliana and differs in some species [Citation33]. Therefore, the conclusions of this review regarding the role of TE relic silencing on genes, largely based on works in A. thaliana, might not fully apply to other species.
3 – One can expect that TE relics are shorter than their active counterparts, due to successive mutations and deletions, and that truncated TEs are less likely to be mobile [Citation31]. Some studies compared the epigenetic silencing of long vs short TEs [Citation15,Citation31,Citation38], which we can then use as a proxy to compare the epigenetic silencing of active TEs vs TE relics. These studies often use the criteria of <500 bp to qualify as ‘short TE,’ which is still much shorter than full-length autonomous elements for all classes, despite TE classes can largely differ in length. For example, class I such as LTR Gypsy or LINE are between 5 and 20 kb and up to 9 kb long, respectively, while class II such as helitrons or MuDR are 5–17 kb and 4–16 kb long in average [Citation39]. Therefore, elements classified as ‘short TEs’ are in general likely to be decayed and immobile, thus qualifying as relics.
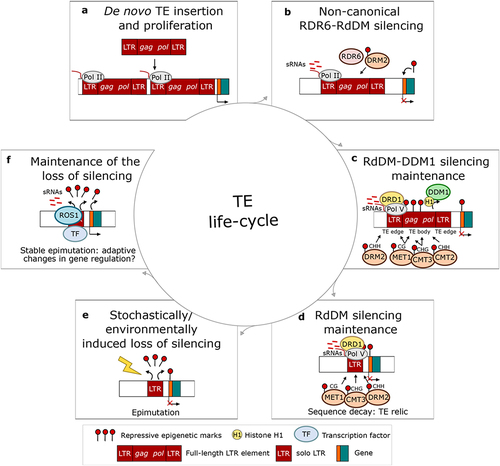
4 – At least in the plant model Arabidopsis, newly inserted active TEs are silenced by different mechanisms compared to older TEs. The body of young TEs is initially silenced by the so-called RDR6-RdDM (see further sections for details) and maintained silenced by the DDM1 pathway, which is not involved in the silencing of old TEs, which instead is performed by the canonical RdDM pathway see further sections for details [Citation15–17,Citation40]. Therefore, we can use the epigenetic marking by one or the other pathway as a proxy for the age of the TE (young TE vs TE relic).
5 – Last but not least, side effects of TE silencing, such as the spread to nearby genes and thus interference with gene transcription, apply to both active TEs and TE relics, as these are independent of their capacity to transpose. Therefore, in this review, we weigh such positive or negative side effects specifically for TE relics, by removing from the benefit/risk balance the prevention of their transposition as an advantage brought by their epigenetic silencing.
We are aware that these are merely indirect proxys for TE relics and their silencing, but in the current state of knowledge, we do not have better, highlighting the lack of studies specifically focusing on TE relic silencing. Thus, this calls for a thorough investigation of the epigenetic fate of TE relics in the future. Nevertheless, whenever possible in specific examples, in this review we give information supporting the idea that the case involves a TE relic rather than an active TE.
TE relics likely retain their potential for chromosome recombination/rearrangements
Non-allelic homologous recombination (NAHR) usually occurs between homologous sequences that are at different loci in a genome. Considering that TEs make up a large fraction of eukaryotic genomes, they can be a powerhouse for NAHR, leading to a variety of dramatic chromosomal rearrangements depending on the location and orientation of the repetitive sequences in the genome [Citation41–43]. In addition, the frequency of TE-mediated NAHR was suggested to increase with TE length and copy number [Citation44]. TE-mediated NAHR has been associated with chromosomal inversions in various species, including Drosophila and rice [Citation45–47]. In humans, somatic TE-mediated NAHR has been associated with several disorders, such as Alzheimer’s disease [Citation48]. Therefore, TE-mediated NAHR poses a real threat for genomes integrity and stability.
Perhaps counterintuitively with their potential to induce NAHR, TEs are generally associated with low recombination, both at large and local scale [Citation49]. This association at large scale is probably explained by selection acting against new TE insertions in gene-rich, highly recombining regions, and thus TEs mostly accumulate in heterochromatic regions, where recombination is rare [Citation50]. At the local scale, TEs are also negatively associated with recombination hotspots, even though there are exceptions, such as L1 or Alu elements in the human genome, which are known recombination hotspots [Citation49,Citation51]. The epigenetic silencing of TEs may be the explanatory mechanism for this negative association. Indeed, mutants lacking DNA methylation or other heterochromatin marks in several eukaryotes see their recombination landscape altered [Citation52–54]. For example, the loss of DNA methylation increased the occurrence of cross-overs in the chromosome arms of A. thaliana [Citation52]. These studies suggested that heterochromatin marks prevent recombination, especially at TE loci [Citation53].
This being said, the question remains whether degenerated TEs – TE relics – represent a NAHR threat for the host or not, and consequently, whether their epigenetic silencing can provide an advantage by preventing TE relic-mediated NAHR ()). On one hand, short and degenerated TE sequences are found more in gene-rich regions, while full-length functional TEs are found in pericentromeric regions [Citation31]. If TE relics have the capacity to induce NAHR, then they are likely to have a large deleterious potential due to their proximity with protein-coding loci, and thus their epigenetic silencing may provide an advantage. However, due to the accumulation of mutations, TE relics, such as solo LTRs, are usually shorter in size, and if the recombination potential increases with TE length as previously suggested [Citation44], TE relics should show low possibility for NAHR and recombination in general. Also, the mutational load of TE relics decreases sequence similarity with other TE copies located elsewhere in the genome, thus decreasing the risk of NAHR.
Some evidence goes against these expectations at first sight. In potatoes, MITE elements are hotspots for recombination and are found in open chromatin state, unlike retrotransposons such as Gypsy elements, which are found in heterochromatic regions with reduced recombination [Citation55]. MITEs are decayed DNA transposons that may retain the capacity to transpose non-autonomously [Citation13]. However, as they are already decayed, we assume that a large part of MITEs are further decayed and become inert, i.e., relics. This assumption is supported by a study in A. thaliana that showed that less than 2% of MITEs have the necessary elements to be mobile non-autonomously [Citation56]. MITEs may be more intact in other species, however we did not find studies characterizing the amount of potentially active MITE copies for other genomes such as potatoes. If we assume that the proportion of active MITE copies is relatively low in potatoes, like in A. thaliana, we can propose that the released MITE silencing may be related to many MITEs being potential TE relics and thus harmless NAHR-wise: non-allelic MITE copies may be too degenerated to be able to recombine with each other, rendering epigenetic silencing as protection against NAHR unnecessary. Nevertheless, released silencing of MITEs may be beneficial by providing a hotspot for allelic recombination and consequent increased genetic diversity. Also, CHH methylation, unlike CG and CHG, was positively associated with MITE recombination hotspots [Citation55], suggesting that MITEs exhibit a fine-tuned chromatin state that might allow recombination and yet suppress the mobility of still potentially active MITE copies. There is, however, evidence for TE relics posing a NAHR threat. L1 elements in humans, for which 99.9% of copies are inactive transposition-wise [Citation32], are found enriched in recombination hotspots, and especially NAHR sites leading to genomic instability associated with pathologies [Citation51]. It thus appears that even though degenerated and shortened, TE relics still have the potential to recombine. This may be explained by the fact that recombination hotspots seem to require a very short consensus sequence, 11-bp to 13-bp long [Citation51,Citation55], and strikingly, this sequence is present in several TE families across plants and animals [Citation51,Citation55]. Therefore, even though highly degenerated, TE relics have probabilistically a high chance to retain the short consensus sequence present in recombination hotspots, and consequently, may pose a significant NAHR threat. L1s in humans and MITEs in potatoes are nevertheless rather the exceptions than the rules, and most TEs are associated with ‘cold’ recombination regions [Citation49]. Further studies are therefore required to provide a broader picture of the NAHR potential of TE relics, and consequently, the role of their epigenetic silencing in preventing genomic instability.
TE relics silencing, a double-edged sword in gene regulation
Multiple lines of evidence suggest that epigenetic targeting of TE relics may affect host fitness, especially because TE relics are located preferentially nearby genes [Citation31]. Especially, numerous studies have focussed on the benefits of keeping gene-associated TE relics silenced. There is a growing body of evidence of TEs bearing functional transcription factor binding sequences (TFBSs), thus serving as an important source of regulatory elements fuelling genome cis-regulatory evolution [Citation57–60]. Epigenetic silencing of TE fragments may thus be a measure to prevent pervasive transcription ()). Indeed, TFBSs inserted upstream of a gene may directly interfere with gene expression regulation by creating an alternative transcription start site (aTSS). For instance, in human neural progenitor cells, loss of activity in the DNA methyltransferase 1 (DNMT1) leads to transcriptional activation of LINE-1 elements, many of which then act as aTSSs [Citation61,Citation62]. In addition to guarding cis-regulation, silencing of intragenic TE relics may be required for proper mRNA splicing ()). Indeed, TE relics may bear spurious splice and polyadenylation signals that, in actively transcribed genes, might interfere with RNA splicing [Citation63]. Epigenetic silencing of such TE relics might then mask the spurious polyadenylation signals. Consistently, in A. thaliana mutants lacking DNA methylation, the loss of methylation of intronic TE fragments leads to premature cleavage and polyadenylation, presumably due to spurious signals at the 5ʹ end of the TE sequence [Citation64].
Also, growing evidence indicates that TE relics may provide transcription factor binding sites [Citation34], and their epigenetic silencing itself may be domesticated (or co-opted) to regulate the expression of adjacent genes ()). In metazoans, one of the most notorious examples is the murine Agouti gene, whose transcription is affected by the methylation level of an upstream TE, which regulates the colour of the fur and the lipid metabolism [Citation65]. In addition, TE relic silencing has been integrated into a self-regulatory loop at REPRESSOR OF SILENCING 1 (ROS1) locus in plants [Citation66]: ROS1 demethylase regulates DNA methylation levels genome-wide, and its expression depends on the methylation status of an upstream helitron-derived methylation sequence. This helitron, classified as ‘transposon fragment’ (TAIR database), is only 316 bp long, which, compared to the 4–17 kb length of active autonomous helitrons in Arabidopsis, suggests that this is a helitron relic [Citation39]. The ROS1 expression <-> helitron relic methylation feedback loop thus allows for fine-tuning methylation levels on a genomic scale. Thus, in some cases, TE relics silencing integrates into gene expression regulation.
However, perhaps a less-studied aspect is the costs associated with silencing TE relics that are located next to genes. For example, methylated TE fragments are depleted around genes and show signs of strong purifying selection as opposed to non-methylated ones, probably due to deleterious effects of TE methylation on the expression of nearby genes [Citation25,Citation67]. Similarly, in the plant Capsella, purifying selection acts against the presence, around genes, of TEs targeted by sRNAs, while purifying selection does not affect gene-associated TEs that are not silenced via the RdDM pathway [Citation68]. As a consequence, a large part of TEs found in chromosome arms lack sRNA silencing, and they happen to be short degenerated TEs, i.e., TE relics [Citation31]. Thus, it seems that the epigenetic silencing ‘friendly fire,’ rather than the mutagenic effect of TEs, is a crucial determinant of the selection pressure acting against their presence around genes [Citation67,Citation68]. The ‘friendly fire’ scenario appears to be even more dramatic in species lacking DNA methylation and relying on histone modifications to silence TEs: in D. melanogaster, the spread of repressive marks reaches up to 20 kb around the target, and this is believed to be the reason for negative selection against the presence of TEs around genes in D. melanogaster [Citation35]. In species showing DNA methylation, the spreading of epigenetic marks from nearby TEs tends to be more controlled by the host. In A. thaliana, this process is performed by the DNA and histone demethylases DEMETER-LIKE and INCREASE IN BONSAI METHYLATION1 (IBM1), respectively, which prevent DNA methylation, especially TE-associated DNA methylation (non-CG context), from entering gene boundaries [Citation69,Citation70].
In addition to changes in the epigenetic status of the locus, silencing of TE relics located within gene bodies may interfere with transcription elongation ()). In A. thaliana and Oryza sativa, IBM2 is presumed to aid RNA pol II in reading through heterochomatinized TE copies within introns. Indeed, mutation of ibm2 leads to premature mRNA termination at introns bearing TE insertions, leading to pleiotropic developmental defects [Citation71,Citation72]. In other words, it appears that the silencing of TEs within genes can hinder transcription elongation, and that the evolutionary response of the host genomes to bypass this problem was not the removal of the silencing of intragenic TEs, but instead the emergence of the IBM2 function. Thus, the maintenance of silencing of intragenic TEs despite its threat for transcription elongation suggests either a beneficial and/or the mechanistic impossibility to selectively release the silencing of these TEs.
All in all, while the silencing of gene-associated TE relics may prove beneficial for the host genome, it can have harmful consequences on the host fitness. The maintenance of such silencing is thus likely to be the consequence of a trade-off between its deleterious and beneficial effects.
TE relics, safekeepers of the germlines?
In animals and plants, TE silencing is relaxed during specific stages of germline development, when, paradoxically, their activity needs to be tightly controlled to preserve the genetic information of the offspring. However, the temporary relaxation of their repression ensures the re-establishment of a tight TE silencing in later stages ([Citation30,Citation73]; reviewed in [Citation74]). Interestingly, many of the unleashed TEs map to relic loci, suggesting the co-option of these copies for the surveillance of TEs in the germline ()).
In most animals, the repression of TEs in the germline is ensured by the metazoan-specific piRNAs, small RNAs deriving from transcripts produced from discrete genomic regions enriched in inactive or truncated TEs, the piRNA clusters [Citation11,Citation12]. These clusters represent a genetic footprint of TEs already encountered by the host: invading copies can insert into existing clusters or create de novo ones [Citation75–77], thus contributing to establish an ‘immunity’ system against TE invasions. This mechanism is highly efficient: evidence suggests that the insertion of a single TE copy in a piRNA cluster might be sufficient to silence all the similar sequences [Citation78–80]. Since piRNA clusters are enriched in old degraded TE copies, these might have been co-opted to mediate the silencing of full-length elements ([Citation81]; reviewed in [Citation82]).
In plants, defence against TEs relies on the RNA-directed DNA methylation (RdDM) pathway (reviewed in [Citation10]). The invading genetic material is first recognized by the non-canonical RNA-dependent RNA polymerase 6 (RDR6)-RdDM pathway, initiated by RNA polymerase II (Pol II)-derived TE mRNAs ()). TE silencing is then reinforced through a combination of canonical RdDM and a siRNA-independent mechanism involving the nucleosome remodeller DECREASE IN DNA METHYLATION1 (DDM1) ()) [Citation15]. As a result, the body of young TEs is generally targeted by the DDM1-dependent pathway, while old TEs and the edges of young TEs are repressed by the canonical RdDM machinery [Citation15–17]. This evidence suggests that TE repression changes during the life-cycle of a TE, and old TE copies – TE relics – become an active source of siRNAs ()) [Citation17,Citation18]. During plant reproduction, despite the absence of piRNA clusters, a similar mechanism ensures tight control of TE expression in the germline. Before fertilization, in the gamete companion cells (central cell and vegetative nucleus), extensive demethylation induces the relaxation of TE silencing, causing a burst of 21–24 nt siRNAs production [Citation38,Citation83]. These siRNAs are thought to be transferred to the adjacent gametes (egg cell and sperm cell), where they direct silencing of homologous TEs [Citation38,Citation84,Citation85]. The demethylation in the companion cells requires the activity of the DNA demethylase DEMETER (DME), which preferentially demethylates short euchromatic TEs (<500 bp), old TE copies located in gene-rich regions [Citation38,Citation83]. Short TEs might thus have a similar function to that of piRNA clusters in animals: they would be a reservoir of siRNAs involved in the genome defence against TEs with some sequence similarity.
In principle, TEs fast decay might cause an excessive sequence divergence among degenerated copies and their ‘native’ relatives, thus limiting the cross-silencing potentially provided by TE relics. However, in both metazoans and plants, recent evidence suggests that the sRNAs defence machinery can tolerate mismatches with the target sequence [Citation86,Citation87]. In other words, TEs quick turnover might be counter-measured by an increased target recognition flexibility of the TE defence machinery. At the same time, however, the broader targeting control for rapidly evolving TEs might increase dangerous off-target effects. The host countermeasures adopted to limit such potential negative impact on genome activity remain, however, still elusive.
TE relics and defence against exogenous threats
Besides being transmitted vertically from parents to offspring, TEs can be transferred between non-mating organisms through horizontal transfer (HT) (reviewed in [Citation88]). HT is a frequent phenomenon, often responsible for the initial colonization of new genomes by TEs. HT of TEs might happen frequently from parasitic organisms, such as triatomine bugs, to their hosts [Citation89], explaining the nearly perfect sequence similarity of certain TEs between hosts and parasites that are distant phylogenetically. In both plants and animals, the sRNAs machinery plays an essential role against invading TEs, by inducing degradation of the invading genetic material and heterochromatin assembly in the insertion site [Citation90]. Nevertheless, the question remains whether TE relics can play any role in the defence against exogenous TEs during HT: the host might be invaded by completely new TE families, leaving little room for cross-silencing from sRNA produced from TE relics, even considering the relative flexibility in the target-sRNA sequence matching explained above [Citation86,Citation87]. This might explain why HT of TEs is relatively common and successful. Nevertheless, cross-silencing from TE relic sRNAs may still help with the invasion of new TE copies belonging to families already present in the host genome. For example, in the mangrove Rhizophora apiculata, the expression level of Gypsy LTR elements increases with their age: young copies are not transcribed, while old insertions are an active source of siRNAs cross‐mapping to young copies. Interestingly, the old elements present low sequence divergence from young ones, but only in the region within the flanking LTRs (sequences required for mobilization) [Citation91]. While this work did not address directly the potential role of old Gypsy elements silencing against the invasion of new TEs by HT, this evidence suggests that old Gypsy elements are selectively retained in the R. apiculata genome, and that their transcriptional activation contributes to the de novo silencing of new insertions. Thus, the co-option of old TE copies for the silencing of full-length insertions has not only evolved to ensure tight repression of TEs in the germline, but it might be crucial also in the defence against invading TEs, as already suggested under the ‘zombie TE’ hypothesis by Lisch and co-authors ()) [Citation92,Citation93]. Under this hypothesis, heterochromatic regions may act as a reservoir of non-functional TE copies, which, under certain conditions such as stress, would be reactivated to participate in the silencing of newly invading TEs with similar sequences [Citation94].
Analogously, since retrovirus-derived sequences (viruses replicating through the reverse transcription of an RNA intermediate) share similar features with TEs, the co-option of truncated viral insertions might be involved in the defence against infectious viruses ()) [Citation90]. In fact, many retroviruses have been acquired by eukaryotic genomes along their evolutionary history, resulting in the formation of so-called endogenous retroviruses (ERVs) [Citation95]. ERVs make up a significant portion of genomes such as the human one, thus largely contributing to the diversity of repetitive sequences. Viral integration into host genomes is, however, not only restricted to retroviruses, since also ssDNA, ssRNA, and dsRNA viruses can integrate into animal and plant genomes, further blurring the line between viruses and TEs ([Citation96,Citation97]; reviewed in [Citation98]). Interestingly, in several metazoans (e.g., mosquitoes and chickens), truncated viral integrations can produce piRNAs involved in the defence against infectious viruses [Citation99,Citation100]. Taken together, this evidence suggests that degraded viral insertions might be co-opted by the host as a defensive strategy against invading viruses, as part of a similar process to that already discussed for TEs. However, this strategy might be restricted only to certain species, for example, piRNAs do not seem to be involved in the antiviral defence in Drosophila [Citation101].
In conclusion, the role of TE relics in the protection against exogenous threats such as horizontally transferred TEs or viruses, while an exciting perspective, remains controversial if not implausible. Future studies will help determine if this phenomenon is substantial or rather anecdotal.
Conclusions: release of TE relic silencing, a ‘Safe’ source of epimutations?
Altogether, current evidence provides different and sometimes opposite expectations of the impact of TE relic silencing on the host fitness, may it be neutral, positive, or negative. This impact may depend on many factors, such as where a specific TE relic is located, e.g., whether it is in genic vs non-genic regions, or if near a gene, whether this gene has a pleiotropic vs low effect on the host fitness. The number of TE copies genetically related to the TE relic is also likely to be important, as this determines the potential for non-allelic recombination, and also for cross-silencing and TE surveillance.
In a word, the fate of TE relic silencing, i.e., whether it is maintained, may be determined on a case-by-case basis, depending on the factors listed above. However, studies suggest that the life-cycle of a TE shows a general trend, from insertion to decay and eventually release of epigenetic silencing (). If such release happens, when it does so in the TE life-cycle, and why or how (complete vs partial release as in the case of MITEs in potato [Citation55]) it does so, remains nevertheless elusive, but it may happen once TEs are decayed, i.e., are TE relics. Indeed, TE relic silencing seems to be relatively fragile: in A. thaliana, euchromatic TEs (largely TE relics) show higher stochastic CG methylation loss than heterochromatic copies [Citation102]. Moreover, a majority of euchromatic TEs (largely TE relics) remain stably unmethylated after a transient release of their epigenetic silencing under laboratory conditions [Citation21]. Therefore, the release of TE relic silencing is likely to be a stable epimutation (i.e., an epigenetic alteration affecting gene activity [Citation103]), and thus transmitted. The release of TE relic silencing may happen as a spontaneous epimutation or triggered by an environmental cue (stress), and it may subsequently be subjected to selective processes if any impact on the host fitness ()) [Citation104]. These characteristics coupled with the incapacity of TE relics for transposition may provide a ‘safe’ ground for adaptive epimutations impacting gene regulation to arise ()).
As an example, the gene HDG3 shows natural variation in its imprinting pattern in A. thaliana, and this results in seed size variation [Citation105]. This phenotypic variation is associated with variation in the methylation of a nearby AtREP10D helitron, considered as a non-autonomous element [Citation106]. This helitron is targeted by the canonical RdDM pathway and not by DDM1 [Citation107], a typical signature for ‘old’ TE silencing [Citation15–17]. Another case of epimutant with phenotypic effect is the colourless non-ripening tomato epimutant: this epimutation involves non-CG DNA methylation – a hallmark of TE silencing – of the LeSPL-CNR gene promoter [Citation108,Citation109]. Interestingly, however, no TE sequence was directly identified as a target of this epigenetic silencing [Citation108,Citation109]. It is tempting to speculate that this variable epimutation targets the relic of a TE too decayed to be identified as such. This is actually a fairly common scenario: after thorough annotation efforts, a substantial proportion of the genomic ‘dark matter’ (unannotated non coding parts) of Arabidopsis genome was shown to be made of old and decayed TE sequences which are challenging to annotate [Citation110].
However, the adaptive potential of epimutations affecting TEs was recently proposed to be mostly related to their potential for TE remobilization rather than their regulatory role [Citation111]. The most likely evolutionary and molecular scenarios behind adaptive TE (relic) epimutations remains therefore to be unravelled.
Figure 1. Beneficial and deleterious effects of the silencing of transposable element relics. (a) The silencing of transposable element (TE) relics might prevent non-allelic recombination. On one hand, such silencing promotes genome stability, on the other hand, it reduces genetic variation. (b) When TE relics bear a Transcription Start Site (TSS), their silencing can have opposite effects on gene expression. The TE relic silencing may prevent the recognition of the ectopic TE-derived TSS by the transcription machinery, thus facilitating the usage of the canonical TSS, and preventing any spurious transcription outside or within a gene. However, with a silenced TE relic within a gene, the transcription machinery may have issues reading ‘through’ the heterochromatized TE relic, resulting in a truncated mRNA. (c) On the other hand, the epigenetic repression of a TE relic inserted within a genic intron might mask spurious polyadenylation signals present in the TE sequence. (d) The spreading of repressive marks from TE relics can affect the expression of nearby genes. While such a spreading usually has deleterious effects on gene activity, it can also be co-opted by the host for the regulation of gene expression. (e) TE relics (e.g., solo LTR) silencing is involved in the surveillance of TEs in both plant and animal germlines, (f) and might be involved in the defence against exogenous TE/viral copies (e.g., LTR elements). On one hand, these defence mechanisms are crucial to maintain genome integrity, on the other hand, they might induce deleterious off-target effects due to the flexibility of the target recognition.
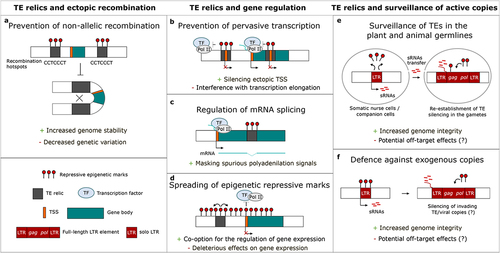
Figure 2. Life-cycle of a transposable element in plants, represented here for the sake of simplicity by a full-length LTR element. (a) The life-cycle of a transposable element (TE) begins with its de novo insertion into the host genome. Such insertions are initially transcribed into messenger RNAs (mRNAs) by the host RNA polymerase II (Pol II). (b) Pol II-derived TE mRNAs initiate the non-canonical RNA-dependent RNA polymerase 6- RNA-directed DNA methylation (RDR6-RdDM) pathway, which relies on the production of small RNAs (sRNAs), and induces the epigenetic silencing of the de novo TE insertion. (c) The maintenance of TE silencing relies on a combination of canonical RdDM and nucleosome remodeller DECREASE IN DNA METHYLATION1 (DDM1). The canonical RdDM targets the edges of young TEs, and requires the nucleosome remodeller DRD1 and the DNA methyltransferases MET1, CMT3 and DRM2, responsible for methylation in CG, CHG and CHH, respectively. The heterochromatic TE body is instead targeted by DDM1 that removes H1 linker histones, thus making the chromatin accessible for the DNA methyltransferases MET1, CMT3 and CMT2, responsible for methylation in CG, CHG and CHH, respectively. (d) Old degenerated TE copies (i.e., solo LTR TE relics) are then repressed only by the canonical RdDM, thus becoming an active source of siRNAs. TE relics tend to accumulate in gene-rich regions. (e) Stochastic or environmental signals might then induce TE relics loss of silencing, thus altering the local chromatin state (i.e., epimutation). (f) The loss of silencing of TE relics might then be actively maintained by the demethylase REPRESSOR OF SILENCING 1 (ROS1), which antagonizes RdDM activity. The loss of silencing might be stable for multiple generations and give rise to a stable epimutation. The TE relic might in fact bear a functional transcription factor binding sequence that could potentially induce adaptive changes in the regulation of nearby genes.
Acknowledgments
We thank Lukáš Fischer for careful reading and improvement of the manuscript. CLP has been supported by the Czech Science Fundation (Project number 22-20240S to CLP). IS has been supported by the European Training Network “EpiDiverse” (EU Horizon 2020 program under Marie Skłodowska-Curie grant agreement No. 764965).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Additional information
Funding
References
- Bousios A, Gaut BS. Mechanistic and evolutionary questions about epigenetic conflicts between transposable elements and their plant hosts. Curr Opin Plant Biol. 2016;30:123–133.
- Cosby RL, Chang N-C, Feschotte C. Host–transposon interactions: conflict, cooperation, and cooption. Genes Dev. 2019;33:1098–1116.
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107.
- Todorovska E. Retrotransposons and their role in plant—genome evolution. Biotechnol Biotechnol Equip. 2007;21:294–305.
- Goodier JL. Restricting retrotransposons: a review. Mob DNA. 2016;7(16). https://doi.org/10.1186/s13100-016-0070-z.
- Grandi N, Tramontano E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol. 2018;9:2039.
- Blumenstiel JP. Birth, school, work, death, and resurrection: the life stages and dynamics of transposable element proliferation. Genes (Basel). 2019;10:336.
- Cogoni C, Macino G. Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10:638–643.
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148.
- Cuerda-Gil D, Slotkin RK. Non-canonical RNA-directed DNA methylation. Nat Plants. 2016;2:16163.
- Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103.
- Weick E-M, A E. Miska, piRNAs: from biogenesis to function. Development. 2014;141:3458–3471.
- Nandini VB. Miniature inverted-repeat transposable elements (MITEs), derived insertional polymorphism as a tool of marker systems for molecular plant breeding. Mol Biol Rep. 2020;47:3155–3167.
- Nuzhdin SV. Transposable elements and genome evolution. In: McDonald JF, editor. Georgia genetics review. Vol. 1. Dordrecht: Springer Netherlands; 2000. p. 129–137.
- Zemach A, Kim MY, Hsieh P-H, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205.
- Stroud H, Do T, Du J, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72.
- Wang Z, Baulcombe DC. Transposon age and non-CG methylation. Nat Commun. 2020;11:1221.
- Bousios A, Diez CM, Takuno S, et al. A role for palindromic structures in the cis-region of maize Sirevirus LTRs in transposable element evolution and host epigenetic response. Genome Res. 2016;26:226–237.
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616.
- Teixeira FK, Heredia F, Sarazin A, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604.
- Li J, Yang D-L, Huang H, et al. Epigenetic memory marks determine epiallele stability at loci targeted by de novo DNA methylation. Nat Plants. 2020;6:661–674.
- Molinier J. To be, or not to be, remethylated. Nat Plants. 2020;6:606–607.
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. PNAS. 2007;104:8597–8604.
- Bird A. The selfishness of law-abiding genes. Trends Genet. 2020;36:8–13.
- Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428.
- Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468.
- Morgan HD, Sutherland HGE, Martin DIK, et al. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318.
- Martin A, Troadec C, Boualem A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138.
- Saito K, Nishida KM, Mori T, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222.
- Hsieh T-F, Ibarra CA, Silva P, et al. Genome-wide demethylation of arabidopsis endosperm. Science. 2009;324:1451–1454.
- Ahmed I, Sarazin A, Bowler C, et al. Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 2011;39:6919–6931.
- Bourque G, Burns KH, Gehring M, et al. Ten things you should know about transposable elements. Genome Biol. 2018;19:199.
- Maumus F, Quesneville H. Ancestral repeats have shaped epigenome and genome composition for millions of years in Arabidopsis thaliana. Nat Commun. 2014;5:4104.
- Batista RA, Moreno-Romero J, Qiu Y, et al. The MADS-box transcription factor PHERES1 controls imprinting in the endosperm by binding to domesticated transposons. eLife. 2019;8:e50541.
- Lee YCG, Karpen GH. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. eLife. 2017;6:e25762.
- Stritt C, Gordon SP, Wicker T, et al. Recent activity in expanding populations and purifying selection have shaped transposable element landscapes across natural accessions of the Mediterranean grass brachypodium distachyon. Genome Biol Evol. 2018;10:304–318.
- Maumus F, Quesneville H. Impact and insights from ancient repetitive elements in plant genomes. Curr Opin Plant Biol. 2016;30:41–46.
- Ibarra CA, Feng X, Schoft VK, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364.
- Zhao D, Ferguson AA, Jiang N. What makes up plant genomes: the vanishing line between transposable elements and genes. Biochim Biophys Acta Gene Regul Mech. 2016;1859:366–380.
- Osakabe A, Jamge B, Axelsson E, et al. The chromatin remodeler DDM1 prevents transposon mobility through deposition of histone variant H2A.W. Nat Cell Biol. 2021;23:391–400.
- Lonnig W-E, Saedler H. Chromosome rearrangements and transposable elements. Annu Rev Genet. 2002;36:389–410.
- Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. BioEssays. 1994;16:269–275.
- Mérel V, Boulesteix M, Fablet M, et al. Transposable elements in Drosophila. Mob DNA. 2020;11:23.
- Petrov DA, Aminetzach YT, Davis JC, et al. Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol. 2003;20:880–892.
- Evgen’ev M, Zelentsova H, Mnjoian L, et al. Invasion of Drosophila virilis by the Penelope transposable element. Chromosoma. 2000;109:350–357.
- Delprat A, Negre B, Puig M, et al. The Transposon Galileo generates natural chromosomal inversions in Drosophila by ectopic recombination. PLOS ONE. 2009;4:e7883.
- Yu C, Han F, Zhang J, et al. A transgenic system for generation of transposon Ac/Ds-induced chromosome rearrangements in rice. Theor Appl Genet. 2012;125:1449–1462.
- Pascarella G, Hashimoto K, Busch A, et al. Recombination of repeat elements generates somatic complexity in human genomes . bioRxiv 2021. DOI:10.1101/2020.07.02.163816.
- Kent TV, Uzunović J, Wright SI. Coevolution between transposable elements and recombination. Philos Trans R Soc B. 2017;372:20160458.
- Termolino P, Cremona G, Consiglio MF, et al. Insights into epigenetic landscape of recombination-free regions. Chromosoma. 2016;125:301–308.
- Myers S, Freeman C, Auton A, et al. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129.
- Mirouze M, Lieberman-Lazarovich M, Aversano R, et al. Loss of DNA methylation affects the recombination landscape in Arabidopsis. PNAS. 2012;109:5880–5885.
- Zamudio N, Barau J, Teissandier A, et al. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 2015;29:1256–1270.
- Okita AK, Zafar F, Su J, et al. Heterochromatin suppresses gross chromosomal rearrangements at centromeres by repressing Tfs1/TFIIS-dependent transcription. Commun Biol. 2019;2:1–13.
- Marand AP, Jansky SH, Zhao H, et al. Meiotic crossovers are associated with open chromatin and enriched with Stowaway transposons in potato. Genome Biol. 2017;18:203.
- Guo C, Spinelli M, Ye C, et al. Genome-wide comparative analysis of miniature inverted repeat transposable elements in 19 arabidopsis thaliana ecotype accessions. Sci Rep. 2017;7:2634.
- Hermant C, Torres-Padilla M-E. TFs for TEs: the transcription factor repertoire of mammalian transposable elements. Genes Dev. 2021;35:22–39.
- Jordan IK, Rogozin IB, Glazko GV, et al. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72.
- Miao B, Fu S, Lyu C, et al. Tissue-specific usage of transposable element-derived promoters in mouse development. Genome Biol. 2020;21:255.
- Sundaram V, Cheng Y, Ma Z, et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976.
- Jönsson ME, Ludvik Brattås P, Gustafsson C, et al. Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat Commun. 2019;10:3182.
- Denli AM, Narvaiza I, Kerman BE, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. 2015;163:583–593.
- Clayton EA, Rishishwar L, Huang T-C, et al. An atlas of transposable element-derived alternative splicing in cancer. Philos Trans R Soc B. 2020;375:20190342.
- Le TN, Miyazaki Y, Takuno S, et al. Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acids Res. 2015;43:3911–3921.
- Dolinoy DC. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr Rev. 2008;66:S7–S11.
- Williams BP, Pignatta D, Henikoff S, et al. Methylation-sensitive expression of a DNA demethylase gene serves as an epigenetic rheostat. PLoS Genet. 2015;11:e1005142.
- Lee YCG. The role of piRNA-mediated epigenetic silencing in the population dynamics of transposable elements in Drosophila melanogaster. PLoS Genet. 2015;11:e1005269.
- Horvath R, Slotte T. The Role of Small RNA-Based Epigenetic Silencing for Purifying Selection on Transposable Elements in Capsella grandiflora. Genome Biol Evol. 2017;9:2911–2920.
- Penterman J, Zilberman D, Huh JH, et al. DNA demethylation in the Arabidopsis genome. PNAS. 2007;104:6752–6757.
- Saze H, Shiraishi A, Miura A, et al. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465.
- Espinas NA, Tu LN, Furci L, et al. Transcriptional regulation of genes bearing intronic heterochromatin in the rice genome. PLoS Genet. 2020;16:e1008637.
- Saze H, Kitayama J, Takashima K, et al. Mechanism for full-length RNA processing of Arabidopsis genes containing intragenic heterochromatin. Nat Commun. 2013;4:2301.
- Walter M, Teissandier A, Pérez-Palacios R, et al. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. eLife. 2016;5:e11418.
- Maupetit-Mehouas S, Vaury C. Transposon reactivation in the germline may Be useful for both transposons and their host genomes. Cells. 2020;9:1172.
- Olovnikov I, Chan K, Sachidanandam R, et al. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol Cell. 2013;51:594–605.
- Shpiz S, Ryazansky S, Olovnikov I, et al. Euchromatic transposon insertions trigger production of novel Pi- and Endo-siRNAs at the target sites in the Drosophila germline. PLoS Genet. 2014;10:e1004138.
- Song J, Liu J, Schnakenberg SL, et al. Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol Evol. 2014;6:2786–2798.
- Ronsseray S, Lehmann M, Anxolabehere D. The maternally inherited regulation of P elements in Drosophila melanogaster can be elicited by two P copies at cytological site 1a on the X chromosome. Genetics. 1991;129:501–512.
- Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27:400–412.
- Zanni V, Eymery A, Coiffet M, et al. Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc Natl Acad Sci USA. 2013;110:19842–19847.
- Ellison CE, Kagda MS, Cao W. Telomeric TART elements target the piRNA machinery in Drosophila. PLoS Biol. 2020;18:e3000689.
- Luo S, Lu J. Silencing of transposable elements by piRNAs in Drosophila: an evolutionary perspective. Genomics Proteomics Bioinformatics. 2017;15:164–176.
- Zemach A, Kim MY, Silva P, et al. Local DNA hypomethylation activates genes in rice endosperm. PNAS. 2010;107:18729–18734.
- Calarco JP, Borges F, Donoghue MTA, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205.
- Martínez G, Panda K, Köhler C, et al. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants. 2016;2:16030.
- Anzelon TA, Chowdhury S, Hughes SM, et al. Structural basis for piRNA-targeting . bioRxiv. 2020. DOI:10.1101/2020.12.07.413112.
- Fei Y, Nyikó T, Molnar A. Non-perfectly matching small RNAs can induce stable and heritable epigenetic modifications and can be used as molecular markers to trace the origin and fate of silencing RNAs. Nucleic Acids Res. 2021;49:1900–1913.
- Gilbert C, Feschotte C. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr Opin Genet Dev. 2018;49:15–24.
- Gilbert C, Schaack S, Pace JK, et al. A role for host-parasite interactions in the horizontal transfer of DNA transposons across animal phyla. Nature. 2010;464:1347–1350.
- Obbard DJ, Gordon KHJ, Buck AH, et al. The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci. 2009;364:99–115.
- Wang Y, Liang W, Tang T. Constant conflict between Gypsy LTR retrotransposons and CHH methylation within a stress-adapted mangrove genome. New Phytol. 2018;220:922–935.
- Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60:43–66.
- Lisch D, Slotkin RK. Strategies for silencing and escape: the ancient struggle between transposable elements and their hosts. Int Rev Cell Mol Biol. 2011;292:119–152.
- Lisch D, Bennetzen JL. Transposable element origins of epigenetic gene regulation. Curr Opin Plant Biol. 2011;14:156–161.
- Moelling K, Broecker F. Viruses and evolution – viruses first? A personal perspective. Front Microbiol. 2019;10. DOI:10.3389/fmicb.2019.00523.
- Chiba S, Kondo H, Tani A, et al. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 2011;7(7):e1002146.
- Horie M, Honda T, Suzuki Y, et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature. 2010;463:84–87.
- Chu H, Jo Y, Cho WK. Evolution of endogenous non-retroviral genes integrated into plant genomes. Curr Plant Biol. 2014;1:55–59.
- Palatini U, Miesen P, Carballar-Lejarazu R, et al. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics. 2017;18:512.
- Sun YH, Xie LH, Zhuo X, et al. Domestic chickens activate a piRNA defense against avian leukosis virus. eLife. 2017;6. DOI:10.7554/eLife.24695.
- Petit M, Mongelli V, Frangeul L, et al. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. PNAS. 2016;113:E4218–E4227.
- Denkena J, Johannes F, Colomé-Tatché M. Region-level epimutation rates in Arabidopsis thaliana. Heredity. 2021. 127: 190–202. DOI:10.1101/2020.08.18.255919.
- Jeggo PA, Holliday R. Azacytidine-induced reactivation of a DNA repair gene in Chinese hamster ovary cells. Mol Cell Biol. 1986;6:2944–2949.
- Ashapkin VV, Kutueva LI, Vanyushin BF. Epigenetic variability in plants: heritability, adaptability, evolutionary significance. Russ J Plant Physiol. 2016;63:181–192.
- Pignatta D, Novitzky K, Satyaki PRV, et al. A variably imprinted epiallele impacts seed development. PLoS Genet. 2018;14:e1007469.
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica. 1999;107:27–37.
- McCue AD, Nuthikattu S, Slotkin RK. Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol. 2013;10:1379–1395.
- Manning K, Tör M, Poole M, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952.
- Chen W, Kong J, Qin C, et al. Requirement of CHROMOMETHYLASE3 for somatic inheritance of the spontaneous tomato epimutation Colourless non-ripening. Sci Rep. 2015;5:9192.
- Maumus F, Quesneville H. Deep investigation of arabidopsis thaliana junk DNA reveals a continuum between repetitive elements and genomic dark matter. PLOS ONE. 2014;9:e94101.
- Baduel P, Colot V. The epiallelic potential of transposable elements and its evolutionary significance in plants. Philos Trans R Soc B. 2021;376:20200123.
