 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Co-occurrence of injection drug use (IDU) and hepatitis C virus infection (HCV) is common in people living with HIV (PLWH) and leads to significantly increased mortality. Epigenetic clocks derived from DNA methylation (DNAm) are associated with disease progression and all-cause mortality. In this study, we hypothesized that epigenetic age mediates the relationships between the co-occurrence of IDU and HCV with mortality risk among PLWH. We tested this hypothesis in the Veterans Aging Cohort Study (n = 927) by using four established epigenetic clocks of DNAm age (i.e., Horvath, Hannum, Pheno, Grim). Compared to individuals without IDU and HCV (IDU-HCV-), participants with IDU and HCV (IDU+HCV+) showed a 2.23-fold greater risk of mortality estimated using a Cox proportional hazards model (hazard ratio: 2.23; 95% confidence interval: 1.62–3.09; p = 1.09E–06). IDU+HCV+ was associated with a significantly increased epigenetic age acceleration (EAA) measured by 3 out of 4 epigenetic clocks, adjusting for demographic and clinical variables (Hannum: p = 8.90E–04, Pheno: p = 2.34E–03, Grim: p = 3.33E–11). Furthermore, we found that epigenetic age partially mediated the relationship between IDU+HCV+ and all-cause mortality, up to a 13.67% mediation proportion. Our results suggest that comorbid IDU with HCV increases EAA in PLWH that partially mediates the increased mortality risk.
Introduction
Injection drug use (IDU) is the principal risk factor for hepatitis C virus (HCV) due to the sharing of needles and significantly contributes to HIV infection [Citation1–5]. Evidence shows that IDU increases mortality for people living with HIV (PLWH) and a substantial proportion of IDU-associated mortality is due to co-infection with HCV [Citation6]. HCV infection can lead to liver disease that may manifest as cirrhosis, liver failure, and hepatocellular carcinoma. The comorbid IDU with HCV infection (IDUHCV) further worsens HIV disease progression and increases the mortality risk. Thus, it is important to establish a biological measure as a proxy for an earlier stage of the disease outcomes. Epigenetic age can serve as a functional indicator for infectious disease and predicts frailty and mortality among PLWH [Citation7,Citation8].
Recently developed epigenetic ‘clocks’ employ DNA methylation (DNAm), mainly measured using the blood-nucleated cell methylome, as a proxy of the ageing process [Citation9]. To date, more than a dozen epigenetic clocks have been developed that serve slightly different purposes. Four clocks have been extensively applied to assess the potential impacts of lifestyle and disease on biological ageing. The ‘Horvath’ clock (Horvath) interrogates 353 CpG sites that estimate the epigenetic age of most tissues and cell types in the human body [Citation10]. The ‘Hannum’ clock (Hannum), comprised of 71 CpG sites, measures the epigenetic age specifically in blood and does not generalize well to other tissues [Citation11]. The ‘Pheno’ clock (Pheno) including 513 CpG sites is significantly associated with healthspan and lifespan [Citation12]. The ‘Grim’ clock (Grim) is a composite biomarker based on the seven protein-based surrogates of DNAm and a DNAm‐based estimator of smoking pack-years using 1030 CpG [Citation13]. These clocks have been applied to detect biological age in a variety of medical and psychiatric conditions [Citation14,Citation15]. For example, PLWH have been shown to age 5 to 10 years older compared to healthy individuals [Citation15–17]. Using a monocyte DNAm-based clock, we recently reported a 10- to 15-year epigenetic age acceleration (EAA) among PLWH [Citation15]. Interestingly, evidence suggests that epigenetic age seems reduced or slowed down following antiretroviral therapy [Citation18,Citation19], indicating that other comorbid conditions or risk factors such as IDU and HCV could contribute to a further acceleration of epigenetic age among PLWH.
Previous studies have shown that both IDU and HCV are associated with aberrant DNAm. Hyper- and hypo-methylation of multiple genes among persons infected with HCV have been reported [Citation20–26]. Aberrant DNAm has also been proposed as a biomarker for HCV-associated hepatocellular carcinoma [Citation22–24]. Compared to individuals who are neither IDU nor infected with HCV, we previously reported six differentially methylated CpG sites associated with IDU among persons living with HIV and HCV co-infection [Citation27]. The CpG sites associated with IDUHCV were found in genes involved in inflammatory functions. As increased inflammation is a hallmark mechanism of the ageing process, epigenetic clocks derived from DNAm may serve as an indicator of the biological outcomes of IDUHCV among people living with HIV. Furthermore, a previous study showed that epigenetic age measured by the Horvath clock increased age acceleration among participants with HCV infection with liver fibrosis or HIV co-infection compared to persons without either viral infection [Citation28]. However, the relationships among IDUHCV, epigenetic age, and mortality remain undefined.
In this study, we examined the mortality risk of IDUHCV among PLWH. Using the four established clocks (i.e., Horvath, Hannum, Pheno, Grim), we tested whether IDUHCV increased EAA. We then conducted a mediation analysis to quantify the effects of IDUHCV on mortality explained by epigenetic age. Given a high prevalence of IDU and HCV among PLWH, it is important to investigate the individuals who have comorbid IDU and HCV (IDU+HCV+) compared to samples without IDU or HCV infection (IDU-HCV-) as an initial step towards establishing the relationship of individual effect of IDU and HCV on epigenetic age. Here, we aimed to examine whether IDUHCV accelerated biological age and tested a potential mediating role of epigenetic age acceleration in IDUHCV-associated mortality risk. However, it is important to note that the study design did not directly assess the impact of drug use (i.e., cannabis, cocaine, stimulant, and opioid use) on epigenetic age for those with HIV and HCV coinfection.
Methods
Population characteristics
The Veterans Aging Cohort Study (VACS) is a longitudinal, prospective, and multisite observational cohort study of US veterans that includes PLWH [Citation29]. Biospecimens including DNA samples derived from whole blood were collected in a subset of the cohort. In this project, we profiled DNAm for 927 PLWH samples (n = 414 IDU+HCV+ and n = 513 IDU-HCV-). A majority of PLWH (63.7%) were virally suppressed on antiretroviral therapy (ART). Survival data were obtained from medical records. Demographic and clinical characteristics are summarized in .
Table 1. Demographic and clinical characterizations.
DNA methylation, data quality control (QC), and epigenetic clock measures
Epigenome-wide CpG methylation was profiled using either Illumina HumanMethylation450 BeadChip (450K) (San Diego, CA, USA) (57.2% of the sample) or Illumina HumanMethylation EPIC BeadChip (EPIC) (San Diego, CA, USA) (42.8% of the sample) in VACS. All samples were processed at the Yale Center for Genomic Analysis [Citation27]. The same QC criteria were used as in our previous studies [Citation27,Citation30,Citation31]. We retrieved methylation raw data using the minfi R package (version 1.18.1) and all probes were normalized. Methylation-inferred sex agreed with self-reported sex (100% of the sample was male).
Epigenetic age was estimated using four epigenetic clocks, the Horvath [Citation10], Hannum [Citation11], Pheno [Citation12], and Grim [Citation13] clocks. EAA was defined as the residuals of regressing epigenetic age on chronological age, which was applied to test the mediation effect on the relationship between IDUHCV and mortality risk [Citation10,Citation14]. We also calculated the mean difference in EAA between IDU+HCV+ and IDU-HCV- and denoted it as the median difference of EAA (MDEAA).
Statistical Analysis
The workflow diagram is presented in Age, self-reported race/ethnicity, body mass index (BMI), smoking status, and alcohol consumption were considered as covariates and were adjusted in each model. Smoking status was assessed by self-report and grouped by smoker and non-smoker. Alcohol consumption was assessed by measuring phosphatidylethanol (PEth) levels, a biomarker for alcohol use [Citation32].
Figure 1. The workflow of mediation analysis. IDUHCV: comorbid injection drug use with hepatitis C virus status. EAA: Epigenetic Age Acceleration.
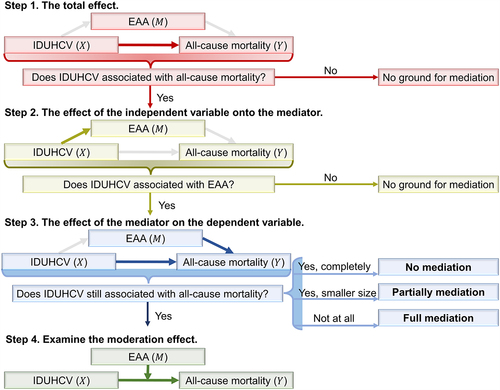
IDUHCV survival analysis among PLWH
Using survival analysis, we tested the total effect of IDUHCV on all-cause mortality in 927 PLWH. The proportional hazard assumption was tested prior to the application of Cox regression analysis. Then, the Cox proportional hazards regression survival analysis was performed to investigate the hazard ratio (HR) of IDUHCV on all-cause mortality, adjusting for relevant covariates [Citation33,Citation34]. The following model was used to calculate the adjusted HR:
where the hazard function is determined by
;
is the independent variable IDUHCV and
is the
covariate;
and
denote the effect size of IDUHCV and covariates in step 1, respectively. The HR was calculated by
. The Z-test (
) was used to determine if the value of
differs significantly from 0.
Association between IDUHCV and EAA
The following linear model was applied to test the association between IDUHCV and EAA after adjusting for relevant covariates:
where denotes the mediator EAA;
and
,
denotes the effect size of IDUHCV in step 2, adjusting for the same covariates as mentioned above. The t-test was used to conduct hypothesis tests on the regression coefficients obtained in linear regression. That is, we tested
vs.
using t-test
, where
is the sample size.
Mediation effect of EAA on IDUHCV associated all-cause mortality
We simultaneously tested the effects of the mediator (EAA quantified by each of the four epigenetic clocks) and the independent variable (IDUHCV) on the dependent variable (mortality). Cox proportional hazards regression survival analysis was used to investigate the HR of IDUHCV, the potential mediators (EAA), and relevant covariates on all-cause mortality. We used the following model to calculate the adjusted HR:
where ,
, and
, denote the effect size of IDUHCV, EAA, and relevant covariates in step 2, respectively. We use
and
to denote the p-values for testing coefficient
and
, respectively.
We estimated various quantities for causal mediation analysis, meaning that we compared the direct to the indirect effect, allowing us additional insight into the data. The effect size, , in step 1 was defined as the Total Effect
in the mediation analysis (
). The effect size,
, in step 3 was defined as the average Direct Effect (
. We also defined the Indirect Effect (
average causal mediation effect) as the multiplication of
in step 2 and
in step 3 (
). The Proportion of Indirect Effect (
, the proportion mediation effect of the independent variable on the dependent variable that goes through the mediator, also called mediation proportion) was defined as
.
For the potential mediating effect of EAA for each epigenetic clock that was significantly associated with IDUHCV in step 2, we further investigated whether it was a full mediation or partial mediation effect. If and
, indicating a full mediation effect; If
,
, and
, that is, the effect of IDUHCV on mortality became smaller when adding the EAA as a mediator compared with the model not including EAA, indicating a partial mediation; otherwise, there was no mediation effect.
Moderation effect of EAA on relationship between IDUHCV and all-cause mortality
For the potential mediators, we further analysed the moderation effect. Rather than mediation analysis attempting to show that a variable is the channel through which independent variable influences dependent variable, moderation analysis attempts to show that a variable changes the relationship between independent variable and dependent variable. We used the following model to further accompany the test for the moderation effect.
where ,
, and
, denote the effect size of IDUHCV, EAA, and relevant covariates, respectively. A significant
coefficient signals the presence of a moderating effect or interaction effect [Citation35,Citation36].
Results
As shown in , most participants were self-reported African American, and all were male. Participants who were IDU+HCV+ were older than those who were IDU-HCV- (t-test p = 8.20E–09). All participants were PLWH on ART. There was no significant difference in the HIV-1 plasma RNA viral load between the two groups (chi-square test p = 2.30E–01). Compared to the IDU-HCV- group, participants in the IDU+HCV+ group had a higher proportion reporting cigarette smoking and higher rates of drug use including cannabis, cocaine, stimulant, and opioid use (P-value2.10E–10).
IDUHCV increases the risk of all-cause mortality among PLWH
Kaplan-Meier curves showed that the group with comorbid IDU+HCV+ was associated with lower survival probability compared to the IDU-HCV- group (). After adjusting for covariates (i.e., age, self-reported race/ethnicity, BMI, smoking status, alcohol consumption), the HR of IDUHCV was 2.23 (95%CI: 1.62–3.09; p = 1.09E–06) (). Of note, self-reported race/ethnicity, BMI, smoking status, and alcohol consumption were not associated with all-cause mortality after adjusting for multiple tests.
Figure 2. The results of mediation analysis. a. Kaplan-Meier curves of IDU+HCV+ and IDU-HCV-. b. Comparison of estimations of Epigenetic Age Acceleration (EAA) between IDU+HCV+ and IDU-HCV-. c. Mediation effect of EAA of IDUHCV on all-cause mortality. IDU+HCV+: People who Inject Drugs Use also co-infected with Hepatitis C Virus; IDU-HCV-: People who without Injection Drugs Use and without Hepatitis C Virus infection.
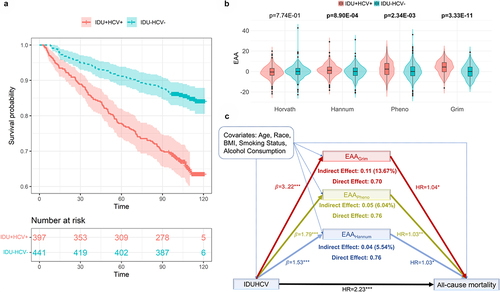
Table 2. Cox proportional hazards regression analysis for assessing the association of comorbid injection drug use with hepatitis C virus (IDUHCV) with survival rate.
IDUHCV is associated with EAA
EAA measured by three of the four epigenetic clocks showed a significant association with IDUHCV adjusting age, self-reported race/ethnicity, BMI, smoking status, and alcohol consumption as covariates (). Each of the three clocks (i.e., Hannum, Pheno, Grim) showed an increased EAA. Median differences in EAA between the two groups were 1.36 years for the Hannum clock, 2.02 years for the Pheno clock, and 4.16 years for the Grim clock. shows the comparisons of EAAs from three clocks between IDU+HCV+ and IDU-HCV-. We found no significant association of Horvath EAA with IDUHCV.
Table 3. Association between comorbid injection drug use with hepatitis C virus (IDUHCV) and Epigenetic Age Acceleration (EAA) among people living with HIV.
Epigenetic ageing mediates the association of IDUHCV with all-cause mortality
We estimated the proportion of IDUHCV on all-cause mortality that was explained by epigenetic ageing via mediation analysis (). The average Direct Effect () was obtained by Cox proportional hazards regression (
,
;
,
;
,
). The Indirect Effect (
) was estimated from the linear model in step 2 and the Cox proportional hazards regression in step 3 (
;
;
). All three clocks showed partial mediation effects (). We also calculated the Proportion of Indirect Effect for each of the three epigenetic clocks (
;
;
). We further examined the moderation effect of these three potential mediators. These three clocks did not show a significant moderation effect or interaction effect, which further confirmed the partial mediation effects of these three clocks between IDUHCV and all-cause mortality.
Table 4. Result of Cox proportional hazards regression analysis for assessing the association between EAA and survival rate with adjustment for the comorbid injection drug use with hepatitis C virus (IDUHCV) status and covariates (age, self-reported race/ethnicity, body mass index, smoking status, alcohol consumption).
Discussion
In this study, we demonstrated that IDUHCV significantly increased EAA among PLWH which contributed to the increased risk of all-cause mortality. We found that IDU+HCV+ had a 2.23-fold higher mortality risk than IDU-HCV-. PLWH with IDU+HCV+ showed an average of up to 4.16 years acceleration compared to the IDU-HCV- group. Importantly, we identified that the impact of IDUHCV on mortality risk was mediated in part by EAA. Our findings provide new insights, which deepen our understanding of the relationship of IDUHCV on mortality among PLWH.
The underlying mechanisms of IDUHCV associated with EAA have not been previously understood. DNAm alterations of the genes involved in the ageing process may play an important role in the age acceleration among IDU+HCV+. Inflammation is one of the hallmarks of ageing and DNAm alterations to inflammatory genes contribute to age acceleration and age-related diseases [Citation37]. We previously reported that hypomethylation of the promoters of NLRC5, TRIM69, and HLA-E was associated with IDU+HCV- compared to IDU-HCV- individuals [Citation27]. We also identified six differentially methylated regions in the major histocompatibility complex. Dysregulation of these genes could lead to disruption of immune functions and changes in inflammasomes. For example, NLRC5 is a negative regulator of the Nuclear Factor Kappa B (NF-κB) signalling pathway-mediated inflammatory response [Citation38]. Hypomethylation of the NLRC5 promoter region results in an increase in gene expression that may reduce the inhibitory effect of NLRC5 on inflammation. Thus, the alteration of DNAm in the inflammasome among IDU+HCV+ in PLWH likely results in an increase of EAA due to chronic inflammation that eventually leads to the increased mortality risk. These findings add new knowledge about the potential consequences of maladaptive DNAm due to high-risk behaviours in the setting of HCV and HIV infections. Early detection of EAA may provide a window of opportunity to slow down the epigenetic clock by changing the behaviour and treating HCV infection.
Our findings provide evidence that an increased mortality risk among individuals with IDU+HCV+ among PLWH is partially mediated by EAA. The mediation effect varies among three estimators, each of them accounting for 5.54% (Hannum), 6.04% (Pheno), and 13.67% (Grim) risk of IDUHCV affecting all-cause mortality, respectively. It is worth noting that the mediation effects of EAA varied among different epigenetic clocks used, with Grim clock showing the largest mediation effects compared to other clocks. Furthermore, a recent study also showed that Grim had the higher mortality discrimination when compared to the other three clocks [Citation39]. This discrepancy may be due to the different sets of CpG sites selected in each clock and their respective utilities. The Hannum and Horvath clocks were built on the CpGs to optimize the prediction of chronological age [Citation10,Citation11], while the CpGs in Pheno and Grim clocks aimed to predict a multi-system proxy of physiological dysregulation [Citation12,Citation13]. The Hannum clock was developed from whole blood, while the Horvath clock was built from multi-tissues and cell types. Additionally, the EAAs estimated by Pheno clock and Grim clock had the largest mediation proportions as both clocks were developed using a Cox penalized regression model that treated mortality hazard as the outcome.
Our results highlight the mediating role of epigenetic ageing in the IDUHCV-associated mortality risk when compared to non-biological risk factors. The clinical factors may be associated with mortality, but do not provide evidence to have mediation effect between IDUHCV and all-cause mortality. For instance, the Grim clock, which showed 13.67% mediation effect between IDUHCV and morality risk, was constructed by selecting CpG sites associated with seven proteins that have been linked to morbidity and mortality (i.e., adrenomedullin, beta-2-microglobulin, cystatin C, growth/differentiation factor-15, leptin, plasminogen activator inhibitor 1, and tissue inhibitor metalloproteinases 1), as well as smoking pack-years. These proteins are associated with immune function and inflammatory process. For example, beta-2-microglobulin (B2M) is associated with the major histocompatibility complex (MHC) class I heavy chain and has been linked to mortality risk in people living with HIV [Citation39–41]Citation39. Several studies have shown that smoking-associated DNA methylation features are predictive of mortality risk. Thus, while the mediation effects of epigenetic clocks are small to moderate, our results provide insights into the dysregulation of the protein that contributes to IDUHCV associated mortality risk among people living with HIV.
Of note, even though epigenetic age was strongly associated with all-cause mortality and the epigenetic age mediates the effect of IDUHCV on mortality risk, a large proportion of IDUHCV-related mortality risk remains unidentified. One possibility is mortality due to overdose among a proportion of intravenous drug users, which may confound the effect of EAA. The direction of confounding could be either negative or positive, but regardless, prior study of the association of the impact of IDU on EAA [Citation40] suggests that the differences in EAA between the IDU+HCV+ and IDU-HCV- groups are not due solely to HCV infection. Secondly, the established clocks are mostly derived from heterogeneous cells, which have a limited capacity to accurately measure organ age acceleration [Citation41]. Tissue/cell type-specific epigenetic clocks are warranted to improve the measurement of epigenetic age effects on mortality among those with IDU+HCV+. Epigenetic clocks including liver enzymes will be helpful in the future.
Several limitations should be considered when interpreting the current findings. Our study was mainly focused on IDU+HCV+ and IDU-HCV- groups because there were only 273 (21.7%) participants of IDU-HCV+ and 42 (3.3%) participants of IDU+HCV- in the study sample. We lack statistical power to detect the effect of IDU or HCV alone on epigenetic age. To further understand the synergic effects of IDU and HCV on epigenetic age among PLWH, further work is needed to include four group comparisons: IDU+HCV+, IDU-HCV+, IDU+HCV-, and IDU-HCV-. Secondly, epigenetic age estimation was based on cross-sectional DNAm data. The results reflect a snapshot of the ageing process. Longitudinal epigenetic age estimation may provide more insights into the mediation role of epigenetic age on risk behavioural contribution to mortality. As discussed earlier, the epigenetic clocks we applied were calculated using whole blood samples that are not the best indicators of cell or organ age. Future analyses using cell-type-specific epigenetic clocks may shed new light on the biological mechanisms.
Conclusion
Our study found evidence of an association between IDUHCV and all-cause mortality in people living with HIV, which was partially mediated by epigenetic age acceleration. These findings provide new insights into the role of epigenetic age in how IDUHCV affects all-cause mortality. Further evaluation of these potential biomarkers is warranted to deepen our understanding of the relationship between epigenetics and mortality in people living with HIV.
Author contributions
XL was responsible for bioinformatics data processing, statistical analyses, and manuscript preparation. ACJ provided DNA samples and clinical data and contributed to the interpretation of findings and manuscript preparation. VCM was involved in data interpretation and manuscript preparation. BEA contributed to the interpretation of findings and manuscript preparation. KX was responsible for the study design, study protocol, sample preparation, interpretation of findings, and manuscript preparation. All authors read and approved the final manuscript.
Availability of data and materials
Demographic variables, clinical variables, and methylation status for the VACS samples are submitted to the GEO dataset (GSE117861) and are available to the public.
Ethics approval and consent to participate
The study was approved by the committee of the Human Research Subject Protection at Yale University and the Institutional Research Board Committee of the Connecticut Veteran Healthcare System. All participants provided written consent.
Acknowledgments
The authors appreciate the support of the Veterans Aging Study Cohort Biomarker Core (VACSBC), the contributions of the VACS participants, and the Yale Center of Genomic Analysis. The views and opinions expressed in this manuscript are those of the authors and do not necessarily represent those of the Department of Veterans Affairs or the United States government. This work uses data provided by patients and collected by the VA as part of their care and support.
Disclosure statement
VCM has received investigator-initiated research grants (to the institution) and consultation fees (both unrelated to the current work) from Eli Lilly, Bayer, Gilead Sciences, and ViiV. The remaining authors declare that they have no competing interests.
Additional information
Funding
References
- Amon JJ, Garfein RS, Ahdieh-Grant L, et al. Prevalence of hepatitis C virus infection among injection drug users in the United States, 1994-2004. Clin Infect Dis. 2008;46(12):1852–11. DOI:10.1086/588297
- Blake A, Smith JE. Modeling hepatitis C elimination among people who inject drugs in new hampshire. JAMA Netw Open. 2021;4(8):e2119092.
- Hagan H, Pouget ER, Des Jarlais DC, et al. Meta-regression of hepatitis C virus infection in relation to time since onset of illicit drug injection: the influence of time and place. Am J Epidemiol. 2008;168(10):1099–1109.
- Nelson PK, Mathers BM, Cowie B, et al. Global epidemiology of hepatitis B and hepatitis C in people who inject drugs: results of systematic reviews. Lancet. 2011;378(9791):571–583. DOI:10.1016/S0140-6736(11)61097-0
- Strathdee SA, Hallett TB, Bobrova N, et al. HIV and risk environment for injecting drug users: the past, present, and future. Lancet. 2010;376(9737):268–284. DOI:10.1016/S0140-6736(10)60743-X
- May MT, Justice AC, Birnie K, et al. Injection drug use and hepatitis C as risk factors for mortality in HIV-Infected individuals: the antiretroviral therapy cohort collaboration. J Acquir Immune Defic Syndr. 2015;69(3):348–354. DOI:10.1097/QAI.0000000000000603
- Oursler KK, Marconi VC, Wang Z, et al. Epigenetic age acceleration markers are associated with physiologic frailty and all-cause mortality in people with HIV. Clin Infect Dis. 2022;76:e638–644.
- Lagathu C, Cossarizza A, Bereziat V, et al. Basic science and pathogenesis of ageing with HIV: potential mechanisms and biomarkers. AIDS. 2017;31(2):S105–19.
- Lopez-Otin C, Blasco MA, Partridge L, et al. The hallmarks of aging. Cell. 2013;153(6):1194–1217.
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115.
- Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. DOI:10.1016/j.molcel.2012.10.016
- Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. DOI:10.18632/aging.101414
- Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–327. DOI:10.18632/aging.101684
- Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384.
- Liang X, Sinha R, Justice AC, et al. A new monocyte epigenetic clock reveals nonlinear effects of alcohol consumption on biological aging in three independent cohorts (N = 2242). Alcohol Clin Exp Res. 2022;46:736–748.
- Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis. 2015;212(10):1563–1573.
- Boulias K, Lieberman J, Greer EL. An epigenetic clock measures accelerated aging in treated HIV infection. Mol Cell. 2016;62(2):153–155.
- Nelson KN, Hui Q, Rimland D, et al. Identification of HIV infection-related DNA methylation sites and advanced epigenetic aging in HIV-positive, treatment-naive U.S. veterans. AIDS. 2017;31(4):571–575. DOI:10.1097/QAD.0000000000001360
- Esteban-Cantos A, Rodriguez-Centeno J, Barruz P, et al. Epigenetic age acceleration changes 2 years after antiretroviral therapy initiation in adults with HIV: a substudy of the NEAT001/ANRS143 randomised trial. Lancet HIV. 2021;8(4):e197–205. DOI:10.1016/S2352-3018(21)00006-0
- Zheng Y, Hlady RA, Joyce BT, et al. DNA methylation of individual repetitive elements in hepatitis C virus infection-induced hepatocellular carcinoma. Clin Epigenetics. 2019;11(1):145. DOI:10.1186/s13148-019-0733-y
- Mekky MA, Salama RH, Abdel-Aal MF, et al. Studying the frequency of aberrant DNA methylation of APC, P14, and E-cadherin genes in HCV-related hepatocarcinogenesis. Cancer Biomark. 2018;22(3):503–509.
- Cai C, Xie X, Zhou J, et al. Identification of TAF1, SAT1, and ARHGEF9 as DNA methylation biomarkers for hepatocellular carcinoma. J Cell Physiol. 2020;235(1):611–618.
- Lambert MP, Paliwal A, Vaissiere T, et al. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J Hepatol. 2011;54(4):705–715. DOI:10.1016/j.jhep.2010.07.027
- Wu HC, Yang HI, Wang Q, et al. Plasma DNA methylation marker and hepatocellular carcinoma risk prediction model for the general population. Carcinogenesis. 2017;38(10):1021–1028.
- Kuramoto J, Arai E, Tian Y, et al. Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis. Carcinogenesis. 2017;38(3):261–270.
- Song MA, Kwee SA, Tiirikainen M, et al. Comparison of genome-scale DNA methylation profiles in hepatocellular carcinoma by viral status. Epigenetics. 2016;11(6):464–474.
- Zhang X, Hu Y, Justice AC, et al. DNA methylation signatures of illicit drug injection and hepatitis C are associated with HIV frailty. Nat Commun. 2017;8(1):2243.
- Gindin Y, Gaggar A, Lok AS, et al. DNA methylation and immune cell markers demonstrate evidence of accelerated aging in patients with chronic Hepatitis B virus or Hepatitis C virus, with or without human immunodeficienct virus co-infection. Clin Infect Dis. 2021;73(1):e184–90.
- Justice AC, Dombrowski E, Conigliaro J, et al. Veterans Aging Cohort Study (VACS): overview and description. Med care. 2006;44(8 Suppl 2):S13–24.
- Xu K, Zhang X, Wang Z, et al. Epigenome-wide association analysis revealed that SOCS3 methylation influences the effect of cumulative stress on obesity. Biol Psychol. 2018;131:63–71.
- Zhang X, Justice AC, Hu Y, et al. Epigenome-wide differential DNA methylation between HIV-infected and uninfected individuals. Epigenetics. 2016;11(10):750–760.
- Liang X, Justice AC, So-Armah K, et al. DNA methylation signature on phosphatidylethanol, not on self-reported alcohol consumption, predicts hazardous alcohol consumption in two distinct populations. Mol Psychiatry. 2021;26(6):2238–2253.
- Cox DR. Regression models and life‐tables. J Royal Stat Soc Ser B (Methodological). 1972;34(2):187–202.
- Therneau TM, Lumley, T. Package ‘survival’. R Top Doc. 2015;128(10):28–33.
- Edwards KD, Konold TR. Moderated mediation analysis: a review and application to school climate research. Pract Assess Res Eval. 2020;25(1):5.
- Saunders DR. Moderator variables in prediction. Educ Psychol Meas. 1956;16(2):209–222.
- Conole ELS, Stevenson AJ, Munoz Maniega S, et al. DNA methylation and protein markers of chronic inflammation and their associations with brain and cognitive aging. Neurology. 2021;97(23):e2340–52.
- Lupfer C, Kanneganti TD. The expanding role of NLRs in antiviral immunity. Immunol Rev. 2013;255(1):13–24.
- Oursler KK, Marconi VC, Wang Z, et al. Epigenetic age acceleration markers are associated with physiologic frailty and all-cause mortality in people with human immunodeficiency virus. Clin Infect Dis. 2023;76(3):e638–44.
- Bachi K, Sierra S, Volkow ND, et al. Is biological aging accelerated in drug addiction? Curr Opin Behav Sci. 2017;13:34–39.
- Horvath, S, Lin DTS, Kobor MS, et al. HIV, pathology and epigenetic age acceleration in different human tissues. Geroscience. 2022
