Abstract
Receptor for Activated C Kinase 1 (RACK1) is a versatile scaffold protein that interacts with a large, diverse group of proteins to regulate various signaling cascades. RACK1 has been shown to regulate hormonal signaling, stress responses and multiple processes of growth and development in plants. However, little is known about the molecular mechanism underlying these regulations. Recently, it has been demonstrated that Arabidopsis RACK1 is phosphorylated by an atypical serine/threonine protein kinase, WITH NO LYSINE 8 (WNK8). Furthermore, RACK1 phosphorylation by WNK8 negatively regulates RACK1 function by influencing its protein stability. These findings promote a new regulatory system in which the action of RACK1 is controlled by phosphorylation and subsequent protein degradation.
Receptor for Activated C Kinase 1 (RACK1) is a versatile scaffold protein that interacts with a large, diverse group of proteins to regulate various signaling cascades in mammalian cells.Citation1–7 RACK1 protein contains a 7 tryptophan–aspartic acid repeat domain (WD40) similar to that in the heterotrimeric GTP-binding protein β subunit. The seven blade propeller structure of RACK1 allows it to function as a signaling hub.Citation4
The protein sequences and the structure of RACK1 are conserved in plants.Citation8,9 Similar to mammalian RACK1, plant RACK1 interacts with a large group of proteins Citation10,11 and is involved in diverse biological processes, ranging from seed germination, leaf and root development to flowering,Citation8,12 and is also involved in responses to plant hormones, Citation8,13 and biotic and abiotic stresses.Citation13–15 At the molecular level, RACK1 protein is associated with ribosomes Citation16-18 and is involved in the regulation of protein translation Citation18 and miRNA abundance.Citation19
It is less clear how RACK1 is able to participate in so many different biological processes. Studies using mammalian cells suggested a few possible mechanisms including translocation of RACK1 protein from one cellular compartment to another, formation of protein complex with other scaffolding proteins and contribution to transcription and translation.Citation4,6 These abilities of RACK1 lead to its temporal and spatial regulation of diverse signal transduction events. Emerging evidence suggest that the transcriptional and translational modification of RACK1 itself also play an important role in regulating its function. For example, the transcription of RACK1 is suppressed by the plant stress hormone abscisic acid (ABA).Citation13 It was suggested that such transcriptional regulation likely contributes to RACK1's negative regulation of ABA responses in the ABA-mediated inhibition of seed germination, early seedling development, and ABA-induced gene expression.Citation13 More recently, it was demonstrated that RACK1 is also regulated at the post-translational level.Citation20 It was found that RACK1 protein is subject to phosphorylation and that phosphorylation of RACK1 renders it unstable.Citation20
Phosphorylation of RACK1 has been previously reported in mammals.Citation21-22 However, a fundamental difference is that mammalian RACK1 is phosphorylated at tyrosine (Tyr) sites whereas plant RACK1 is phosphorylated at serine (Ser) and threonine (Thr) sites. Therefore, mammalian RACK1 is a substrate of tyrosine kinase whereas plant RACK1 is a substrate of serine/threonine kinase. This finding may not be surprising since for plant proteins, most phosphorylation occurs on Ser and Thr and to a less extent on Tyr.Citation23
It should be noted that the Tyr phosphorylation site of mammalian RACK1 is also required for its binding to its Tyr kinase. Specifically, Tyr228 and Tyr246 are required for RACK1 binding to Src kinase Citation21 and Tyr52 of RACK1 is required for its binding to c-Abl kinase.Citation22 Urano et al. (2015) Citation20 showed that Ser122 and Thr162 (Thr162 of RACK1A, Thr161 of RACK1B and RACK1C) of Arabidopsis RACK1 are phosphorylated by an atypical Ser/Thr kinase, WITH NO LYSINE 8 (WNK8). However, it remains unknown whether Ser122 and Thr162 are also required for RACK1 binding to WNK8. The study by Urano et al. (2015) Citation20 also does not rule out the possibility that other phosphorylation sites may exist in RACK1 protein. Amino acid sequence alignment indicated that Tyr phosphorylation sites identified in mammalian RACK1 (Tyr52, Tyr228 and Tyr246) are conserved in plant RACK1 proteins.Citation20 Whether RACK1 is phosphorylated by a yet unidentified Tyr kinase in plants remains an open question.
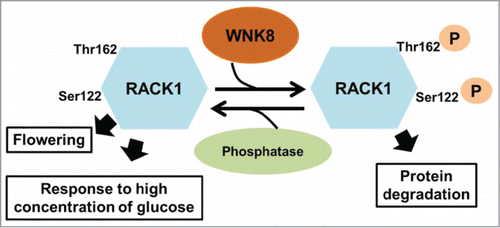
An important finding of the study by Urano et al. (2015) Citation20 is that phosphorylation of RACK1 affects its protein stability. While the post-translational modification events have previously reported for mammalian RACK1 protein, Citation21,22 its impact on protein stability has not. The finding by Urano et al. (2015) Citation20 promotes a new regulatory system in which the action of RACK1 is controlled by phosphorylation and subsequent protein degradation (). Specifically, nonphosphorylated RACK1 positively regulate flowering and negatively regulate the response to high concentration of glucose. Upon phosphorylation by WNK8, RACK1 protein becomes unstable and undergoes protein degradation. Presumably, RACK1 is degraded through the 26S proteasome but this has not been experimentally examined. The phosphatase that specifically de-phosphorylates phosphorylated RACK1 by WNK8, however, remains to be identified. It is speculated that such a phosphatase counteracts the phosphorylation of RACK1 by WNK8, providing a positive feedback loop. The assumption of presence of other forms of posttranslational modification events for RACK1 is consistent with the fact that no difference in RACK1 protein abundance was detected between wnk8 mutant and the wild type.Citation20
Figure 1. Phosphorylation of RACK1 by WNK8. WNK8 interacts with and phosphorylates RACK1 at Ser122 and Thr162. Phosphorylation of RACK1 promotes protein degradation. An unidentified phosphatase counteracts RACK1 phosphorylation by WNK8. Nonphosphorylated RACK1 protein acts downstream of WNK8 to regulate flowering and response to high concentration of glucose.
The finding by Urano et al. (2015) Citation20 provides new insights into the molecular mechanism underlying the regulatory actions of RACK1. This also opens up a few more questions: (1) How RACK1 phosphorylation by WNK8 is regulated? (2) What is the phosphatase that counteracts RACK1 phosphorylation by WNK8? (3) Is RACK1 protein subject to Tyr phosphorylation by an unidentified Tyr kinase? (4) Is RACK1 protein subject to other forms of posttranslational modifications? Addressing these questions will drastically enhance our understanding of the molecular mechanism of action of RACK1 in plants.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Plant-Microbe Interfaces Scientific Focus Area in the Genomic Science Program, United States Department of Energy, Office of Science, Biological and Environmental Research. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the United States Department of Energy under contract DE-AC05–00OR22725.
References
- McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharm 2002; 62:1261–73; http://dx.doi.org/10.1124/mol.62.6.1261
- Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep 2004; 5:1137–41; PMID:15577927; http://dx.doi.org/10.1038/sj.embor.7400291
- Sklan EH, Podoly E, Soreq H. RACK1 has the nerve to act: structure meets function in the nervous system. Prog Neurobiol 2006; 78:117–34; PMID:16457939; http://dx.doi.org/10.1016/j.pneurobio.2005.12.002
- Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun Signal 2011; 9:22; PMID:21978545; http://dx.doi.org/10.1186/1478-811X-9-22
- Gandin V, Senft D, Topisirovic I, Ronai ZA. RACK1 function in cell motility and protein synthesis. Genes Cancer 2013; 4:369–77; PMID:24349634; http://dx.doi.org/10.1177/1947601913486348
- Ron D, Adams DR, Baillie GS, Long A, O'Connor R, Kiely PA. RACK(1) to the future - a historical perspective. Cell Commun Signal 2013; 11:53; PMID:23915285; http://dx.doi.org/10.1186/1478-811X-11-53
- Li JJ, Xie D. RACK1, a versatile hub in cancer. Oncogene 2014; PMID: 24882575; http://dx.doi.org/10.1038/onc.2014.127.; Epub ahead of print
- Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot 2006; 57:2697–708; PMID:16829549; http://dx.doi.org/10.1093/jxb/erl035
- Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci 2008; 17:1771–80; PMID:18715992; http://dx.doi.org/10.1110/ps.035121.108
- Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR, Carpita NC, Carr T, Chen JG, Cooke TR, et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol 2011; 7: 532; PMID:21952135; http://dx.doi.org/10.1038/msb.2011.66
- Kundu N, Dozier U, Deslandes L, Somssich IE, Ullah H. Arabidopsis scaffold protein RACK1A interacts with diverse environmental stress and photosynthesis related proteins. Plant Signal Behav 2013; 8:e24012; PMID:23435172; http://dx.doi.org/10.4161/psb.24012
- Guo J, Chen JG. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol 2008; 8:108; PMID:18947417; http://dx.doi.org/10.1186/1471-2229-8-108
- Guo J, Wang J, Xi L, Huang WD, Liang J, Chen JG. RACK1 is a negative regulator of ABA responses in Arabidopsis. J Exp Bot 2009; 60: 3819–33; PMID:19584117; http://dx.doi.org/10.1093/jxb/erp221
- Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, Umemura K, Shirasu K, Kawasaki T, Shimamoto K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 2008; 20:2265–79; PMID:18723578; http://dx.doi.org/10.1105/tpc.107.054395
- Wang B, Yu J, Zhu D, Chang Y, Zhao Q. Maize ZmRACK1 is involved in the plant response to fungal phytopathogens. Int J Mol Sci 2014; 15:9343–59; PMID:24865494; http://dx.doi.org/10.3390/ijms15069343
- Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J. Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol 2005; 137:848–62; PMID:15734919; http://dx.doi.org/10.1104/pp.104.053637
- Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P. High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol 2005; 57:577–91; PMID:15821981; http://dx.doi.org/10.1007/s11103-005-0699-3
- Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, Weston DJ, Ellis BE, Chen JG. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol 2011; 155:370–83; PMID:21098678; http://dx.doi.org/10.1104/pp.110.160663
- Speth C, Willing EM, Rausch S, Schneeberger K, Laubinger S. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J 2013; 76:433–45; PMID:23941160; http://dx.doi.org/10.1111/tpj.12308.
- Urano D, Czarnecki O, Wang X, Jones AM, Chen JG- Arabidopsis receptor of activated C kinase1 phosphorylation by WITH NO LYSINE8 KINASE. Plant Physiol 2015; 167:507–16; PMID:25489024; http://dx.doi.org/10.1104/pp.114.247460
- Chang BY, Harte RA, Cartwright CA. RACK1: a novel substrate for the Src protein-tyrosine kinase. Oncogene 2002; 21:7619–29; PMID:12400005; http://dx.doi.org/10.1038/sj.onc.1206002
- Kiely PA, Baillie GS, Barrett R, Buckley DA, Adams DR, Houslay MD, O'Connor R. Phosphorylation of RACK1 on tyrosine 52 by c-Abl is required for insulin-like growth factor I-mediated regulation of focal adhesion kinase. J Biol Chem 2009; 284:20263–74; PMID:19423701; http://dx.doi.org/10.1074/jbc.M109.017640
- Ghelis T. Signal processing by protein tyrosine phosphorylation in plants. Plant Signal Behav 2011; 6:942–951; PMID:21628997; http://dx.doi.org/10.4161/psb.6.7.15261
