ABSTRACT
Although A-to-I RNA editing leads to similar effects to A-to-G DNA mutation, nonsynonymous RNA editing (recoding) is believed to confer its adaptiveness by ‘epigenetically’ regulating proteomic diversity in a temporospatial manner, avoiding the pleiotropic effect of genomic mutations. Recent discoveries on the evolutionary trajectory of Ser>Gly auto-editing site in insect Adar gene demonstrated a selective advantage to having an editable codon compared to uneditable ones. However, apart from pure observations, quantitative approaches for justifying the adaptiveness of individual RNA editing sites are still lacking. We performed a comparative genomic analysis on 113 Diptera species, focusing on the Adar Ser>Gly auto-recoding site in Drosophila. We only found one species having a derived Gly at the corresponding site, and this occurrence was significantly lower than genome-wide random expectation. This suggests that the Adar Ser>Gly site is unlikely to be genomically replaced with G during evolution, and thus indicating the advantage of editable status over hardwired genomic alleles. Similar trends were observed for the conserved Ile>Met recoding in gene Syt1. In the light of evolution, we established a comparative genomic approach for quantitatively justifying the adaptiveness of individual editing sites. Priority should be given to such adaptive editing sites in future functional studies.
Introduction
The metazoan ADAR (adenosine deaminase acting on RNA) enzyme, which mainly expresses in the nervous system, catalyses adenosine-to-inosine (A-to-I) RNA editing [Citation1]. To date, A-to-I editing is found to be one of the most widely observed RNA modifications in animals, with millions of adenosines being potentially editable in animal transcriptome [Citation1–3]. Since inosine is read as guanosine, A-to-I RNA editing shares similar properties with A-to-G DNA mutations (). As a result, editing within the coding sequences (CDS) has the potential to alter the genetically encoded amino acids, resulting in nonsynonymous mutations, often referred to as ‘recoding’ [Citation4]. Nonetheless, a fundamental difference between A-to-I RNA editing and A-to-G DNA mutations lies in the flexibility of RNA editing that could be regulated in a temporal-spatial manner, whereas DNA mutations are permanently integrated into the genome and may trigger pleiotropic effects. For example, a DNA mutation might benefit the adult fruitfly but be deleterious or lethal to the larva, and then this mutation could not be maintained during evolution as antagonism appeared between different developmental stages. But RNA editing provides an epigenetic approach for regulating the proteome.
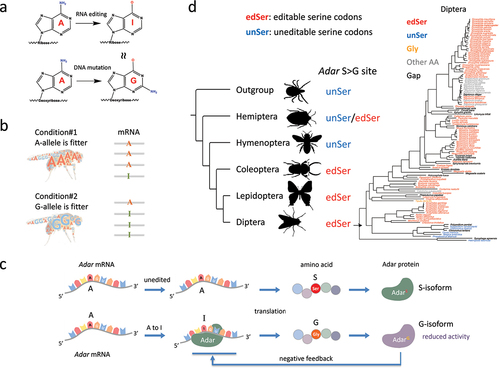
Figure 1. A-to-I RNA editing and the evolution of adar S > G auto-recoding site in insects. (a) A-to-I RNA editing mediated by ADAR protein. I is recognized as G by cellular machineries. (b) Predictions made by the adaptive RNA editing hypothesis. A and G alleles are fitter under different conditions while RNA editing could adjust the relative proportions of two alleles. When a is fitter under a particular condition, the editing level decreases to represent more A, and vice versa. (c) The adar S > G auto-recoding site forms a negative feedback loop that stabilizes the RNA editing efficiency. (d) Evolution of adar S > G site in insects. Editable serine codons are colored in red; uneditable serine codons are colored in blue; glycine codons are colored in orange. The phylogenetic tree of all diptera species was colored with the codon classification of adar S > G site.
Considering the advantages of RNA editing over DNA mutation, early studies posited that nonsynonymous RNA editing serves as a driving force for adaptive evolution. This is because the editing mechanism can diversify the transcriptome and proteome when needed, aiding organisms in adapting to variable environments [Citation5,Citation6]. Numerous studies have reported the overrepresentation of nonsynonymous RNA editing events in the transcriptomes of various species, such as insects [Citation7–10], cephalopods [Citation4,Citation11,Citation12], microbes [Citation13–16], and seed plants [Citation17,Citation18], suggesting the positive selection on recoding events. This prompts us to believe that there is a selective advantage of the seemingly adaptive RNA editing mechanism. However, it remains unanswered how can we support the notion that ‘having the potential to be edited’ (editable allele) is superior to ‘having only one version of the A- or G-allele’ (uneditable allele or fixed-G)? Under the adaptive RNA editing hypothesis, it is expected that the A- and G-alleles should have higher fitness under different conditions, respectively (e.g., A is better under condition#1 and G is better under condition#2), and the RNA editing level could be regulated accordingly (more A under condition#1 and more G under condition#2) so that the editable state gains an advantage due to this flexibility ().
Genome-wide studies and debates on the adaptation of nonsynonymous RNA editing focus on cephalopods where recoding events are prevalent. Earlier studies first found that the numbers and editing levels of recoding sites in cephalopods exceeded the neutral expectation, therefore prompting that recoding events were overall adaptive [Citation4,Citation11]. Then, a non-adaptive explanation was proposed based on restorative and harm-permitting theories [Citation19]. Later, new genomic evidences were found to support the adaptive and diversifying hypothesis [Citation20,Citation21]. These comparative genomic studies have the following common features: (1) they essentially investigate the selection force acting on the overall (rather than individual) nonsynonymous editing sites, such as observing the overrepresentation of recoding sites, or analysing the fraction of recoding sites being replaced with other nucleotides during evolution; (2) the number of species used was usually restricted to the species with high-quality transcriptome data. For example, four coleoids species were used in Liscovitch-Brauer et al. 2017 paper [Citation11] and two more species were added in Shoshan et al. 2021 paper [Citation21]. Given a single nonsynonymous editing site, there still lacks an in silico approach to judge whether this recoding site is adaptive or not. Particularly, we expect an approach that analyses the genome evolution alone and thus avoids the requirements on transcriptome data, then more species could be used. This notion will be mentioned later again.
For justifying the adaptiveness of individual RNA editing sites, a recent research in fungi has taken significant steps towards addressing ‘how the editable state is better than the uneditable state or hardwired G’ by constructing mutant strains [Citation22]. In the case of a specific A-to-I RNA editing site in Fusarium graminearum, solid observations proved that the genomically uneditable mutant exhibits higher fitness during the asexual stage, while the genomically fixed-G mutant is fitter during the sexual stage. In contrast, the wild-type (being editable) could smartly regulate the editing level and achieves a higher average fitness across both stages [Citation22]. This is the first experimental evidence showing the real adaptiveness of an individual RNA editing site. Then, similar conclusions were made based on observations of differential protein dynamics of edited and unedited isoforms under different temperatures [Citation23–25]. However, in addition to these case studies requiring functional experiments, there is a growing anticipation for the development of a comparative genomics approach capable of deducing the advantages (or necessity) of RNA editing over the uneditable state or hardwired G-allele. The previous genome-wide studies largely focused on the global trend of selection pattern on nonsynonymous editing sites, but it is relatively difficult to tell exactly which editing sites are adaptive [Citation11,Citation21]. This is because for an individual editing site, the evolutionary transcriptomic methodology will be constrained by the limited number of species with qualified transcriptome data. Therefore, given a particular recoding site, we ask how could we know the necessity (adaptiveness) of RNA editing from comparative genomic approaches and without conducting experiments on the uneditable and fully edited mutants? This in silico approach, once established, could significantly expand the catalogue of adaptive RNA editing sites, enhancing our understanding of the evolutionary significance of the RNA editing mechanism.
In this article, we will first present our recent discoveries concerning the evolutionary trajectory of S>G auto-recoding site in the insect Adar gene, asserting that this represents the initial phase of an experiment-free approach for identifying potential adaptive RNA editing sites. Then, we tried to propose additional comparative genomic approach to infer the adaptiveness of RNA editing. Particularly, for the Adar S>G auto-recoding site in Drosophila, we only found 1 out of 113 Diptera species had a derived Gly at the corresponding site, and this extremely low fraction was significantly lower than the random expectation. This suggests that the A-to-I editing sites, at least for this particular S>G site, are unlikely to be genomically replaced with G during evolution, and thus indicating the advantage of editable sites over hardwired genomic alleles. In the light of evolution, we established a comparative genomic approach for quantitatively justifying the adaptiveness of RNA editing. We provide novel and interesting perspectives for the RNA editing community.
Results
Gain of Adar S>G auto-recoding site in insects indicates the selective advantage of RNA editing
Early studies uncovered that the sole Adar gene in insect genome, which is orthologous to mammalian ADAR2 [Citation26], possesses an auto-recoding site (S>G site) in Drosophila melanogaster and this editing event is likely to be highly conserved in (at least) the Drosophila genus [Citation27]. Notably, the function of Drosophila S>G site is well studied. Editing at this S>G site converts a serine codon (AGC) into a glycine codon (GGC), leading to the edited AdarG isoform with reduced catalytic activity compared to the unedited AdarS isoform, establishing a negative feedback loop (as shown in ). Despite the deep conservation of the editing event on this editable serine codon (AGC/T, denoted as edSer) in several Drosophila species [Citation27], the sequence alignment and phylogeny of all 400 high-quality arthropod genomes revealed that the common ancestor of insects had an uneditable serine codon (TCN, denoted as unSer) at this position () [Citation27]. This evolutionary transition from unSer-to-edSer highlights the potential advantage of having the ability to be edited (editable) compared to being constantly limited to only one protein version (uneditable or hardwired G). Moreover, at the corresponding position of this Adar S>G site, our studies suggest that the unSer-to-edSer transition independently occurred for multiple times during insect evolution, reinforcing the selective advantage of RNA editing [Citation27,Citation28]. Notably, the analyses are not restricted to the species with qualified transcriptome, which largely broaden the evolutionary scale. Meanwhile, we also stress that the analysis on Adar Ser>Gly site largely benefits from the fact that Ser has six codons and two of which were editable at the 1st codon position (AGC/T) and four were uneditable at the 1st codon position (TCN). The other qualified recoding sites are (1) Arg>Gly (AGA/G>GGA/G): Arg has six codons and two of which were editable at the 1st codon position (AGA/G) and four were uneditable at the 1st codon position (CGN); (2) Ile>Met (ATA>ATG): Met has three codons one of which was editable at the 3rd codon position (ATA) and two were uneditable at the 3rd codon position (ATC/T). For other nonsynonymous editing codon, they do not have an uneditable counterpart that could prevent the nonsynonymous editing and maintain the pre-edited amino acid.
Thus, using the Adar Ser>Gly auto-recoding site with known function as an example, our recent works followed the previous efforts in judging the adaptation of RNA editing from an evolutionary angle [Citation11,Citation19,Citation21], and continued to look for an experiment-free methodology for identifying potentially adaptive and functional RNA editing sites (at single site level). The key innovative concept is to first find a recoded codon with an uneditable counterpart and then make full use of the phylogeny and evolutionary history of the editing site to illustrate the importance of having an editable status compared to an uneditable ancestral state.
A new prediction for adaptive RNA editing based on comparative genomics
Notably, the previously proposed observations of the uneditable-to-editable transition during evolution [Citation27,Citation28] were ‘pure descriptive’ although it might suggest a selective advantage of having the editable codon. A quantitative approach with neutral expectation is still lacking. Here, we propose another quantitative genomic criterion to judge whether an RNA editing site is adaptive. Take the Drosophila Adar S>G site for instance, if this editing event is advantageous due to its flexibility (editability), then its genomic sequence should not be replaced with G during evolution because a hardwired G would abolish the flexibility (). As a consequence, genomic G at this corresponding position is deleterious and should be suppressed across the phylogeny (). The strength of suppression is absolutely quantifiable based on the following workflow.
Figure 2. Putative evidence for adaptive RNA editing based on comparative genomic analysis. (a) For a potentially adaptive A-to-I RNA editing site, the editable status is fitter than hardwired G. (b) The genomic A-to-G mutations should be depleted at adaptive RNA editing sites because the hardwired G-allele would abolish the flexibility conferred by RNA editing. (c) As a control, unedited adenosines do not have a clear (predictable) fitness change after A-to-G mutation. (d) For the unedited adenosines in D. melanogaster. The genomic replacement of A-to-G should be frequently observed in the phylogeny.
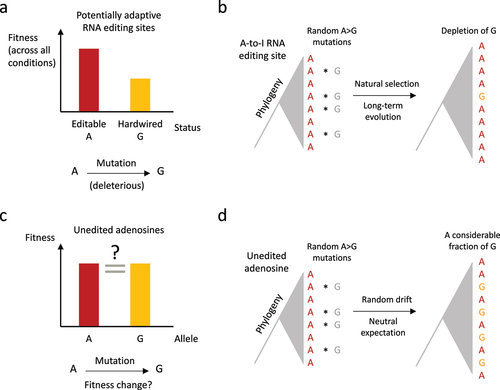
For the numerous unedited adenosines in CDS, there is no clear evidence for the advantage of genomic A over genomic G (), and therefore we should not expect a suppression of A-to-G transition in the phylogeny (). With such a clear prediction, we could test whether a particular (nonsynonymous) RNA editing site is adaptive by looking at the A-to-G transition rate in the phylogeny: if the editing site has significantly lower transition rate than the unedited adenosines (e.g., unedited adenosines within the same CDS), then this might serve as supporting evidence for the flexible advantage of RNA editing mechanism, at least for the particular editing site.
Adar alignment in diptera species
The ancestral state of the Adar S>G auto-recoding site was an uneditable serine codon (TCN, unSer) at the common ancestor of all insects, while in Diptera the editable serine codons (AGC/T, edSer) are dominant (). This suggests that the unSer-to-edSer transition occurred in the ancestor of Diptera (but not necessarily the most recent common ancestor of this order). In addition, the presence, conservation, and essentiality of this S>G editing site have been confirmed in at least the Drosophila genus.
According to the aforementioned prediction, if this RNA editing site is adaptive due to the flexibility of editing mechanism, then the genomic A-to-G mutation should be suppressed at the corresponding position in other species. For Adar S>G site, the A version encodes Ser and G version encodes Gly, so we searched this orthologous site in 113 Diptera species. We obtained 84 Ser, 1 Gly, 17 gaps, and 11 other amino acids (AAs) (). For the gaps and other AAs, they usually differ by at least two nucleotides from the original codon. They might either (1) come from the inaccurate assembly or annotation of some genomes; or (2) reflect the genetic drift that fixes some non-functional sequences. These gaps or AAs are unlikely to derive from a single point mutation during evolution. Since they only make up a small fraction of the species at this particular S>G site, we will only focus on species with Ser and Gly. The frequency of derived AA (Gly) is 1/(84 + 1) = 1.18% in Diptera. In other words, at this functional RNA editing site, the genomic sequence seems unlikely to be replaced with G due to the abolishment of flexibility by A-to-G mutation.
Figure 3. The basic concepts used for downstream analyses. (a) In 113 diptera species, the codons and AAs corresponding to the adar S > G site were displayed. (b) Classification of unedited adenosines in D. melanogaster adar gene. (c) Definition of original codon/AA and derived codon/AA. Transition rates were calculated using the original and derived codons/AAs.
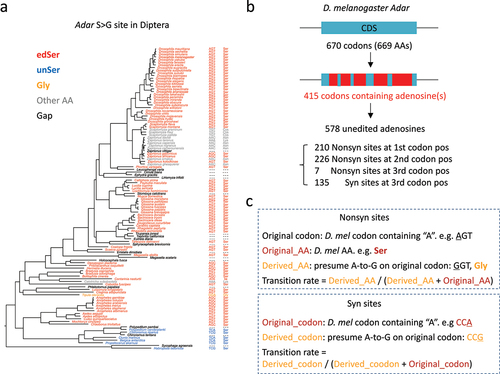
Next, we wonder whether this ‘transition rate’ of 1.18% at S>G site is lower than random expectation. To achieve a quantitative estimation of the expected transition rate, we parsed all the codons in the D. melanogaster Adar CDS and performed similar comparative genomic analyses. Adar CDS has 670 codons (669 AAs excluding the stop codon), among which 415 codons contain at least one adenosine (). S>G site is the only RNA editing site in Adar CDS of D. melanogaster and therefore all the other codons could be regarded as ‘unedited control.’ Totally 578 unedited adenosine sites were found among these 415 codons. By presuming an A-to-G mutation, we annotated the 578 adenosines and classified them as 135 synonymous sites and 443 nonsynonymous sites (including 210 nonsynonymous sites at the 1st codon position, 226 nonsynonymous sites at the 2nd codon position, and 7 nonsynonymous sites at the 3rd codon position) ().
For each of the 443 nonsynonymous adenosine sites in Adar of D. melanogaster, let ():
Original_codon = this adenosine-containing codon in D. melanogaster. Such as codon AGT at the Adar S>G site.
Original_AA = the AA encoded by the Original_codon. Such as Ser for the Adar S>G site. Note that the Original_AA could also be encoded by several other synonymous codons, not only the Original_codon.
Derived_AA = presume an A-to-G mutation in the Original_codon and predict the AA outcome. Such as Gly for the Adar S>G site. Note that the Derived_AA could also be encoded by several other synonymous codons, not only the A-to-G version of the Original_codon.
Other_AA = AA encoded by other codons at the corresponding position in other species.
The transition rate at a nonsynonymous site = Derived_AA/(Derived_AA + Original_AA).
For each of the 135 synonymous adenosine sites, we only define Original_codon, G_version_codon, and Other_codon because A-to-G does not change AAs.
The transition rate at a synonymous site = G_version_codon/(G_version_codon + Original_codon).
All these parameters will be compared between the edited and unedited adenosines in Adar CDS.
Derived genomic G at nonsynonymous editing site is significantly avoided
For each of the 578 unedited adenosines in Adar CDS, we found that the transition rates for nonsynonymous sites were 6.13 ± 0.75%, 4.02 ± 0.48%, and 2.88 ± 2.1% for the 1st, 2nd, and 3rd codon positions, respectively (). The S>G auto-recoding site (the only editing site in Adar CDS) is located at the 1st codon position with a transition rate of 1.18%, which is significantly lower than random expectation (observed < mean − 3 × S.E. of expectation). In contrast, for synonymous sites, the A-to-G substitution was much more common than nonsynonymous sites (), and this is expected due to the relaxed selection on silent sites. However, considering that synonymous codon usage bias prefers G/C-ending codons compared to A/T-ending codons due to differential translation rates (decoding rates) [Citation29,Citation30], the synonymous A-to-G mutations at the 3rd codon positions are intuitively favoured by natural selection. We therefore looked at the synonymous A-to-C and A-to-T mutations at the 3rd codon positions. The synonymous transition rates were 24.5 ± 1.95% for A-to-C and 26.4 ± 1.82% for A-to-T, which were still significantly higher than the missense transition rates, suggesting the overall deleterious nature of missense mutations. Our results demonstrated that the nonsynonymous editing site is less likely to be genomically replaced with G in sibling species, reflecting the advantage of RNA editing over the hardwired G-allele.
Figure 4. A-to-G transition in other diptera species is significantly avoided for the RNA editing site in drosophila Adar. (a) Mean ± S.E. (standard error) of the expected transition rate from original AA (codon) to derived AA (codon). Unedited adenosines were used and shown as squares and whiskers. The observed transition rate at adar S > G site was labeled in the plot. (b) Definition of gapped diptera species and histogram showing the numbers of gaps at each site. (c) Mean ± S.E. (standard error) of the expected transition rate using unedited adenosines with > 18 gapped species in diptera.
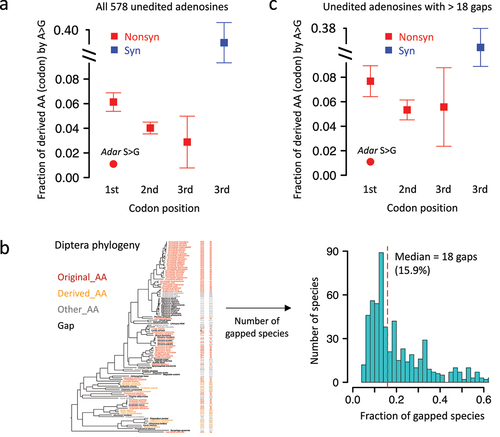
Since our calculation of transition rate did not consider the ‘gapped’ species at a particular site, we need to exclude the potential bias caused by gaps (). On average, about 18 (15.9%) of the 113 Diptera species had gaps at a particular site corresponding to the adenosines in D. melanogaster (). To test whether the number of gaps affect the transition rate from Original_AA to Derived_AA, we retrieved the sites with > 18 (median) gaps. The results showed that the distribution of transition rates () was similar to what we observed using all sites, where the expected rates from unedited adenosines were much higher than the observed 1.18% transition rate at S>G site. Then, the synonymous A-to-C and A-to-T transition rates at the 3rd codon positions were 23.4 ± 2.66% and 28.6 ± 2.63%, which were again remarkably higher than nonsynonymous transition rates, excluding the effect of codon usage bias. This suggests that our conclusion is robust under different cut-offs. Here, we illustrate several examples to show that the unedited adenosines indeed had much higher transition rate to be replaced with G version in Diptera species ().
Figure 5. Examples of unedited nonsynonymous adenosines in adar CDS with high transition rate. The information of each site (CDS position, AA position, AA change of the site and the numbers of each category) was displayed in the plot.
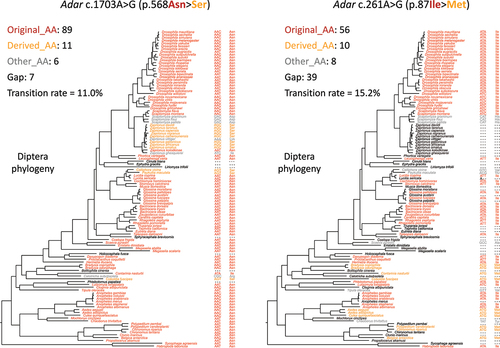
For example, site c.1703A>G (p.568Ile>Val) had a transition rate of 11/(11 + 89) = 11.0% in Diptera, 9 times higher than the transition rate of S>G editing site (). If we reasonably allowed more gapped species (allowing >20 gaps), we would find sites like c.261A>G (p.87Ile>Met) that had a transition rate of 10/(10 + 56) = 15.2% in Diptera, 12 times higher than the transition rate of S>G editing site (). More strikingly, this transition rate could be as high as 53/(53 + 34) = 60.9% for site c.706A>G (p.236Ile>Val). Compared to Adar S>G editing sites, these cases all exhibited a dramatically higher fraction of derived AA in the phylogeny.
Notably, for Adar S>G site, the ancestral AA is known to be Ser. For the many other unedited (nonsynonymous) adenosines, the ancestral AAs remained unclear. Nevertheless, one could infer from the fact that for each site, the majority (average = 98.5%) of the AA in Diptera belonged to the original AA (encoded by D. melanogaster), so that this original AA was likely to be the ancestral state while the derived AA appeared during evolution (just like the examples shown in ).
Taken together, by using the unedited adenosines as a control, we quantitatively proved that the editing site(s) is unlikely to be genomically mutated to G in the phylogeny, indicating the potential evolutionary adaptiveness (which is, the flexibility) of RNA editing mechanism.
Conditional regulation of Adar S>G auto-recoding
Following this line of thought, we would expect such adaptive mechanism to be regulated by environmental conditions. To validate this expectation, we retrieved the RNA editomes of Drosophila brains under different temperatures (Materials and Methods). In three Drosophila species, the Adar S>G recoding level decreased with temperature (). Moreover, since this recoding site affects the activity of ADAR, we would expect that such flexibility will also be reflected by overall editing levels or specific target sites. First, we found that the overall editing levels were down-regulated at high temperature ().
Figure 6. Editing levels under different conditions. (a) S > G auto-recoding level in three different drosophila species under 25°C and 30h. (b) Global editing level of shared editing sites across three species. (c) and (d) Display two examples of elevated editing level under high temperature. The trend is conserved across three species. D. mel, D. melanogaster; D. sim, D. simulans; D. pse, D. pseudoobscura. (e) S>G auto-recoding level in 10 samples of different developmental stages and head/body of male/female adults. Data were retrieved from our previous study [Citation8].
![Figure 6. Editing levels under different conditions. (a) S > G auto-recoding level in three different drosophila species under 25°C and 30h. (b) Global editing level of shared editing sites across three species. (c) and (d) Display two examples of elevated editing level under high temperature. The trend is conserved across three species. D. mel, D. melanogaster; D. sim, D. simulans; D. pse, D. pseudoobscura. (e) S>G auto-recoding level in 10 samples of different developmental stages and head/body of male/female adults. Data were retrieved from our previous study [Citation8].](/cms/asset/c1416aff-254b-4d29-9fa7-fcb1c83cc810/kepi_a_2333665_f0006_oc.jpg)
This seems paradoxical with the negative effect of S>G auto-recoding on editing activity. However, the global down-regulation of editing level was likely caused by the well-known effect that high temperature unravels dsRNA structure and reduces editing level. In fact, the fold-change of down-regulation for Adar S>G site was 0.21, 0.37, 0.18 for D. melanogaster, D. simulans, and D. pseudoobscura (mean of fold-changes in female and male), while this fold-change for the global editing level was 0.90, 0.89, 0.90 for three Drosophila species. This suggests that the reduced Adar recoding level has actually ‘buffered’ the decrease of overall editing efficiency, echoing the notion that auto-edited Adar isoform has lower editing activity. Moreover, for individual editing sites, one can always find outliers that show the opposite trend to the globally decreased editing level, such as Tyr>Cys site in gene Pi4KIIIα () and Ser>Gly site in gene CG2747 (). These results support the conditional regulation of Adar auto-recoding sites and the effect on overall editing activity.
Moreover, given the putative adaptiveness of Adar Ser>Gly site, one would expect such site to be regulated in time and space. To address this question, we retrieved the transcriptomes of different developmental stages (time) and heads/bodies (space) of D. melanogaster [Citation8] and found that Adar Ser>Gly editing level varies extensively across these samples (). These results generally support the active and regulatory role of this editing site, but we also recognize that more transcriptomes from various conditions (like circadian time, social experience, motivational states) are needed to fully prove the editing to be an adaptive mechanism.
Evolution of Ile>Met recoding site in Syt1: derived genomic G is absent
As we have clarified, the comparison between editable versus uneditable codon is the direct reflection of the advantage of RNA editing. The Adar Ser>Gly site is a typical case where the ancestral sequence in Diptera was uneditable and in later clades the editable codon emerged. This comparison benefits from the fact that Ser has six codons and two of which were editable at the 1st codon position and four were uneditable. Similar analysis is applicable to Arg>Gly recoding at the 1st codon position and Ile>Met recoding at the 3rd codon position (see the previous subsections for details). We wonder whether we could find evidence for the potential adaptiveness of other recoding sites.
A well-known neuronal gene with extensive recoding events is Syt1 (Synaptotagmin 1). Notably, an Ile>Met recoding site is highly conserved across insects [Citation31] and this site meets our criteria to test the adaptiveness of ‘editability.’ In Drosophila brains, this Ile>Met site (FBtr0077726_CDS:405) has editing levels of 0.7 ~ 0.8 in different fly species (Materials and Methods). By looking at the orthologous site in all 113 Diptera species (), we found the following patterns: (1) this site does not have derived genomic G (encoding Met) in all tested Diptera species; (2) the ancestral state of this codon, although not for sure, was likely an uneditable Ile codon (). The results support the adaptation of Syt1 Ile>Met recoding site. This site was originally uneditable and then gained editability during evolution, once gained an editable codon, the replacement with G would be deleterious and was suppressed in this clade. Note that according to the codons observed in Diptera () together with the editable codons present in other insect orders, we reserve the possibility that the ancestor of Diptera had an editable Ile codon and the few uneditable Ile codons were actually the derived ones. Anyway, this occasion does not deny the potential advantage of an editable status reflected by the absence of derived G (Met) in all species.
Figure 7. A-to-G transition in other diptera species is significantly avoided for the Ile>Met (I > m) recoding site in Syt1. (a) In 113 diptera species, the codons and AAs corresponding to the Syt1 I > M site were displayed. (b) Mean ± S.E. (standard error) of the expected transition rate from original AA (codon) to derived AA (codon). Unedited adenosines were used and shown as squares and whiskers. The observed transition rate at Syt1 I > M site (rate = 0) was labeled in the plot. (c) Mean ± S.E. of the expected transition rate using unedited adenosines with > 17 (median value for all sites) gapped species in diptera.
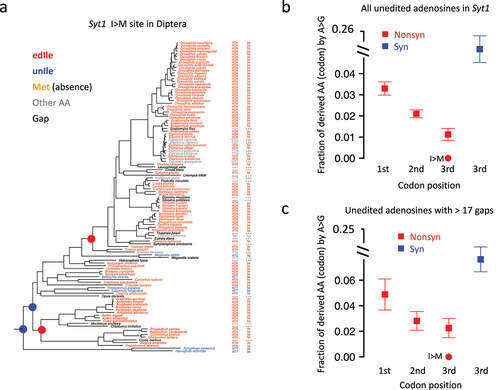
Then, we parsed all the unedited adenosines in the Syt1 CDS in D. melanogaster. By looking for the species where the original AA in D. melanogaster were replaced with the post-edited AA in the genome of a target species, we calculated the fraction of derived AA for each codon (). The Ile>Met recoding site at the 3rd codon position had remarkably lower transition rates than the unedited (missense) adenosines at the 3rd codon position (). This pattern held true when we considered the unedited sites with many gapped species in the alignment (). These results again highlight the unlikelihood of Syt1 Ile>Met site to be replaced with G during evolution, supporting its advantage of being editable.
Discussion
Judging the adaptiveness of individual editing sites in the light of evolution: an attempt to narrow down the list of candidate sites for functional validation
In this work, by analysing CDS alignment of Adar gene in 113 Diptera species, we found evidence suggesting that the A-to-I editing sites, at least for this particular Adar S>G auto-recoding site, significantly avoid to be genomically replaced with G during evolution, and thus indicating the advantage of editable sites (flexible) over hardwired genomic alleles (not regulatable).
For a long time, although the positive selection on nonsynonymous RNA editing has been observed for several representative species [Citation8,Citation11,Citation12], it is unclear how exactly RNA editing is better than the hardwired G-allele, and could we find other genomic evidence to prove the necessity of RNA editing. Moreover, the global adaptive signals for nonsynonymous RNA editing sites does not represent the functional importance of every single RNA editing site. It is still challenging to narrow down the candidates of functional RNA editing sites. Given an individual RNA editing site, only experimental instead of in silico approach is available to infer its adaptiveness. Based on the ideas of previous evolutionary genomics studies, our current study provides a step towards solving these issues with in silico approaches, without the need to conduct experiments with mutant animals. Using our comparative genomic methods, one could quantitatively judge how unexpected it is to observe an AA transition in the phylogeny, and the type of AA transition is determined by the information of an editing site in the target species (like D. melanogaster). The abundant unedited adenosines within the same gene could be used as control (background) to judge the extent of transition rate of the RNA editing site. Conceivably, not every editing site had a significantly lower transition rate compared to random expectation. Thus, we propose that the significant ones are more likely to be truly adaptive. Our work provides an approach to putatively judge whether a given editing site is adaptive so that the selected candidate sites might be given the priority for functional studies in the future.
Limitations and cautions
Indeed, in the comparison with unedited adenosines, a hidden hypothesis is that the AA replacements (by A-to-G) at the corresponding sites are completely random so that their occurrences could be used as a neutral expectation. However, many of the background unedited adenosines were mainly nonsynonymous sites. The relative fitness of the two protein isoforms resulted from A- and G-alleles depends on the shift in protein function after the A-to-G substitution. This sheds uncertainty to the use of unedited adenosines as random control. Nevertheless, although it is currently unfeasible to predict the functional impact of every nonsynonymous A-to-G substitution, we believe that at under a large scale of random mutations, we should have unbiased chances to obtain beneficial, neutral, or deleterious mutations. Therefore, the overall unedited adenosines could be used as a neutral control to measure the selection force acting on Adar S>G site.
More interestingly, while we observed differential A-to-G transition rates between edited (1.18%) and unedited (mean = 6.13%) adenosines at the 1st codon position (nonsynonymous), this difference was not seen for A-to-T transition (as a control). In detail, the A-to-T transition rate for Adar S>G editing site (which means, observing a Cys in the Diptera phylogeny) was 3/(84 + 3) = 3.45%, but this ratio for unedited adenosines was 3.49 ± 0.65% (mean ± S.E.) at the 1st codon position, suggesting that no constraint is added to the A-to-T transition of this editing site, further supporting the unique suppression of A-to-G DNA mutation at this position. For A-to-C mutation, there are cases where A-to-C at the 1st codon position leading to synonymous changes, complicating the comparison, and therefore this type of mutation was not considered. Nevertheless, we hold the view that the most convincing evidence for advantageous editing is still the differential A-to-G transition rates seen in our results, and the comparison on A-to-T mutations only serves as auxiliary evidence. Then, considering the existence of synonymous codon usage bias, one should be cautious to compare the mutation rates at the 3rd codon positions as we have already performed in the Results section. Taken together, we fully demonstrated that in Diptera species, the genomic mutations abolishing Adar auto-editing site is suppressed, and this suppression is unique to A-to-G mutations, indicating the adaptiveness of A-to-I RNA editing mechanism.
The complexity of Adar S>G site in diptera
Nevertheless, we should be cautious since the selection force acting on the Adar S>G site might be complicated. Because S>G recoding changes Adar activity in a plastic way, it has the potential to affect editing at all editing sites. Thus, editing at any of the other downstream sites could be driving the selection of editing at the S>G site. This does not disprove the importance of Adar S>G site, however one should be aware that (1) the selection on S>G site might be indirect. It is possible that the constraint on some downstream editing site has constrained the S>G site itself; (2) one could also envision that the selection on this S>G site is particularly strong due to the need to regulate Adar activity, but this situation maybe not applicable to many other sites.
Next, another issue worth thinking is that is this Adar S>G site truly edited outside the Drosophila genus although the genome sequences show editable codons? To answer this question, we searched and downloaded the head transcriptomes of several Diptera species: Anopheles gambiae, Aedes aegypti, Culex quinquefasciatus, and Bactrocera tryoni. The first three species are mosquitoes, and the last species is close to fruitfly. We directly mapped the RNA-Seq reads to the corresponding Adar sequence in that species to see whether the S>G site had signals of A-to-G variation (Materials and Methods). It turned out that a reliable editing signal was only observed in fly Bactrocera tryoni (). This suggests that although the unSer-to-edSer transition at genome level occurred very early during Diptera evolution, the RNA editing events at edSer codon only emerged lately, at least after the split between mosquito and fly.
Figure 8. Checking the editing status of adar S > G site in representative diptera species. Editing event at this position was already observed in the drosophila genus. For non-drosophila clades, head transcriptomes of four species were downloaded and mapped to the adar sequence. Accession IDs of the transcriptome data were given above each sample. Screenshots of IGV visualization at S > G site was displayed. Not all RNA-Seq reads were displayed. The detected editing event and the editing level were highlighted.
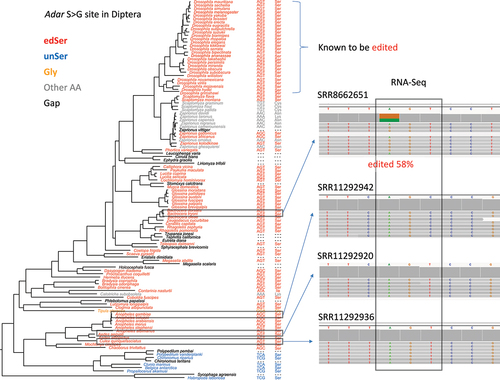
However, the absence of S>G editing events in some early Diptera clades (mosquito) does not negate the benefit of S>G recoding in flies. When we only focus on the monophyly of all kinds of flies, the fraction of transition to genomic Gly is exactly zero (), confirming the essentiality for maintaining the editable status.
Summary
In the light of evolution, we established a comparative genomic approach for quantitatively justifying the adaptiveness of RNA editing. We provide novel and interesting perspectives for the RNA editing community. With this experiment-free methodology, one could narrow down the adaptive RNA editing sites and such sites should be given a priority in the functional studies.
Materials and Methods
Data availability
The analysis of the Adar auto-recoding codon involves the reference sequences of 113 Diptera species which were downloaded from NCBI https://www.ncbi.nlm.nih.gov/. The detailed links of the data were supplemented in our previous works [Citation27, Citation28]. The phylogeny of Diptera was also retrieved from our previous study [Citation27].
Sequence alignment and codon extraction
The amino acid sequences of the Adar gene were aligned using G-INS-i strategy in MAFFT v7.310 [Citation32], default parameters were used. Then, using TranslatorX v1.1 [Citation33], the nucleotide sequences of Adar gene were aligned based on the pre-aligned amino acid sequences. Default parameters were used. The entire alignment file was split by codon (tri-nucleotide) and then the alignment of each codon was extracted. The alignment of Syt1 CDS (D. melanogaster transcript ID FBtr0077726) in Diptera species was done with an identical approach.
Using public data to calculate the editing levels at particular sites
To validate whether the Adar S>G site is edited in other non-Drosophila species in Diptera, we downloaded the head transcriptomes of Anopheles gambiae (SRR11292942), Aedes aegypti (SRR11292920), Culex quinquefasciatus (SRR11292936), and Bactrocera tryoni (SRR8662651) from NCBI (https://www.ncbi.nlm.nih.gov/). We directly mapped the RNA-Seq reads to the Adar CDS sequence of that species. BWA [Citation34,Citation35] with default parameters was used and the editing status at S>G site was visualized by IGV.
For the brain transcriptomes of Drosophila under different temperatures (25h and 30h), RNA editing sites, levels, and annotations were directly retrieved from our previous work. The raw data were uploaded to NCBI SRA (https://www.ncbi.nlm.nih.gov/sra) with accession ID SRP074828. For three Drosophila species D. melanogaster, D. simulans, and D. pseudoobscura, we obtained the samples of normal condition (25h) and heat stress (30h for 14h). Female and male samples were available for each condition. The editing site information was summarized in Supplementary Table S1 (D. melanogaster, samples B2 and B3 refer to female normal and 30h 14h, samples B6 and B7 refer to male normal and 30h 14h), Supplementary Table S2 (D. simulans, samples S2 and S3 refer to female normal and 30h 14h, samples S6 and S7 refer to male normal and 30h 14h), and Supplementary Table S3 (D. pseudoobscura, samples P2 and P3 refer to female normal and 30h 14h, samples P6 and P7 refer to male normal and 30h 14h). The developmental transcriptomes and RNA editomes of D. melanogaster were retrieved from our previous study [Citation8].
Statistics and visualization
The statistics like mean and S.E. were performed in R studio (version 3.6.3). The graphic works were done in R studio or Adobe Illustrator version 2023.
Abbreviations
| AA | = | amino acid. |
| A-to-I | = | adenosine-to-inosine. |
| ADAR | = | adenosine deaminase acting on RNA. |
| CDS | = | coding sequence. |
| S.E. | = | standard error |
| edSer | = | editable serine codon |
| unSer | = | uneditable serine codon |
Authors’ contributions
Conceptualization & supervision: Y.D. Data analysis: Y.D., L.M., and C.Z. Writing – original draft: Y.D., C.Z., L.M., F.S., L.T., W.C., and H.L. Writing – review & editing: Y.D., C.Z., L.M., F.S., L.T., W.C., and H.L.
Acknowledgments
We thank the members in Cai Lab for their help and suggestions to this work. We thank Dr Jiyao Liu for the help in data download and analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The analysis of the Adar auto-recoding codon involves the reference sequences of 113 Diptera species which were downloaded from NCBI https://www.ncbi.nlm.nih.gov/. The detailed links of the data were supplemented in our previous works [Citation27,Citation28]. The phylogeny of Diptera was also retrieved from our previous study [Citation27]. The head transcriptomes of four Diptera species were downloaded from NCBI with the following accession IDs: Anopheles gambiae (SRR11292942), Aedes aegypti (SRR11292920), Culex quinquefasciatus (SRR11292936), and Bactrocera tryoni (SRR8662651). The brain transcriptomes of three Drosophila species D. melanogaster, D. simulans, and D. pseudoobscura were downloaded from NCBI with accession number SRP074828. The developmental transcriptomes and RNA editomes of D. melanogaster were retrieved from our previous study [Citation8]
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2024.2333665.
Additional information
Funding
References
- Eisenberg E, Levanon EY. A-to-I RNA editing - immune protector and transcriptome diversifier. Nat Rev Genet. 2018;19(8):473–16. doi: 10.1038/s41576-018-0006-1
- Zhang P, Zhu Y, Guo Q, et al. On the origin and evolution of RNA editing in metazoans. Cell Rep. 2023;42(2):112112. doi: 10.1016/j.celrep.2023.112112
- Xu Y, Liu J, Zhao T, et al. Identification and interpretation of A-to-I RNA editing events in insect transcriptomes. Int J Mol Sci. 2023;24(24):17126. doi: 10.3390/ijms242417126
- Alon S, Garrett SC, Levanon EY, et al. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. Elife. 2015;4:4. doi: 10.7554/eLife.05198
- Gommans WM, Mullen SP, Maas S. RNA editing: a driving force for adaptive evolution? BioEssays. 2009;31(10):1137–1145. doi: 10.1002/bies.200900045
- Zhan D, Zheng C, Cai W, et al. The many roles of A-to-I RNA editing in animals: functional or adaptive? Front Biosci (Landmark Ed). 2023;28(10):256. doi: 10.31083/j.fbl2810256
- Yablonovitch AL, Deng P, Jacobson D, et al. The evolution and adaptation of A-to-I RNA editing. PloS Genet. 2017;13(11):e1007064. doi: 10.1371/journal.pgen.1007064
- Duan Y, Xu Y, Song F, et al. Differential adaptive RNA editing signals between insects and plants revealed by a new measurement termed haplotype diversity. Biol Direct. 2023;18(1):47. doi: 10.1186/s13062-023-00404-7
- Zhang Y, Duan Y. Genome-wide analysis on driver and passenger RNA editing sites suggests an underestimation of adaptive signals in insects. Genes (Basel). 2023;14(10):1951. doi: 10.3390/genes14101951
- Zhao T, Ma L, Xu S, et al. Narrowing down the candidates of beneficial A-to-I RNA editing by comparing the recoding sites with uneditable counterparts. Nucleus (Calcutta). 2024;15(1):2304503. doi: 10.1080/19491034.2024.2304503
- Liscovitch-Brauer N, Alon S, Porath HT, et al. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell. 2017;169(2):191–202 e111. doi: 10.1016/j.cell.2017.03.025
- Duan Y, Li H, Cai W. Adaptation of A-to-I RNA editing in bacteria, fungi, and animals. Front Microbiol. 2023;14:1204080. doi: 10.3389/fmicb.2023.1204080
- Bian Z, Ni Y, Xu JR, et al. A-to-I mRNA editing in fungi: occurrence, function, and evolution. Cell Mol Life Sci. 2019;76(2):329–340. doi: 10.1007/s00018-018-2936-3
- Liu H, Wang Q, He Y, et al. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016;26(4):499–509. doi: 10.1101/gr.199877.115
- Liu H, Li Y, Chen D, et al. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc Natl Acad Sci USA. 2017;114(37):E7756–E7765. doi: 10.1073/pnas.1702591114
- Qi Z, Lu P, Long X, et al. Adaptive advantages of restorative RNA editing in fungi for resolving survival-reproduction trade-offs. Sci Adv. 2024;10(1):eadk6130. doi: 10.1126/sciadv.adk6130
- Edera AA, Gandini CL, Sanchez-Puerta MV. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol Biol. 2018;97(3):215–231. doi: 10.1007/s11103-018-0734-9
- Duan Y, Cai W, Li H. Chloroplast C-to-U RNA editing in vascular plants is adaptive due to its restorative effect: testing the restorative hypothesis. RNA. 2023;29(2):141–152. doi: 10.1261/rna.079450.122
- Jiang D, Zhang J. The preponderance of nonsynonymous A-to-I RNA editing in coleoids is nonadaptive. Nat Commun. 2019;10(1):5411. doi: 10.1038/s41467-019-13275-2
- Moldovan M, Chervontseva Z, Bazykin G, et al. Adaptive evolution at mRNA editing sites in soft-bodied cephalopods. PeerJ. 2020;8:e10456. doi: 10.7717/peerj.10456
- Shoshan Y, Liscovitch-Brauer N, Rosenthal JJC, et al. Adaptive proteome diversification by nonsynonymous A-to-I RNA editing in coleoid cephalopods. Mol Biol Evol. 2021;38(9):3775–3788. doi: 10.1093/molbev/msab154
- Xin K, Zhang Y, Fan L, et al. Liu H: experimental evidence for the functional importance and adaptive advantage of A-to-I RNA editing in fungi. Proc Natl Acad Sci U S A. 2023;120(12):e2219029120. doi: 10.1073/pnas.2219029120
- Birk MA, Liscovitch-Brauer N, Dominguez MJ, et al. Temperature-dependent RNA editing in octopus extensively recodes the neural proteome. Cell. 2023;186(12):2544–2555. doi: 10.1016/j.cell.2023.05.004
- Rangan KJ, Reck-Peterson SL. RNA recoding in cephalopods tailors microtubule motor protein function. Cell. 2023;186(12):2531–2543. doi: 10.1016/j.cell.2023.04.032
- Garrett S, Rosenthal JJ. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science. 2012;335(6070):848–851. doi: 10.1126/science.1212795
- Palladino MJ, Keegan LP, O’Connell MA, et al. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA. 2000;6(7):1004–1018. doi: 10.1017/S1355838200000248
- Ma L, Zheng C, Xu S, et al. A full repertoire of hemiptera genomes reveals a multi-step evolutionary trajectory of auto-RNA editing site in insect adar gene. RNA Biol. 2023;20(1):703–714. doi: 10.1080/15476286.2023.2254985
- Duan Y, Ma L, Song F, et al. Autorecoding A-to-I RNA editing sites in the adar gene underwent compensatory gains and losses in major insect clades. RNA. 2023;29(10):1509–1519. doi: 10.1261/rna.079682.123
- Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19(1):20–30. doi: 10.1038/nrm.2017.91
- Qian W, Yang JR, Pearson NM, et al. Balanced codon usage optimizes eukaryotic translational efficiency. PloS Genet. 2012;8(3):e1002603. doi: 10.1371/journal.pgen.1002603
- Reenan RA. Molecular determinants and guided evolution of species-specific RNA editing. Nature. 2005;434(7031):409–413. doi: 10.1038/nature03364
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38(suppl_2):W7–W13. doi: 10.1093/nar/gkq291
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324
- Li B, Duan Y, Du Z, et al. Natural selection and genetic diversity maintenance in a parasitic wasp during continuous biological control application. Nat Commun. 2024;15(1):1379. doi: 10.1038/s41467-024-45631-2
