Abstract
Ubiquitin-mediated protein degradation is involved in various cellular processes including plant–microbe interactions and defense responses. Although there are many E3 ubiquitin ligases in rice, the functions of their targets in defense responses are unclear. We recently found that SPIN6 (SPL11-interacting Protein 6) is a Rho GTPase-activating protein and acts as the target of the E3 ligase SPL11, a negative regulator of plant cell death and innate immunity. Our results showed that SPIN6 serves as a link between the SPL11-mediated ubiquitination pathway and the OsRac1-associated defense system. Here, we show that SPIN6 interacts with OsHUB1 and OsHUB2, the homologous proteins of Arabidopsis RING finger E3 ligases HUB1 and HUB2. OsHub1 and OsHub2 are down-regulated in the Spin6 RNAi plants and during the compatible interaction between rice and Magnaporthe oryzae. OsHub1 and OsHub2 are induced by hormone treatments. Like HUB1 and HUB2 in Arabidopsis, OsHUB1 and OsHUB2 in rice form homo- and hetero-dimers. Our results suggest that OsHUB1 and OsHUB2 may be associated with the SPIN6/OsRac1 pathway in rice immunity.
The Rho GTPase-Activating Protein SPIN6 Negatively Regulates Plant Cell Death and Innate Immunity in Rice
Rice diseases are a severe threat to global food production and social stability. The most effective and environmentally friendly approach to control these diseases is to develop rice cultivars with broad-spectrum resistance.Citation1,2 In the last 2 decades, scientists have used various genetic and genomic approaches to understand the molecular basis of host resistance to rice pathogens, and they have identified many important genes involved in rice immunity.Citation2 They determined, for example, that E3 ligases play important roles in plant immunity.Citation3,4 Although there are more than 1300 E3 ubiquitin ligases in the rice genome, only a few of them are involved in rice immunity.Citation5-7 One of these, the U-box-type E3 ligase SPL11, is involved in the regulation of plant defense, cell death, and flowering time.Citation8
We recently showed that SPIN6, a plant-specific Rho GTPase-activating protein, interacts with SPL11 in vitro and in vivo.Citation9 We found that SPL11 ubiquitinated SPIN6 in vitro and promoted its degradation in planta. Silencing of Spin6 in transgenic rice and knockout of the gene in the T-DNA mutant led to the formation of lesion mimic cell death phenotypes and to enhanced resistance to fungal and bacterial pathogens. RNAi silencing and mutant plants showed enhanced PAMP-triggered ROS production. Interestingly, SPIN6 also interacted with the small GTPase OsRac1 in vitro and in vivo. Fluorescence resonance energy transfer (FRET) analysis showed that SPIN6 catalyzes the GTP-bound active form of OsRac1 into the GDP-bound inactive form of OsRac1 in rice protoplasts, suggesting that SPIN6 is the RhoGAP of OsRac1 in rice. Our previous results have demonstrated that SPIN6 negatively regulates cell death and innate immunity and is the link between the E3 ligase SPL11 and the small GTPase OsRac1 in the rice defense signaling pathway.Citation9
SPIN6 Interacts with OsHUB1 and OsHUB2, the Homologous Proteins of Arabidopsis RING Finger E3 Ligases HUB1 and HUB2
To understand the signaling pathway mediated by SPIN6, we identified several proteins that interact with SPIN6 in rice using the yeast 2-hybrid screen. Two of the identified SPIN6-interacting proteins are OsHUB1 and OsHUB2, which are the homologous proteins of HUB1 and HUB2 in Arabidopsis.Citation10,11 Co-transformation of OsHUB1 and OsHUB2 with SPIN6 confirmed their interaction in yeast (). To further confirm their interaction, we conducted LCI (luciferase complementation imaging) assay in Nicotiana benthamiana.Citation12 We detected strong LUC activity in the NLuc-OsHUB1/2 and CLuc-SPIN6 combinations but not in the control combinations (), indicating that SPIN6 interacts with OsHUB1 and OsHUB2 in vivo. To confirm the relationship between SPIN6 and OsHUB1/2, we measured the expression level of OsHub1 and OsHub2 in the Spin6 RNAi transgenic plants. Real-time PCR analysis showed that OsHub1 and OsHub2 transcriptional levels were reduced in the Spin6 RNAi transgenic lines (), suggesting that OsHUB1/2 may participate in the SPIN6-mediated signaling pathway in rice.
Figure 1. The interaction between SPIN6 and OsHUB1/2 in vitro and in vivo. (A) The interaction between SPIN6 and OsHUB1/2 in yeast. The full-length CDS of Spin6 and OsHub1/2 were cloned into pDBleu and pPC86, respectively. The derived constructs were transformed into yeast strain MaV203. The transformed yeast cells were plated on SD-L-W-H medium with or without 3AT. (B) The interaction between SPIN6 and OsHUB1/2 in N. benthamiana leaves. The full-length CDS of Spin6 and OsHub1/2 were cloned into cLUC and nLUC, respectively. The leaves of N. benthamiana were infiltrated with Agrobacterium strains containing the indicated construct pairs. The top panels show quantification of LUC activity, and the bottom panels show the fluorescence in infiltrated leaves as detected by a low-light, cooled, CCD imaging apparatus.
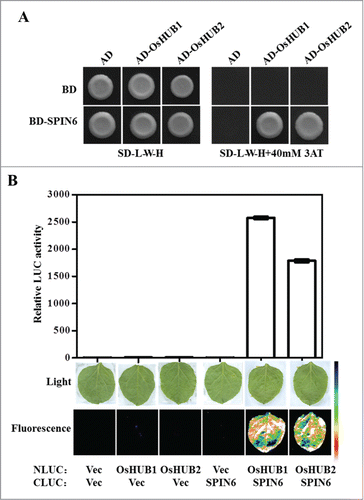
Figure 2. OsHub1/2 expression pattern after M. oryzae infection and hormone treatments. (A) Total RNA of 6-week-old Spin6 RNAi plants and control plants was used for real-time PCR. (**t test, with P < 0.01; *t test, with P < 0.05) (B) Total RNA of 3-week-old Nipponbare seedlings after inoculation with M. oryzae was used for real-time PCR. (C, D) 3-week-old Nipponbare seedlings were treated with 100 μM JA, 100 μM SA, 200 μM ABA and 100 μM ethephon and the leaves were sampled at 0, 4, 8, 12, and 24 h after the treatments. Total RNA was extracted and analyzed by real-time PCR.
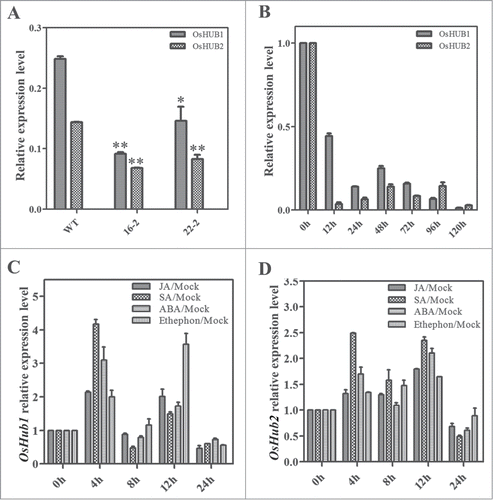
OsHub1 and OsHub2 Expression Patterns in Response to Pathogen Infection and Hormone Treatments
To elucidate the role of OsHUB1 and OsHUB2 in the interaction between rice and Magnaporthe oryzae, we inoculated Nipponbare seedlings with a virulent M. oryzae isolate, RO1–1, and collected leaf samples at different times after infection. The real-time PCR results showed that the expression of both OsHub1 and OsHub2 was significantly downregulated after RO1–1 infection and that the down-regulation was first evident at 12 h after inoculation (). Next, we analyzed the transcription of OsHub1 and OsHub2 in seedlings after the treatments with the plant hormones including jasmonic acid (JA), salicylic acid (SA), ethylene (ET) and abscisic acid (ABA). The analysis showed that the transcriptional level of OsHub1 peaked at 4 h after treatment with JA, SA, or ABA treatments and at 12 h after treatment with ethephon (). In contrast, the transcriptional level of OsHub2 peaked at 4 h after treatment with SA and at 12 h after treatment with JA, ABA, or ethephon (). In Arabidopsis, Hub1 is induced by inoculation with Alternaria brassicicola, Botrytis cinerea, Erysiphe cichoracearum, or Pseudomonas syringae pv. tomato (strain DC3000) but not by treatment with MeJA, SA, or ACC.Citation13,14 These results suggested that OsHub1 and OsHub2 might be involved in a different regulatory pathway in rice relative to HUB1 and HUB2 in Arabidopsis. In addition, the difference in their expression patterns in response to hormone treatments suggests that OsHub1 and OsHub2 may play different roles in hormone signaling pathways in rice.
Formation of OsHUB1/2 Homo- and Hetero-Dimers
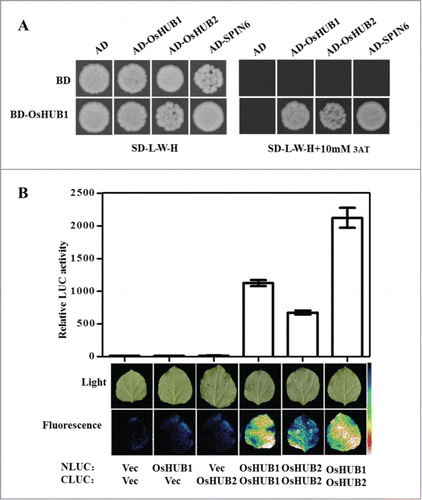
In Arabidopsis, HUB1 and HUB2 can form homo- and hetero-dimers and monoubiquitinate histone H2B to regulate flowering time.Citation15,16 To determine whether OsHUB1 and OsHUB2 can form homo- and hetero-dimers in rice, we used the yeast 2-hybrid assay The analysis showed that OsHUB1 interacted with OsHUB1 and OsHUB2 (). We further used the LCI system to confirm that these interactions also occur in vivo. High LUC activity was detected in the OsHUB1/OsHUB1, OsHUB1/OsHUB2, and OsHUB2/OsHUB2 combinations compared to the controls (), indicating that OsHUB1 and OsHUB2 can form homo- and hetero-dimers in rice.
Figure 3. OsHUB1 and OsHUB2 form homo- and hetero-dimers in vitro and in vivo. (A) The interaction between OsHUB1 and OsHUB2 in yeast. The full-length CDS of OsHub1 and OsHub2 were cloned into pDBleu and pPC86, respectively. The derived constructs were transformed into yeast strain MaV203. The transformed yeast cells were plated on SD-L-W-H medium with or without 3AT. (B) The interaction between OsHUB1 and OsHUB2 in N. benthamiana leaves. The leaves of N. benthamiana were infiltrated with Agrobacterium strains containing the indicated plasmid pairs. The top panels show quantification of LUC activity and the bottom panels show the fluorescence in infiltrated leaves as detected by low-light, cooled, CCD imaging apparatus.
In Arabidopsis, HUB1 is a key regulator of various cellular processes including flowering time, seed dormancy, the cell-cycle, and root growth.Citation10,15-18 Studies have shown that HUB1 and HUB2 can monoubiquitinate histone H2B to regulate SNC1 expression and thereby to modulate the immunity response.Citation13 In rice, OsHub1 is involved in tiller development.Citation11 Whether OsHUB1 and OsHUB2 are also involved in flowering time control, seed dormancy, the cell cycle, root growth and other cellular processes remains to be determined.
In this study, we found that OsHUB1 and OsHUB2 can form homo- and hetero-dimers and can interact with SPIN6 in vivo and in vitro. OsHub1 and OsHub2 are down-regulated in Spin6 RNAi plants and in response to M. oryzae infection. In addition, OsHub1 and OsHub2 are induced by hormone treatments. These results suggest that OsHUB1 and OsHUB2 may form a complex to regulate SPIN6/OsRac1-mediated plant defense responses and may also have functions in hormone signaling pathways.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Transgenic Crop Initiative to G.L.W. (Grant No. 2012ZX08009001) and the National Natural Science Foundation of China to Y.S.N. (Grant No. 31101405).
References
- Liu W, Liu J, Ning Y, Ding B, Wang X, Wang Z, Wang GL. Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol Plant 2013; 6(3):605-20; PMID:23340743; http://dx.doi.org/10.1093/mp/sst015
- Liu W, Liu J, Triplett L, Leach JE, Wang GL. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol 2014; 52:213-41; PMID:24906128; http://dx.doi.org/10.1146/annurev-phyto-102313-045926
- Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell 2014; 54(2):263-72; PMID:24766890; http://dx.doi.org/10.1016/j.molcel.2014.03.028
- Duplan V, Rivas S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci 2014; 5:42; PMID:24592270; http://dx.doi.org/10.3389/fpls.2014.00042
- Marino D, Peeters N, Rivas S. Ubiquitination during plant immune signaling. Plant Physiol 2012; 160(1):15-27; PMID:22689893; http://dx.doi.org/10.1104/pp.112.199281
- Li W, Zhong S, Li G, Li Q, Mao B, Deng Y, Zhang H, Zeng L, Song F, He Z. Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defence. Cell Res 2011; 21(5):835-48; PMID:21221134; http://dx.doi.org/10.1038/cr.2011.4
- Du Z, Zhou X, Li L, Su Z. plantsUPS: a database of plants' Ubiquitin Proteasome System. BMC Genomics 2009; 10:227; PMID:19445698; http://dx.doi.org/10.1186/1471-2164-10-227
- Liu J, Li W, Ning Y, Shirsekar G, Cai Y, Wang X, Dai L, Wang Z, Liu W, Wang GL. The U-Box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol 2012; 160(1):28-37; PMID:22659522; http://dx.doi.org/10.1104/pp.112.199430
- Liu J, Park CH, He F, Nagano M, Wang M, Bellizzi M, Zhang K, Zeng X, Liu W, Ning Y, et al. The RhoGAP SPIN6 Associates with SPL11 and OsRac1 and Negatively Regulates Programmed Cell Death and Innate Immunity in Rice. PLoS Pathog 2015; 11(2):e1004629; PMID:25658451; http://dx.doi.org/10.1371/journal.ppat.1004629
- Liu Y, Koornneef M, Soppe WJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 2007; 19(2):433-44; PMID:17329563; http://dx.doi.org/10.1105/tpc.106.049221
- Sun F, Zhang W, Xiong G, Yan M, Qian Q, Li J, Wang Y. Identification and functional analysis of the MOC1 interacting protein 1. J Genet Genomics 2010; 37(1):69-77; PMID:20171579; http://dx.doi.org/10.1016/S1673-8527(09)60026-6
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 2008; 146(2):368-76; PMID:18065554; http://dx.doi.org/10.1104/pp.107.111740
- Zou B, Yang DL, Shi Z, Dong H, Hua J. Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol 2014; 165(1):309-18; PMID:24664204; http://dx.doi.org/10.1104/pp.113.227801
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T. HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 2009; 21(3):1000-19; PMID:19286969; http://dx.doi.org/10.1105/tpc.108.062364
- Xu L, Menard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH. The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J 2009; 57(2):279-88; PMID:18798874; http://dx.doi.org/10.1111/j.1365-313X.2008.03684.x
- Cao Y, Dai Y, Cui S, Ma L. Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 2008; 20(10):2586-602; PMID:18849490; http://dx.doi.org/10.1105/tpc.108.062760
- Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J 2009; 57(3):522-33; PMID:18980658; http://dx.doi.org/10.1111/j.1365-313X.2008.03709.x
- Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, et al. The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 2007; 19(2):417-32; PMID:17329565; http://dx.doi.org/10.1105/tpc.106.041319
