Abstract
Members of the SEPALLATA (SEP) gene sub-family encode class E floral homeotic MADS-domain transcription factors (MADS TFs) that specify the identity of floral organs. The Arabidopsis thaliana genome contains 4 ancestrally duplicated and functionally redundant SEP genes, SEP1–4. Recently, a gene family of unique effectors, phyllogens, was identified as an inducer of leaf-like floral organs in phytoplasmas (plant pathogenic bacteria). While it was shown that phyllogens target some MADS TFs, including SEP3 for degradation, it is unknown whether the other SEPs (SEP1, SEP2, and SEP4) of Arabidopsis are also degraded by them. In this study, we found that all 4 SEP proteins of Arabidopsis are degraded by a phyllogen using a transient co-expression assay in Nicotiana benthamiana. This finding indicates that phyllogens may broadly target class E MADS TFs of plants.
Many angiosperms have flowers composed of 4 whorls of floral organs: sepals, petals, stamens, and carpels. The identity of these floral organs is regulated by a specific combination of floral homeotic MADS-domain transcription factors (MADS TFs) constituting the ABCE model.Citation1-3 Among these MADS TF genes, class E genes of the SEPALLATA (SEP) gene sub-family are essential for the development of all 4 floral organs as well as for floral meristem determinacyCitation1,4 because they function as the “glue” for MADS TF tetrameric complex formation.Citation5,6 The Arabidopsis thaliana genome contains 4 functionally redundant SEP genes, SEP1–4. This is suggested by the findings that the triple mutant (sep1/2/3) and the quadruple mutant (sep1/2/3/4) exhibit abnormal flowers with green sepal- or leaf-like organs in all 4 whorls, and also exhibit a loss of floral meristem determinacy,Citation1,4 however, a mutant of one of SEP genes produces only subtle phenotypes, consistent with the possibility of functional redundancy. Citation1
Phytoplasmas are obligate plant pathogenic bacteria with highly reduced genomes.Citation7,8 They inhabit both plant phloem sieve cells and insect cells altering their own gene expression,Citation9 and cause drastic morphological changes in infected plants, including witches' broom (proliferation of small branches resulting in a characteristic bushy growth) and phyllody (a loss of floral meristem determinacy and the transformation of floral organs into leaves).Citation10,11 In recent publications, witches' broom and phyllody symptoms of plants were shown to be associated with aberrant expression patterns of auxin-related genes and the genes encoding MADS TFs, respectively.Citation12,13 Moreover, an inducer of witches' broom, designated TENGU, and a gene family of phyllody-inducing effectors, designated phyllogens, were identified from phytoplasmas.Citation12,14,15
Phyllogens interact with and degrade some MADS TFs, including SEP3 of Arabidopsis.Citation15,16 However, it is unknown whether the other 3 class E proteins (SEP1, SEP2, and SEP4) of Arabidopsis are also degraded by phyllogens, though their interactions with a phyllogen were shown in a yeast 2-hybrid assay.Citation16 It is also interesting from the perspective of the phylogenetic differences between SEP genes: in an evolutionary analysis of SEP gene sub-family, SEP3 and the other 3 SEP genes of Arabidopsis were divided into 2 different clades.Citation17
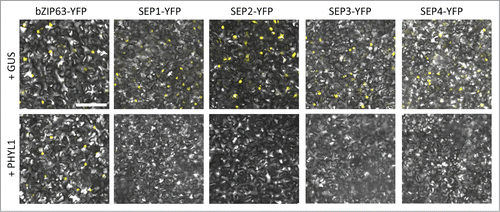
In this study, we examined whether a phyllogen degrades SEP1, SEP2, and SEP4 of Arabidopsis. We transiently expressed YFP-fused SEP1, SEP2, SEP3, SEP4, or bZIP63, a basic leucine zipper transcription factor involved in the glucose-abscisic acid interaction network of Arabidopsis,Citation18 in Nicotiana benthamiana leaves by agroinfiltration in combination with either GUS protein (control) or a phyllogen (PHYL1) of OY-W phytoplasma,Citation19 and monitored their accumulation and subcellular localization by confocal microscopy. As expected, all of the YFP-fused transcription factors localized to the nucleus when GUS was co-expressed (, upper panels). While co-expression with PHYL1 did not affect the accumulation or nuclear localization of YFP-fused bZIP63, it reduced the fluorescence derived from YFP-fused SEP1, SEP2, and SEP4 as well as SEP3-YFP (, lower panels), indicating that all 4 SEP proteins of Arabidopsis can be degraded in the presence of phyllogens in vivo. This result agrees well with the fact that transgenic Arabidopsis plants expressing phyllogens show floral homeotic phenotypes similar to those of triple or quadruple mutants of sep genes.Citation1,4,14,15 Moreover, SEP3- and SEP1/2/4-like genes are highly conserved among various angiosperms as genes of the SEP sub-family encoding class E MADS TFs.Citation17 Therefore, it is likely that phyllogens broadly target class E proteins of angiosperms, including eudicots, monocots, and other taxa, for degradation. Future studies investigating the target specificity of phyllogens will shed further light on the relationships between phyllody caused by phytoplasmas and the function of class E genes in angiosperms.
Figure 1. PHYL1 induces the degradation of SEP1–4 proteins. Agrobacterium cultures (OD600 = 1.0) for transient expression of YFP-fused proteins (bZIP63 and SEP1–4) and GUS or PHYL1 protein were mixed at a ratio of 1:10 and infiltrated into Nicotiana benthamiana leaves. The accumulation and subcellular localization of the transiently expressed YFP-fused proteins were observed 50 h after infiltration. Scale bar = 100 µm. The Agrobacterium strain and plasmids were the same as in the previous study, respectively.Citation15 The composition of the infiltration buffer is as follows: 10 mM morpholinepropanesulfonic acid (MES, pH 5.6), 10 mM MgCl2, and 150 µM acetosyringone. Nuclear localization of the transcription factors used in this study was confirmed by DAPI staining (data not shown).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (category “S” of Scientific Research Grant 25221201), by the Funding Program for Next Generation World-Leading Researchers (project: GS005) initiated by the Council for Science and Technology Policy (CSTP), and by the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN).
References
- Pelaz S, Ditta SG, Baumanm E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000; 405:200-3; PMID:10821278; http://dx.doi.org/10.1038/35012103
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001; 409:525-9; PMID:11206550; http://dx.doi.org/10.1038/35054083
- Theissen G, Saedler H. Floral quartets. Naure 2001; 409:469-71; PMID:11206529; http://dx.doi.org/10.1038/35054172
- Ditta G, Pinyopich A, Robels P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 2004; 14:1935-40; PMID:15530395; http://dx.doi.org/10.1016/j.cub.2004.10.028
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC. SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 2009; 10:R24; PMID:19243611; http://dx.doi.org/10.1186/gb-2009-10-2-r24
- Jetha K, Theissen G, Melzer R. Arabidopsis SEPALLATA proteins differ in cooperative DNA-binding during the formation of floral quartet-like complexes. Nucl Acids Res 2014; 42:10927-42; PMID:25183521; http://dx.doi.org/10.1093/nar/gku755
- Oshima K, Miyata S, Sawayanagi T, Kakizawa S, Nishigawa H, Jung HY, Furuki K, Yanazaki M, Suzuki S, Wei W, Kuboyama T, Ugaki M, Namba S. Minimal set of metabolic pathways suggested from the genome of onion yellows phytoplasma. J Gen Plant Pathol 2002; 68:225-36; http://dx.doi.org/10.1007/PL00013081
- Oshima K, Kakizawa S, Nishigawa H, Jung HY, Wei W, Suzuki S, Arashida R, Nakata D, Miyata S, Ugaki M, Namba S. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat Genet 2004; 36:27-9; PMID:14661021; http://dx.doi.org/10.1038/ng1277
- Oshima K, Ishii Y, Kakizawa S, Sugawara K, Neriya Y, Himeno M, Minato N, Miura C, Shiraishi T, Yamaji Y, Namba S. Dramatic transcriptional changes in an intracellular parasite enable host switching between plant and insect. PLoS ONE 2011; 6:e23242; PMID:21858041; http://dx.doi.org/10.1371/journal.pone.0023242
- Hogenhout SA, Oshima K, Ammar ED, Kakizawa S, Kingdom HN, Namba S. Phytoplasmas: bacteria that manipulate plants and insects. Mol Plant Pathol 2008; 9:403-23; PMID:18705857; http://dx.doi.org/10.1111/j.1364-3703.2008.00472.x
- Maejima K, Oshima K, Namba S. Exploring the phytoplasmas, plant pathogenic bacteria. J Gen Plant Pathol 2014; 80:210-21; http://dx.doi.org/10.1007/s10327-014-0512-8
- Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, Namba S. A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci U S A 2009; 106:6416-21; PMID:19329488; http://dx.doi.org/10.1073/pnas.0813038106
- Himeno M, Neriya Y, Minato N, Miura C, Sugawara K, Ishii Y, Yamaji Y, Kakizawa S, Oshima K, Namba S. Unique morphological changes in plant pathogenic phytoplasma-infected petunia flowers are related to transcriptional regulation of floral homeotic genes in an organ-specific manner. Plant J 2011; 67:971-9; PMID:21605209; http://dx.doi.org/10.1111/j.1365-313X.2011.04650.x
- MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Tóth R, Nicolaisen M, Hogenhout SA. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol 2011; 157:831-41; PMID:21849514; http://dx.doi.org/10.1104/pp.111.181586
- Maejima K, Iwai R, Himeno M, Komatsu K, Kitazawa Y, Fujita N, Ishikawa K, Fukuoka M, Minato N, Yamaji Y, Oshima K, Namba S. Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J 2014; 78:541-54; PMID:24597566; http://dx.doi.org/10.1111/tpj.12495
- MacLean AM, Orlovskis Z, Kowitwanich K, Zdziarska AM, Angenent GC, Immink RG, Hogenhout SA. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol 2014; 12:e1001835; PMID:24714165; http://dx.doi.org/10.1371/journal.pbio.1001835
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 2005; 169:2209-23; PMID:15687268; http://dx.doi.org/10.1534/genetics.104.037770
- Matiolli CC, Tomaz JP, Duarte GT, Prado FM, Del Bem LEV, Silveira AB, Gauer L, Corrêa LGG, Drumond RD, Carvalho Viana AJ, Di Mascio P, Meyer C, Vincentz M. The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiol 2011; 157:692-705; PMID:21844310; http://dx.doi.org/10.1104/pp.111.181743
- Oshima K, Shiomi T, Kuboyama T, Sawayanagi T, Nishigawa H, Kakizawa T, Miyata S, Ugaki M, Namba S. Isolation and characterization of derivative lines of the onion yellows phytoplasma that do not cause stunting or phloem hyperplasia. Phytopathology 2001; 91:1024-9; http://dx.doi.org/10.1094/PHYTO.2001.91.11.1024
